Back to Journals » OncoTargets and Therapy » Volume 12
Upregulated expression of ACTL8 contributes to invasion and metastasis and indicates poor prognosis in colorectal cancer
Authors Han Q, Sun ML, Liu WS, Zhao HS, Jiang LY , Yu ZJ, Wei MJ
Received 30 August 2018
Accepted for publication 24 January 2019
Published 1 March 2019 Volume 2019:12 Pages 1749—1763
DOI https://doi.org/10.2147/OTT.S185858
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr William C. Cho
Qiang Han,1 Ming-Li Sun,1 Wen-Si Liu,1 Hai-Shan Zhao,1 Long-Yang Jiang,1 Zhao-Jin Yu,1 Min-Jie Wei1,2
1Department of Pharmacology, School of Pharmacy, China Medical University, Shenyang 110122, Liaoning, China; 2Department of Pharmacology, Liaoning Engineering Technology Research Center for the Research, Development and Industrialization of Innovative Peptide Drugs, China Medical University, Shenyang 110122, Liaoning, China
Background: ACTL8 is a member of the CT antigens. There are only few studies on the role of ACTL8 in malignant tumors. The aim of this study is to investigate the expression and clinical significance of ACTL8 protein in colorectal cancer (CRC).
Materials and methods: Human CRC tissues and cell lines, and paired adjacent non-tumor tissues and human intestinal epithelial cell lines were obtained to evaluate the expression of ACTL8. The association between protein expression of ACTL8 and clinicopathological parameters and prognosis of CRC patients was examined. The biological functions of ACTL8 in the invasion and metastasis of CRC were determined by wound healing and transwell invasion assays after silencing of ACTL8 in CRC cell lines. The potential target genes of ACTL8 were also identified by quantitative reverse transcription PCR and Western blotting after silencing of ACTL8 in CRC cell lines.
Results: It was found that ACTL8 was upregulated in human CRC tissues and cell lines. The expression of ACTL8 was positively associated with poor differentiation, invasion and metastasis, postoperative infection, and poor prognosis, but negatively associated with proximal margin length. In addition, silencing of ACTL8 significantly decreased the capacity of invasion and migration in HT29 and SW620 CRC cell lines. Moreover, silencing of ACTL8 significantly decreased the expression of TRIM29 in HT29 and SW620 CRC cell lines.
Conclusion: These results suggest that ACTL8 plays a key role in the invasion and metastasis of CRC, and TRIM29 may be involved in the ACTL8-mediated poor prognosis of CRC.
Keywords: ACTL8, CRC, prognosis, invasion and metastasis, TRIM29
Introduction
Colorectal cancer (CRC) is one of the most common malignancies in humans, with a high incidence rate, and ranks third and second among the most common cancers in men and women, respectively.1 A total of 376,300 new CRC cases and 191,000 deaths in China were estimated in 2015,2 and 135,430 new CRC cases and 50,266 deaths in the US were estimated in 2017.3 Despite the major advancement in comprehensive therapy of CRC, the mortality rate of CRC patients with advanced distant metastasis remains high.4,5 Unfortunately, there is still a lack of effective treatment for highly invasive CRC. Therefore, a deep investigation of the molecular mechanism of distant metastasis is crucial for the identification of new therapeutic strategies for patients with metastatic CRC.
CT antigens are a group of proteins with normal expression restricted to adult testicular germ cells, but are aberrantly activated and expressed in a proportion of human cancer cells. Recent studies have found that the expression of CT antigens is also closely related to the invasion and metastasis of a variety of malignant tumors,6,7 including CRC.8
ACTL8, also known as CT57, is a member of the sugar kinase/heat shock protein 70/actin superfamily. It was initially identified as a member of the CT antigens by Chen et al,9 and later verified by others.10,11 So far, there are only few studies on the role of ACTL8 in malignant tumors. Freitas et al12 found the expression of ACTL8 was increased in malignant gliomas, but there was no direct correlation with survival of patients. Buhrke et al found that the downregulation of ACTL8 was associated with cytoskeletal reorganization and higher differentiation of colon cancer cells,13 suggesting that overexpression of ACTL8 may help to maintain poorer differentiation of colon cancer cells. However, no relevant studies have been found on the expression and clinical significance of ACTL8 in CRC.
In the initial research, we found ACTL8 mRNA was overexpressed in a variety of malignancies, including colon and rectal cancer, by big data analysis. In the following study, we detected the levels of ACTL8 protein in 132 CRC tissues, 42 paracancerous tissues, and 97 normal tissues, and analyzed the correlation between ACTL8 expression and clinicopathological parameters and prognosis of CRC patients, to investigate the clinical significance of ACTL8 expression in CRC. Furthermore, we investigated the effect of ACTL8 on the invasion and metastasis of CRC and its possible mechanisms, based on the information provided by big data analysis.
Materials and methods
Big data analysis
The specific expression sites and differential expression of ACTL8 were analyzed using The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) data from Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/). Proteins that interact with ACTL8 were predicted by using STRING (https://string-db.org/cgi/input.pl) and Gene-Cloud of Biotechnology Information (GCBI) (https://www.gcbi.com.cn/gcanalyze/html/generadar/index). The prognostic value of ACTL8 was determined using UALCAN (http://ualcan.path.uab.edu/index.html).
Patients and tissue specimens
The tissue samples were collected from patients with CRC in the Department of Gastrointestinal Surgery, the Fourth Affiliated Hospital of China Medical University, between 2008 and 2011 which included 132 cancer tissue specimens, 42 paracancerous tissue specimens, and 97 normal tissue specimens. The 132 cases of CRC included 65 cases of colon cancer and 67 cases of rectal cancer; all of them were confirmed by pathological examination. Among the patients with CRC, 69 were male and 63 were female. The average age of patients was 63.8±11.6 years (range 39–84 years). The patients did not receive chemotherapy, radiotherapy, or immunotherapy before surgery. The average age of the patients from whom paracancerous and normal tissue specimens were collected was 62.7±12.4 years (range 17–88 years).
For the 132 cases of CRC included in this study, clinical data on family history, concurrent infection, differentiation level, depth of invasion, percentage of intestine occupied, expression of creatine kinase level (CKL), proximal margin length (PML), and distal margin length (DML) were collected. In addition, data on the expression of CA125, CEA, and AFP in blood were synchronously collected. Some cases were found to have missing data, as shown in Table 1.
The study collected survival data for a total of 70 cases. The follow-up time was 1–113 months, during which 29 patients died. The 5-year survival rate was 65.6%. The average survival time was 82.1±4.8 months.
This study was approved by the Medical Ethics Committee of China Medical University. The Ethics Committee waived the need for written informed consent from the patients included in the study due to the retrospective nature of the study, and because the patients who offered tissue samples had already signed the consent form after being informed that their tissue samples would be used for diagnosis and scientific research during their clinical treatment in the hospital.
Tissue microarray (TMA) and immunohistochemistry (IHC)
All the 271 tissues were embedded in paraffin, and the representative locations were marked by H&E staining. Tissue sample (1.5 mm) was extracted from each representative location and accurately transferred by the Organization Microarrayer (Pathology Devices, San Diego, CA, USA) to another paraffin block to establish the TMA. TMA sections of 4 μm thickness were adhered to poly-L-lysine-coated glass slides and prepared for IHC.
The microarray sections were deparaffinized with xylene for 30 minutes and hydrated in graded ethanol and distilled water. The microarray sections were placed in 0.01 M citrate buffer (pH 6.0), and antigen retrieval was performed for 8 minutes at a high pressure of 80 kPa. The sections were then treated with 3% H2O2 for 10 minutes at room temperature to remove endogenous peroxidase. After blocking the nonspecific protein-binding site with normal goat serum, rabbit antihuman ACTL8 polyclonal antibody (1:50, ab36030; AbSci, Vancouver, WA, USA) was added and incubated at 4°C overnight. On the next day, followed by incubation with biotinylated goat anti-rabbit IgG and streptavidin–horseradish peroxidase complex (KIT-9710; MXB biotechnologies, Fuzhou, Fujian, China) at 37°C for 30 minutes, the sections were stained with 3,3′-diaminobenzidine for 25 seconds. PBS replaced the anti-ACTL8 primary antibody as a negative control.
Immunohistochemical evaluation
TMA staining was assessed blindly by two experienced clinicopathologists (HSZ and MLS). ACTL8 protein was located mainly in the cytoplasm. The percentage of stained cells (0%–100%) and the score of staining intensity (no stain, 0; weakly positive, 1; medium intensity, 2; and strongly positive, 3) were multiplied as immunoreactive score (IRS), and ranged from 0% to 300%. Receiver operating characteristic (ROC) curves were used to define the low and high expression of ACTL8 in accordance with IRS.
Cell lines and culture
Human intestinal epithelial cells were cultured in DMEM containing 10% FBS (37°C, 5% CO2). Human CRC cells HT29 and HCT116 were cultured in McCoy’s 5a medium and RPMI-1640 medium, respectively, containing 10% FBS (37°C, 5% CO2). Human CRC cells SW480 and SW620 were cultured in L15 medium containing 10% FBS (37°C, air). The above cells were purchased from BeNa Culture Collection (Beijing, China). All cell experiments were performed during the logarithmic growth phase of the cells.
Western blotting
The total protein was extracted from the CRC cells and epithelial cells by total protein extraction kit (BB-3101; Bestbio, Shanghai, China), and then the proteins were quantified by BCA protein assay kit (Beyotime Biotechnology, Shanghai, China). For Western blotting analysis, an equal quantity of total protein was loaded, and separated by SDS-PAGE. Following electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane and blocked with 5% (w/v) nonfat milk for 1 hour at room temperature. The membrane was incubated with primary antibodies against ACTL8 (1:2,000, ab36030; AbSci), TRIM29 (1:2,000, 35641; Signalway Antibody Co., College Park, MD, USA), or β-actin (1:1,000, 8457; Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C overnight. The blots were subsequently incubated with a horseradish peroxidase-conjugated secondary antibody (1:10,000, E030120; EarthOx, Millbrae, CA, USA) for 1 hour at room temperature. Proteins were visualized by ECL detection system (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The target protein band was detected, and the optical density was analyzed using ImageJ2x software (2.1.4.7).
Cell transfection
For analyzing the function of ACTL8 in CRC, specific siRNAs against ACTL8 and a scrambled negative control were used (Ribobio Co., Ltd., Guangzhou, China). For transfection, the HT29 and SW620 cells were seeded in six-well plates at a density of 2×105 cells per well. After cultured in serum-free medium for 2 hours, the cells were transfected with complete medium plus 50 nM Lipofectamine™ 2000 (Thermo Fisher Scientific), which was used as the transfection reagent, according to the manufacturer’s instructions. After transfection for 72 hours, mRNA and protein were collected.
Real-time qRT-PCR
Total RNA was extracted using TRIzol (R1100; Solarbio, Beijing, China) according to the manufacturer’s protocol. For quantitative reverse transcription PCR (qRT-PCR), RNA was reverse transcribed to cDNA using FastQuant RT Kit (KR106; Tiangen, Beijing, China). Real-time PCR analyses were performed using SuperReal PreMix Plus (FP205; Tiangen) in the Applied Biosystems 7300 Real-Time PCR System (Thermo Fisher Scientific). The expression levels of ACTL8 were normalized to β-actin (B661102; Sangon, Shanghai, China). Relative expression levels between samples were calculated using the 2−ΔΔCT method. The primer sequences for PCR were as follows: TRIM29: sense, 5′-ACATCATACCAGCCCTCGTC-3′ and reverse, 5′-AGCCTTTCAGGGAGAAGGAG-3′; CFL1: sense, 5′-GCCGCTATGCCCTCTA-3′ and reverse, 5′-GCCGCTATGCCCTCTA-3′.
Cell migration and invasion assay
Cell migration was detected by wound healing assay. HT29 and SW620 cells transfected with si-ACTL8 and si-NC were seeded in six-well plate at a density of 5×105 cells per well, and cultured to 100% confluency before transfection. Then, a wound was created with a pipette tip. After washing the shedding cells, the remaining cells were cultured in medium without FBS. At 0 and 48 hours after injury, images were taken using Nikon Eclipse TS100 Microscope (Nikon, Tokyo, Japan). The assay was independently repeated three times.
To obtain more convincing results, transwell cell invasion assay was performed as follows. After spreading basement membrane (356234; Corning, Corning, NY, USA) in the upper chamber, 1×105 cells/well were incubated with serum-free medium. Medium containing 10% FBS was added into the lower chamber. After 48 hours, cells attached to the lower surface were stained with 0.1% crystal violet (C8470; Solarbio). The migrated cells were counted under a light microscope in five predetermined fields (magnification, ×200). The assay was independently repeated three times.
Statistical analyses
The correlation between the expression of ACTL8 and clinicopathological features of CRC patients was analyzed by Pearson’s chi-squared test or Fisher’s exact test. The correlation between the two parameters was analyzed by Pearson correlation or Spearman rank correlation analysis. Wilcoxon rank-sum test was used to analyze the differences between groups with non-normally distributed variables. The independent-samples t-test was used to compare the differences between groups with normally distributed variables. The cutoff values of low and high expression of ACTL8 were defined by ROC curve analysis. Kaplan–Meier curves were plotted to assess the association between ACTL8 and survival, and statistical differences in the survival time between patient subgroups were compared using log-rank test. Univariate and multivariate survival analyses were performed using a Cox regression model. One-way ANOVA was used in GEPIA to test the difference between normal and cancer samples. P<0.05 was defined as statistically significant. The statistical analyses were conducted using the SPSS software package, version 17.0 (SPSS Inc, Chicago, IL, USA).
Results
ACTL8 was upregulated in CRC tissues and cell lines
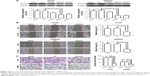
We analyzed the mRNA expression of ACTL8 by using TCGA and GTEx data from GEPIA (http://gepia.cancer-pku.cn/), and found that ACTL8 was overexpressed in CRC tissues compared to that in normal tissues (Figure S1). Then, we examined ACTL8 protein expression in 132 CRC tissue specimens, 42 paracancerous tissue specimens, and 97 normal tissue specimens by IHC, and in four malignant CRC cell lines and one normal intestinal epithelial cell line by Western blotting. ACTL8 showed high expression in CRC tissues compared to normal tissues (median [IQR]: 200 [140–284.25] vs 40 [4.5–150], P=2.85e-17) and tumor-adjacent tissues (median [IQR]: 200 [140–284.25] vs 61 [18.75–157], P=5.89e-9) (Figure 1A and B). ACTL8 also showed high expression in CRC tissues compared to paired normal tissues (n=89, median [IQR]: 186 [150–283.5] vs 40 [4–153], P=1.55e-12; Figure 1C) and paired tumor-adjacent tissues (n=33, median [IQR]: 210 [145–288] vs 62 [12.5–149], P=5.79e-6; Figure 1D). Western blotting analysis showed that ACTL8 protein levels in malignant cell lines (HT29, HCT116, SW480, SW620) were significantly increased compared with those in the relatively normal intestinal epithelial cell line (Figure 1E). Thus, our data showed that ACTL8 was overexpressed in CRC tissues and cell lines.
Overexpression of ACTL8 conferred worse prognosis in CRC patients
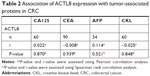
To determine the clinical significance of ACTL8 overexpression, we first selected the cutoff value (173%) of high and low expression of ACTL8 by ROC curve based on overall survival (OS) of patients (Figure S2). Then, we performed further analysis to investigate the correlation of ACTL8 overexpression with clinicopathological features. As shown in Table 1, upregulation of ACTL8 was significantly related to poorer differentiation (P=0.036), deeper invasion of intestinal wall (P=0.029), and concurrent infection (P=0.024), but not to age, sex, tumor position, family history, percentage of intestine occupied, and CKL level. Next, we analyzed the correlation between the expression of ACTL8 protein and PML and DML. As shown in Figure 2, ACTL8 expression was significantly negatively correlated with PML (r=−0.269, P=0.011), but not with DML (r=−0.115, P=0.285). In addition, we analyzed the correlation between the expression of ACTL8 protein and the expression of tumor-associated protein of CRC, but found no statistically significant correlation (P>0.05; Table 2).
Next, we performed the survival analysis. OS was evaluated via log-rank tests. Kaplan–Meier curves revealed that overexpression of ACTL8 was associated with poorer prognosis of CRC patients (P=0.003; Figure 3). Moreover, in univariate Cox regression analysis (Table 3), ACTL8 levels, concurrent infection, and invasion depth were notably associated with OS. In multivariate Cox regression analysis (Table 3), when age, concurrent infection, invasion depth, and ACTL8 expression were included, ACTL8 expression (P=0.145) was not found to act as an independent risk factor affecting prognosis. Thus, these analyses supported a role for ACTL8 in aggressiveness and progression of CRC, and indicated the value of ACTL8 as a prognostic risk factor.
ACTL8 silencing decreased the migration and invasion of CRC cells
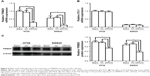
According to the above results that higher ACTL8 expression in tumor tissues was associated with poorer differentiation and deeper infiltration of intestinal wall, it was believed that ACTL8 might be important in enhancing the migration and invasion of CRC cells. To verify this hypothesis, we inhibited ACTL8 expression in HT29 and SW620 CRC cell lines using siRNA, and then performed wound healing and transwell invasion assays to assess the ability of migration and invasion of cells. As shown in Figure 4, we selected siRNAb to knock down ACTL8 in HT29 and SW620 cells, and found that the cell-covered area was significantly increased in the ACTL8-silenced groups compared with si-NC groups in the migration assays (P=0.024). Consistent with the migration assay results, the number of invading cells in the invasion assays was significantly reduced in the ACTL8-silenced groups compared with si-NC groups (P=0.0024). Thus, both the migration and invasion assays supported a role for ACTL8 in the migration and invasion of CRC cells.
ACTL8 silencing reduced the expression of TRIM29 in CRC cells
To explore the mechanisms by which ACTL8 mediates aggressiveness and poor prognosis, we tried to identify proteins interacting with ACTL8 using GCBI and STRING (Figure S3A and C). Differential expression and correlation analysis were performed to further analyze the targets, and the results showed that TRIM29 and CFL1 were the factors that significantly interact with ACTL8 and were concurrently involved in cell invasion and migration. GEPIA data showed that mRNA expression of TRIM29 and CFL1 was significantly higher in CRC tissues than in adjacent tissues (Figure S3B and D). Furthermore, ACTL8 showed a modest relationship with TRIM29 (r=0.168, P=2.51e-4, n=645; Figure S3E) and CFL1 (r=0.313, P=1.26e-15, n=645; Figure S3F) in CRC, based on the TCGA data. These data suggested that TRIM29 and CFL1 might be important target genes of ACTL8. To verify this hypothesis, we inhibited ACTL8 expression in HT29 and SW620 cells using siRNA, and then detected the expression of TRIM29 and CFL1 by qRT-PCR to assess the effect of ACTL8 silencing on the mRNA expression of TRIM29 and CFL1. As shown in Figure 5, we found the mRNA expression of TRIM29 was significantly reduced in the ACTL8-silenced groups compared with si-NC groups (P=0.0059 and 0.0083 for HT29 and SW620, respectively), but found no change in CFL1 mRNA expression (P=0.8280 and 0.5341 for HT29 and SW620, respectively). We then examined the protein expression of TRIM29, and found that consistent with the results of mRNA expression, the protein expression of TRIM29 in the ACTL8-silenced groups was also reduced significantly compared with that in si-NC groups (P=0.0289 and 0.0105 for HT29 and SW620, respectively). Thus, these data supported that ACTL8 may mediate migration and invasion by regulating TRIM29 expression in CRC.
Discussion
ACTL8 is one of the members of the actin superfamily. The current studies show that ACTL8 is a kind of CT antigen, which may be a potential biomarker and immunotherapeutic target in cancers.12,14 However, there have been no studies on the expression and clinical significance of ACTL8 in CRC. In this study, we first detected the expression of ACTL8 in CRC tissue samples and analyzed its clinical significance. We found that the expression of ACTL8 was significantly elevated in CRC tissues compared to normal or paracancerous tissues, suggesting that ACTL8 may be involved in the development of CRC.
We further analyzed the correlation between ACTL8 expression and clinical data of CRC patients, and found that higher expression of ACTL8 was associated with poor CRC differentiation. Buhrke et al found that downregulation of ACTL8 in human intestinal cell line Caco-2 promoted cytoskeletal remodeling of colon cancer cells and induced differentiation,13 which is consistent with our results. In addition, we also found that the high expression of ACTL8 was closely related to the depth of intestinal wall infiltration. These data suggested that ACTL8 may be related to the malignant phenotype of CRC cells. Furthermore, we further verified this effect on the cell model. We found that invasive and migration abilities were significantly decreased when ACTL8 protein was silenced in the CRC cell line model, further illustrating that overexpression of ACTL8 may mediate the invasion and migration of CRC cells.
Postoperative complications in patients with CRC are important factors influencing the outcome of surgery and prognosis. Our analysis found that overexpression of ACTL8 was associated with postoperative infection in CRC patients. This indicates that overexpression of ACTL8 may affect the immune environment in the patient, or the living environment of the pathogen. Previous studies have shown that the two single nucleotide polymorphisms (SNP) loci of ACTL8 gene rs1339934 and rs6686929 are associated with susceptibility to diabetes,15 and an increased risk of development of diabetes was recognized,16,17 indicating that ACTL8 is related to infection. In addition, many researchers have suggested that ACTL8 is a potential therapeutic target in tumor immunotherapy,14,18 suggesting that expression of ACTL8 may be related to immunity levels. However, the above studies do not accurately support the correlation between overexpression of ACTL8 and concurrent infection in patients; therefore, further research is still necessary.
In addition, PML is one of the indicators of the effect of tumor surgery. Reports on the clinical significance of PML in digestive tumors are rare. Kim et al19 found that PML was negatively correlated with poor differentiation and lymph node metastasis in gastric cancer, but positively correlated with 5-year survival rate, indicating that PML can be a potential protective factor for gastric cancer. In the present study, we found that ACTL8 expression was significantly negatively correlated with PML in CRC cases, as reported by Kim et al, suggesting that ACTL8 expression may have an inverse effect on PML in CRC, suggesting that it may play a role in promoting CRC progression.
Based on the above analysis, we found that overexpression of ACTL8 may be associated with poor differentiation, invasion and metastasis, and postoperative infection, and negatively correlated with PML, which are all indicators related to the prognosis of patients. Therefore, we further analyzed the effect of ACTL8 expression on the survival of CRC patients, and found that the OS of CRC patients with ACTL8 overexpression was significantly shortened. Univariate Cox regression analysis showed that concurrent infection, depth of invasion, and ACTL8 expression were significantly associated with OS, but multivariate Cox regression analysis showed that ACTL8 expression was not an independent risk factor of prognosis. This may be explained by the fact that ACTL8 expression affects OS of CRC patients by affecting the level of tumor cell differentiation, promoting invasion and metastasis, or causing postoperative infection.
Studies on the correlation between ACTL8 expression and prognosis in cancers are rarely reported. Freitas et al12 found high expression of ACTL8 in malignant glioma (glioblastoma multiforme) is associated with improved prognosis, which is contrary to the results of this study. However, we found that high mRNA expression of ACTL8 was significantly associated with shortened survival in patients with invasive breast carcinoma, head and neck squamous cell carcinoma, clear cell renal cell carcinoma, papillary renal cell carcinoma, liver cancer, sarcoma, and mesothelioma in TCGA data (Figure S4), supporting the results of this study. The difference in different research may be related to different tumors. Therefore, the influence of ACTL8 expression on the prognosis of tumor patients needs further study and confirmation, but the results of this study suggest that overexpression of ACTL8 is associated with poor prognosis of CRC.
To explore the mechanisms by which ACTL8 mediates poor prognosis of CRC, we first analyzed the correlation between the expression of ACTL8 and the expression of tumor-associated proteins in CRC, but found no significant results. Then, by big data analysis, we found two proteins, TRIM2920–22 and CFL1,23,24 which might interact with ACTL8 and are involved in cell invasion and migration. Furthermore, TCGA data of CRC showed mRNA expression levels of TRIM29 and CFL1 were significantly higher in CRC than in adjacent tissues, but modestly positively correlated with ACTL8 expression. We further found that silencing of ACTL8 expression caused a decrease in TRIM29 mRNA and protein expression levels, while CFL1 expression did not change significantly in CRC cell models. Previous studies20–22 have shown that higher expression of TRIM29 was an independent predictor of tumor metastasis and poorer survival in malignant tumors such as gastric cancer, osteosarcoma, and non-small-cell lung cancer. There are several ways in which the high expression of TRIM29 leads to enhanced metastasis and poor prognosis, including inducing epithelial–mesenchymal transition in osteosarcoma, activating P13K/AKT signaling pathway in thyroid carcinoma, activating NF-κB signaling pathway in bladder cancer, and activating Wnt/β-catenin pathway in cervical cancer.21,25–28 Also, we speculated that the modest correlation between ACTL8 and TRIM29 may be due to the fact that TRIM29 is located on the downstream of ACTL8 and controlled by other factors, and not only ACTL8. Meanwhile, the expression of CFL1 did not decrease with the downregulation of ACTL8, which may be due to that CFL1 is the upstream protein of ACTL8 which when silenced does not affect the expression of CFL1 and that ACTL8 does not directly regulate the expression of CFL1. Therefore, the results of this study suggest that TRIM29 may be involved in the process of ACTL8-mediated CRC invasion and metastasis and poor prognosis. However, the accurate mechanisms need further study.
Conclusion
This study was the first to find that the expression of ACTL8 was elevated in CRC tissue samples, and was positively associated with poor differentiation, invasion and metastasis, postoperative infection, and poor prognosis, but negatively associated with PML. Furthermore, TRIM29 was found to be a key factor in the process of CRC pathogenesis mediated by ACTL8, as in TCGA data, and in ACTL8-silenced CRC cell lines. These results indicate that overexpression of ACTL8 is associated with the development of CRC, and promotes CRC invasion and metastasis, leading to poor prognosis of CRC patients, and TRIM29 may be involved in the ACTL8-mediated poor prognosis of CRC.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of the People’s Republic of China (Nos 81601370 and 81501346) and NSFC-Liaoning Joint Fund Key Program (No. U1608281).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Siegel RL, Miller KD, Fedewa SA, et al. Colorectal Cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. | ||
Vatandoust S, Price TJ, Karapetis CS. Colorectal cancer: metastases to a single organ. World J Gastroenterol. 2015;21(41):11767–11776. | ||
Rahbari NN, Carr PR, Jansen L, et al. Time of metastasis and outcome in colorectal cancer. Ann Surg. 2019;269(3):494–502. | ||
Gjerstorff MF, Terp MG, Hansen MB, Ditzel HJ. The role of GAGE cancer/testis antigen in metastasis: the jury is still out. BMC Cancer. 2016;16:7. | ||
Song X, Hao J, Wang J, et al. The cancer/testis antigen MAGEC2 promotes amoeboid invasion of tumor cells by enhancing STAT3 signaling. Oncogene. 2017;36(11):1476–1486. | ||
Chen Z, Li M, Yuan Y, Wang Q, Yan L, Gu J. Cancer/testis antigens and clinical risk factors for liver metastasis of colorectal cancer: a predictive panel. Dis Colon Rectum. 2010;53(1):31–38. | ||
Chen YT, Scanlan MJ, Venditti CA, et al. Identification of cancer/testis-antigen genes by massively parallel signature sequencing. Proc Natl Acad Sci U S A. 2005;102(22):7940–7945. | ||
Hofmann O, Caballero OL, Stevenson BJ, et al. Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci U S A. 2008;105(51):20422–20427. | ||
Almeida LG, Sakabe NJ, Deoliveira AR, et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37(Database issue):D816–D819. | ||
Freitas M, Malheiros S, Stávale JN, et al. Expression of cancer/testis antigens is correlated with improved survival in glioblastoma. Oncotarget. 2013;4(4):636–646. | ||
Buhrke T, Lengler I, Lampen A. Analysis of proteomic changes induced upon cellular differentiation of the human intestinal cell line Caco-2. Dev Growth Differ. 2011;53(3):411–426. | ||
Yao J, Caballero OL, Yung WK, et al. Tumor subtype-specific cancer-testis antigens as potential biomarkers and immunotherapeutic targets for cancers. Cancer Immunol Res. 2014;2(4):371–379. | ||
Comuzzie AG, Cole SA, Laston SL, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7(12):e51954. | ||
Peleg AY, Weerarathna T, Mccarthy JS, Davis TM. Common infections in diabetes: pathogenesis, management and relationship to glycaemic control. Diabetes Metab Res Rev. 2007;23(1):3–13. | ||
Price CL, Hassi HO, English NR, Blakemore AI, Stagg AJ, Knight SC. Methylglyoxal modulates immune responses: relevance to diabetes. J Cell Mol Med. 2010;14(6B):1806–1815. | ||
Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100(11):2014–2021. | ||
Kim MG, Lee JH, Ha TK, Kwon SJ. The distance of proximal resection margin dose not significantly influence on the prognosis of gastric cancer patients after curative resection. Ann Surg Treat Res. 2014;87(5):223–231. | ||
Kosaka Y, Inoue H, Ohmachi T, et al. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann Surg Oncol. 2007;14(9):2543–2549. | ||
Zeng SX, Cai QC, Guo CH, et al. High expression of TRIM29 (ATDC) contributes to poor prognosis and tumor metastasis by inducing epithelial-mesenchymal transition in osteosarcoma. Oncol Rep. 2017;38(3):1645–1654. | ||
Zhou ZY, Yang GY, Zhou J, Yu MH. Significance of TRIM29 and β-catenin expression in non-small-cell lung cancer. J Chin Med Assoc. 2012;75(6):269–274. | ||
Yang ZL, Miao X, Xiong L, et al. CFL1 and Arp3 are biomarkers for metastasis and poor prognosis of squamous cell/adenosquamous carcinomas and adenocarcinomas of gallbladder. Cancer Invest. 2013;31(2):132–139. | ||
Castro MA, Dal-Pizzol F, Zdanov S, et al. CFL1 expression levels as a prognostic and drug resistance marker in nonsmall cell lung cancer. Cancer. 2010;116(15):3645–3655. | ||
Xu J, Li Z, Su Q, Zhao J, Ma J. TRIM29 promotes progression of thyroid carcinoma via activating P13K/AKT signaling pathway. Oncol Rep. 2017;37(3):1555–1564. | ||
Tan ST, Liu SY, Wu B. TRIM29 overexpression promotes proliferation and survival of bladder cancer cells through NF-κB signaling. Cancer Res Treat. 2016;48(4):1302–1312. | ||
Xu R, Hu J, Zhang T, Jiang C, Wang HY. TRIM29 overexpression is associated with poor prognosis and promotes tumor progression by activating Wnt/β-catenin pathway in cervical cancer. Oncotarget. 2016;7(19):28579–28591. | ||
Zhou XM, Sun R, Luo DH, et al. Upregulated TRIM29 promotes proliferation and metastasis of nasopharyngeal carcinoma via PTEN/AKT/mTOR signal pathway. Oncotarget. 2016;7(12):13634–13650. |
Supplementary materials
  | Figure S4 The correlation between ACTL8 mRNA expression and survival of patients with various cancers. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.