Back to Journals » Cancer Management and Research » Volume 11
Upregulated circ RNA hsa_circ_0000337 promotes cell proliferation, migration, and invasion of esophageal squamous cell carcinoma
Authors Song H, Xu D, Shi P, He B, Li Z , Ji Y , Agbeko CK, Wang J
Received 22 November 2018
Accepted for publication 31 January 2019
Published 4 March 2019 Volume 2019:11 Pages 1997—2006
DOI https://doi.org/10.2147/CMAR.S195546
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Huan Song,1,* Dian Xu,1,* Peiyi Shi,1 Biyu He,1 Zhongqi Li,1 Ye Ji,1 Charles Kwaku Agbeko,1 Jianming Wang1,2
1Department of Epidemiology, Center for Global Health, School of Public Health, Nanjing Medical University, Nanjing 211166, People’s Republic of China; 2Key Laboratory of Infectious Diseases, School of Public Health, Nanjing Medical University, Nanjing 211166, People’s Republic of China
*These authors contributed equally to this work
Background: As a new class of endogenous ncRNAs, circRNAs have been recently verified to be involved in the carcinogenesis and progression of human cancers. In the current study, we attempted to explore the potential function of a candidate circRNA (hsa_circ_0000337) in esophageal squamous cell carcinoma (ESCC).
Patients and methods: The altered expression of hsa_circ_0000337 was validated in clinical samples from 48 patients with ESCC. The human esophageal carcinoma cell lines KYSE-150 and TE-1, and the normal human esophageal epithelial cell line (HET-1A) were applied for functional analysis of hsa_circ_0000337. Cell proliferation was measured using the Cell Counting Kit-8 assay and the colony formation assay. Cell invasion and migration were detected by Transwell and wound healing assays, respectively. We further performed bioinformatic analysis and luciferase reporter assays to explore the role of hsa_circ_0000337 as a miRNA sponge.
Results: hsa_circ_0000337 was significantly upregulated in ESCC tissues compared to adjacent normal-appearing tissues (P<0.0001). In our in vitro experiment, the expression of hsa_circ_0000337 was higher in TE-1 compared to the normal human esophageal epithelial cell line HET-1A (P<0.001), but was not significantly different in KYSE-150 (P>0.05). Knockdown of hsa_circ_0000337 significantly inhibited cell proliferation, migration, and invasion in TE-1 and KYSE-150 cell lines. Bioinformatics predicted and luciferase reporter assay verified that hsa_circ_0000337 could bind to miR-670-5p, a ncRNA involved in carcinogenesis. It is estimated that 21 genes are regulated by miR-670-5p.
Conclusion: hsa_circ_0000337 was found to be an upregulated circRNA that is related to ESCC and promotes the progression of disease by regulating cell proliferation, migration, and invasion. These findings suggest that this circRNA could be a promising diagnostic biomarker and potential therapeutic target.
Keywords: esophageal squamous cell carcinoma, RNA, untranslated RNA, small untranslated, biomarker
Introduction
ncRNAs are a novel type of RNAs that are mainly expressed in eukaryotes.1 With the rapid development of high-throughput sequencing technology and bioinformatics, accumulating evidence has demonstrated that ncRNAs are involved in the pathophysiology of various diseases, including Alzheimer’s disease,2 cardiovascular disease,3 and cancers.4–8 Abundant and functionally important types of ncRNAs include tRNAs, rRNAs, miRNAs, siRNAs, lncRNAs, circRNAs, etc. ncRNAs play an important role in the epigenetic regulation of gene expression. For example, miRNAs affect the pathopoiesis and progression of angiogenesis-related diseases by regulating angiogenesis, cholesterol metabolism, and the inflammatory response,9 and lncRNAs regulate the proliferation, invasion, migration, and apoptosis of various cancers.10–12 In comparison to miRNAs and lncRNAs, the expression and functions of the newly discovered circRNAs remain to be elucidated.
circRNAs are generally considered to be ncRNAs, but increasing evidence has confirmed the capacity of some specific circRNAs to encode proteins.13 Unlike traditional linear RNAs, circRNAs are characterized by covalently closed loop structures with neither 5′ caps nor 3′ tails and are free from digestion by RNA exonucleases.14 Recently, a growing number of studies have indicated that circRNAs might present with greatly different expression in cells, tissues, and developmental stages,15,16 and could function in the following roles: serving as competitive endogenous ncRNAs or miRNA sponges, interacting with RNA-binding proteins, regulating parental gene expression, or encoding proteins.17 The dysregulation of circRNAs in breast cancer, gastric cancer, and colorectal cancer have been reported and implicate the possibility of circRNAs as novel biomarkers.7,18,19 However, the functions of circRNAs in esophageal carcinoma remain to be elucidated.
Esophageal carcinoma is one of the most common malignant tumors, with an estimated 572,000 new cases and over 509,000 deaths in 2018 globally.20 Esophageal carcinoma may be due to either esophageal squamous cell carcinoma (ESCC) or esophageal adenocarcinoma. Histologically, ESCC is the most prevalent worldwide type. Surgical intervention, radiotherapy, chemotherapy, and molecular targeted therapy have all substantially contributed to treatment, but the overall 5-year survival rate of patients with ESCC is still <20% and even lower in developing countries,21 primarily due to the majority of patients being diagnosed at an advanced stage. Previous studies have revealed the potential of epigenetic traits as biomarkers for the early diagnosis and therapeutic targeting of human cancers, but the role of circRNAs in ESCC has not been well explored.
In the current study, we had a great interest in a candidate circRNA (hsa_circ_0000337, also known as hsa_circRNA_100872) in ESCC, which was identified through a high-throughput Arraystar Human circRNA Microarray.8 hsa_circ_0000337 was one of the top ten upregulated circRNAs with a fold change (FC) =6.255 and P=0.006. To evaluate the feasibility of hsa_circ_0000337 as a diagnostic biomarker and therapeutic target, we verified its expression in ESCC tissues and cell lines and explored the functions of this specific circRNA in vitro by Cell Counting Kit-8 (CCK-8), colony formation, wound healing, and Transwell assays. Moreover, we used bioinformatic analysis and luciferase reporter assay to predict and identify the circRNA–miRNA–mRNA axis in ESCC.
Patients and methods
Clinical specimens
We recruited ESCC patients from the First People’s Hospital of Yancheng City from October 2016 to September 2017. No patients had received chemotherapy or radiotherapy before enrollment. Forty-eight pairs of cancer tissues and matched adjacent normal-appearing tissues were collected during esophagectomy. The cancer tissues were obtained from the center of the cancer lesion and the corresponding adjacent normal-appearing tissues were located >5 cm away from the edge of tumors. All of the tissue specimens were confirmed by two experienced pathologists and were kept in RNAsafety (Shanghai Biotechnology Corporation, Shanghai, People’s Republic of China) and stored at −80°C until use. The characteristics of the study subjects are shown in Table 1. Written informed consent was obtained from all patients. This study was approved by the Institutional Review Board of Nanjing Medical University (Nanjing, People’s Republic of China). After informed consent was obtained from all participants, questionnaires were used to collect demographic data.
  | Table 1 Characteristics of study subjects with ESCC for validation Abbreviation: ESCC, esophageal squamous cell carcinoma. |
Cell lines
The human esophageal carcinoma cell lines KYSE-150 and TE-1 (Chinese Academy of Sciences, Shanghai, People’s Republic of China) and the normal human esophageal epithelial cell line (HET-1A; Shanghai Yu Bo Biological Technology Co., Ltd., Shanghai, People’s Republic of China) were used for functional analysis in Suzhou Healthcare Biology Science Co. Ltd (Suzhou, People’s Republic of China). Cell lines were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA), with 10% FBS (Thermo Fisher Scientific) and antibiotics containing 100 U/mL penicillin and 100 U/mL streptomycin (Sigma-Aldrich Co., St Louis, MO, USA) at 37°C with 5% CO2.
RNA extraction and quantitative reverse transcription (qRT)-PCR
Total RNA was extracted from clinical tissue specimens using a RNeasy Mini Kit (Qiagen, Hilden, Germany) and from cells with Trizol (Thermo Fisher Scientific). cDNAs were synthesized using the PrimeScript™ RT reagent Kit (Qiagen, Hilden, Germany). The qRT-PCR was performed with SYBR® Premix Ex Taq™ (Takara Bio Inc.) on a Roche Applied Science LightCycler 480II (Hoffman-La Roche Ltd., Basel, Switzerland). Divergent primers of circRNAs were designed through primer 5.0 and synthesized by Realgene (Nanjing, People’s Republic of China). The primer sequences for hsa_circ_0000337 were 5′-TCAGGATGCCTTGGGACT-3′ and 5′-TGTGCAGCCGGTCTAACC-3′. The relative expression of hsa_circ_0000337 was calculated by 2−ΔΔCT and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primer sequences for GAPDH were 5′-CGCTCTCTGCTCCTCCTGTTC-3′ and 5′-ATCCGTTGACTCCGACCTTCAC-3′.
Oligonucleotide transfection
We constructed three types of siRNAs targeting hsa_circ_0000337 to inhibit its expression in the TE-1 and KYSE-150 cell lines. The sequences were 5′-GAUGACAAGACAACCAUAATT-3′ and 5′-UUAUGGUUGUCUUGUCAUCTT-3′ for siRNA1, 5′-CAAGACAACCAUAAAGUGUTT-3′ and 5′-ACACUUUAUGGUUGUCUUGTT-3′ for siRNA2, and 5′-GCUUGUUUGGCAAGAAAGATT-3′ and 5′-UCUUUCUUGCCAAACAAGCTT-3′ for siRNA3. We used Lipofectamine 2000 (Thermo Fisher Scientific) to transfect the siRNAs into the cell lines according to the manufacturer’s instructions.
Cell proliferation
Cell proliferation was measured with the CCK-8 assay (Dojindo Laboratories, Kumamoto, Japan) and the colony formation assay. For the CCK-8 assay, we seeded cells into a 96-well plate at a density of 200 cells per well, and then added CCK-8 solution (10 µL) to each well and incubated at 37°C. The absorbance value at 450 nm was measured using a microplate reader (BioTek, Winooski, VT, USA). For the colony formation assay, we plated cells in a 24-well plate at a density of 1×104 cells per well and incubated at 37°C. Colonies were fixed with 4% paraformaldehyde (Sigma-Aldrich Co.) and stained with crystal violet (Sangon Biotech [Shanghai] Co. Ltd., Shanghai, People’s Republic of China) for 20–30 minutes. Cell colonies were then counted and recorded.
Cell invasion
We used a 24-well Transwell chamber (8-µm pore size, Corning® Costar®; Sigma-Aldrich Co.) coated with Matrigel to measure invasive cell ability. In brief, 4×105cells per well were placed into the upper chamber with 200 µL serum-free medium, while medium containing 10% FBS (HyClone, Logan, Utah, USA) was added into the lower chamber. After incubation at 37°C for 24 hours, noninvasive cells in the upper membrane were scraped and the invasive cells in the bottom were then fixed with 95% ethanol (Sinopharm Chemical Reagent Co., Ltd., Shanghai, People’s Republic of China) and stained with 0.5% crystal violet. The stained cells were counted under a microscope.
Cell migration
We performed a wound healing assay by monitoring and quantifying cell growth in a defined area. Cells were cultured in serum-free medium for 24 hours and then mitomycin (1 µg/mL) was added in order to inhibit cell division. We used a 200-µL pipette tip to scrape the center of the well and washed the residual cells with PBS thrice, and then cultured cells in serum-free DMEM at 37°C with 5% CO2. The edge of the scratch was separately photographed at 0 and 24 hours to calculate the relative distance of cell migration. The edge of the scratch was separately photographed at 0 and 24 hours to calculate the relative unhealed area.
Luciferase reporter assay
The luciferase reporter assay was used to test the effect of miRNAs targeted by hsa_circ_0000337. The 3′-UTR sequences of hsa_circ_0000337 with the predicted miRNA binding sites were amplified using PCR and then inserted into a psiCHECK-2 luciferase vector. The plasmid with wild-type or mutated hsa_circ_0000337 and miR-670-5p mimics were transfected into TE-1 and KYSE-150 cell lines using Lipofectamine 2000. After 48 hours of cotransfection, the relative luciferase reporter activity was detected using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Bioinformatics and statistical analysis
We predicted the circRNA/miRNA interaction by miRNA target prediction software (Arraystar’s homemade) established from TargetScan and miRanda. Three bioinformatics databases, including TargetScan (http://www.targetscan.org/vert_72/), miRDB (http://www.mirdb.org/), and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) were used to predict the target genes of related miRNAs. Then, we took the intersection of the three databases using Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html). The Shapiro–Wilk test was used to detect the normality of quantitative data. Comparisons between groups were analyzed with the Wilcoxon signed-rank test. All statistical analyses were performed using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered as statistically significant.
Ethics statement
This study was approved by the Ethics Committee of Nanjing Medical University. After informed consent was obtained from all participants, questionnaires were used to collect demographic data. This study was conducted in accordance with the Declaration of Helsinki.
Results
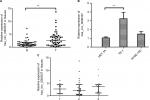
hsa_circ_0000337 overexpression in ESCC tissues and cell lines
In the chip-based discovery stage,8 we observed that hsa_circ_0000337 was one of the top ten upregulated miRNAs in ESCC with a fold change of over 6.2 and P=0.006. In the current study, we further validated this finding by comparing the expression level of hsa_circ_0000337 between cancer lesions and matched adjacent normal-appearing tissues from 48 patients with ESCC. As shown in Figure 1A, it was expressed at a high level in ESCC clinical samples. Additionally, we further explored the altered expression of hsa_circ_0000337 using in vitro cell lines. Compared to HET-1A (normal human esophageal epithelial cell line), it was significantly overexpressed in TE-1 cell lines (well-differentiated squamous carcinoma), but with no significant difference in KYSE-150 cell lines (poorly differentiated squamous cell carcinoma) (Figure 1B). However, no significant relationship was observed between hsa_circ_0000337 and differentiation in ESCC clinical samples (Figure 1C).
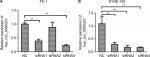
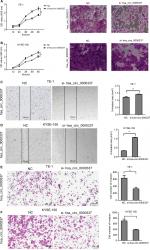
hsa_circ_0000337 knockdown inhibits proliferation and invasion, and promotes apoptosis in ESCC
We constructed three types of siRNAs targeting hsa_circ_0000337 to inhibit its expression in the TE-1 and KYSE-150 cell lines. As shown in Figure 2, siRNA3 significantly silenced the expression of hsa_circ_0000337 compared to the negative control. We then selected the siRNA3 interference cell (si-hsa_circ_0000337) as a competitive inhibitor to observe the function of hsa_circ_0000337 on the proliferation, migration, and invasion of ESCC. As shown in Figure 3A and B, knockdown of hsa_circ_0000337 significantly inhibited the growth of TE-1 and KYSE-150 cell lines in both the CCK-8 assay and colony formation assay. According to the wound healing assay, inhibition of hsa_circ_0000337 could slow the migration rate of the TE-1 and KYSE-150 cell lines (Figure 3C and D). Silencing of hsa_circ_0000337 significantly limited cell invasion capacity in accordance with the Transwell assay (Figure 3E and F).
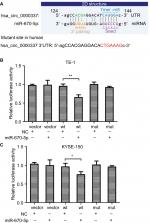
hsa_circ_0000337 serves as a sponge for miR-670-5p
Considering the sequence of the miRNA response element, we defined five miRNAs as having potential binding sites of hsa_circ_0000337. These miRNAs were miR-155-5p, miR-193b-5p, miR-494-5p, miR-670-5p, and miR-766-5p. Then, we used the luciferase reporter assay to verify whether these miRNAs could directly target hsa_circ_0000337. Our results suggested that luciferase activity declined following cotransfection with hsa_circ_0000337 wild-type and miR-670-5p mimics in TE-1 and KYSE-150 cell lines, while cotransfection with miR-670-5p mimics and the mutant hsa_circ_0000337 had no obvious effect on the relative luciferase activity (Figure 4). However, there was no significant difference with the other four miRNAs (data not shown). This observation indicated that hsa_circ_0000337 may directly serve as a sponge for miR-670-5p in ESCC.
To date, the well-accepted functional pattern of circRNAs is the circRNA-miRNA-mRNA axis.22,23 In the current study, we also predicted the target genes of miR-670-5p using the TargetScan, miRDB, and miRTarBase databases (Figure 5). We took the intersection from these databases and predicted 21 target genes, including GJB7, PCTP, STX6, FAM174B, PPP2CA, TOR2A, YOD1, ZSWIM6, UBR7, PROX1, SEMA4B, DHX33, PPP1R37, PLXDC1, ANAPC16, ZNF385A, SP1, SH3BP5L, MKNK2, SEMA4C, and EHD1.
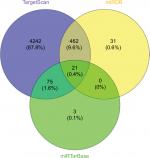  | Figure 5 Predicted genes targeted by miR-670-5p. |
Discussion
circRNAs represent a new subtype of endogenous ncRNAs involved in carcinogenesis, and have shown promise as biomarkers for the diagnosis and prognosis of human cancers.24–26 The circRNA hsa_circ_0000337 has been reported to be upregulated in ESCC,8 but its underlying mechanism has not yet been discovered. In the current study, we validated the upregulation of hsa_circ_0000337 in both ESCC cell lines and clinical samples. Moreover, we performed in vitro experiments, including CCK-8 and colony formation assays, as well as Transwell, wound healing, and luciferase reporter assays and revealed that hsa_circ_0000337 could promote the proliferation, migration, and invasion of ESCC cancer cells. Bioinformatics analysis and luciferase reporter assay suggested that miR-670-5p was the target miRNA of hsa_circ_0000337.
Compared to miRNA and lncRNA, circRNA is highly abundant, stable, and specifically expressed, which makes it an ideal molecular biomarker. For example, hsa_circ_002059, which is downregulated in gastric cancer tissues, has been suggested to be a prognostic biomarker since its expression in plasma was associated with tumor grade and distal metastasis.27 circFBLIM1 plays a vital role in HCC progression.28 circNT5E functions as an important tumor promoter in glioblastoma tumorigenesis by stimulating tumor proliferation, invasion, migration, and apoptosis,29 while the circITGA7 can inhibit colorectal cancer growth and metastasis.30 In terms of potential molecular mechanisms, studies have indicated that circRNAs containing miRNA target sites can serve as miRNA sponges and participate in posttranscriptional regulation of expression. For example, hsa_circ_0072995 acts as the sponge of miR-30c-2-3p and promotes breast cancer cell migration and invasion,29 while circMTO1 suppresses HCC progression by binding to miR-9.31
hsa_circ_0000337 is located on chr11: 70200406–70202360 with a length of 419 bp and its gene symbol is PPFIA1 (PTPRF interacting protein alpha 1). Studies have reported that PPFIA1 amplification was frequently observed in breast cancer, especially, in patients with liver metastasis, and seemed to be a promising prognostic indicator of metastatic relapse.32–34
The exact mechanism of circRNA functions in ESCC remains unclear. We hypothesized that hsa_circ_0000337 could bind to specific miRNAs and regulate gene transcription through the circRNA–miRNA–mRNA axis.35,36 Bioinformatics analysis and luciferase reporter assay showed that hsa_circ_0000337 interacts with miR-670-5p, which is a key miRNA that targets multiple genes. It has been reported that miR-670-5p was associated with hepatocellular carcinoma by modulating PROX1 expression at the posttranscriptional level.37 We predicted 21 candidate genes as the possible targets of miR-670-5p, of which PPP2CA, YOD1, PROX1, SEMA4B, DHX33, and SP1 were associated with tumor proliferation, migration, or invasion according to the published literature.38–43
hsa_circ_0000337 was observed to be upregulated in ESCC in a chip-based discovery study,8 and it was also validated in the current study by expanding the sample size. We further investigated the expression level of hsa_circ_0000337 in ESCC cell lines. We found that it was overexpressed in TE-1 cells, but was not significantly different in KYSE-150 cells. The TE-1 and KYSE-150 cell lines were isolated from ESCC patients with different differentiation pathology. Therefore, we assumed that hsa_circ_0000337 might be associated with tumor differentiation. In the current study, due to the limited number of samples with poor differentiation, we did not observe a significant relationship between hsa_circ_0000337 and ESCC differentiation.
There are several limitations to our study. First, although we performed a validation study by expanding the sample size, the number of study subjects was still low and insufficient to prove the clinical value of the candidate circRNA as a biomarker. Recruitment of more ESCC patients across multiple centers with different grades and stages will help to validate our current findings. Second, we measured the function of hsa_circ_0000337 by knocking down its expression in vitro and predicted its target genes by bioinformatic analysis, but we did not perform overexpression analysis for further validation. Finally, the downstream functional proteins regulated by hsa_circ_0000337 in vivo should be explored in the future.
Conclusion
hsa_circ_0000337 was upregulated in ESCC and promoted cancer progression by regulating cell proliferation, migration, and invasion. Our findings indicate that hsa_circ_0000337 may act as a promising biomarker and provide a new therapeutic target in ESCC.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (81673249), National Key R&D Program of China (2017YFC0907000), Social Development Project in Jiangsu Province (BE2015694), Scientific Research Innovation Project for Graduate Students in Jiangsu Province (KYCX17_1293), and Priority Academic Program Development of Jiangsu Higher Education Institutions. The funding agencies had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author contributions
HS and JW conceived, initiated, and led the study. HS, DX, PS, BH, ZL, YJ, and CKA collected and analyzed the data with input from all authors. HS, DX, PS, and JW prepared the manuscript. All authors contributed to the data analysis, drafted and revised the paper, and agreed to be accountable for all aspects of the work. All authors reviewed and approved the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Kaikkonen MU, Adelman K. Emerging roles of non-coding rna transcription. Trends Biochem Sci. 2018;43(9):654–667. | ||
Tan L, Yu JT, Hu N, Tan L. Non-coding RNAs in Alzheimer’s disease. Mol Neurobiol. 2013;47(1):382–393. | ||
Romaine SP, Tomaszewski M, Condorelli G, Samani NJ. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart. 2015;101(12):921–928. | ||
Li PF, Chen SC, Xia T, et al. Non-coding RNAs and gastric cancer. World J Gastroenterol. 2014;20(18):5411–5419. | ||
Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. | ||
Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521. | ||
Lu L, Sun J, Shi P, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8(27):44096–44107. | ||
Shi P, Sun J, He B, et al. Profiles of differentially expressed circRNAs in esophageal and breast cancer. Cancer Manag Res. 2018;10:2207–2221. | ||
Sun LL, Li WD, Lei FR, Li XQ. The regulatory role of microRNAs in angiogenesis-related diseases. J Cell Mol Med. 2018;22(10):4568–4587. | ||
Huang Y, Xiang B, Liu Y, Wang Y, Kan H. lncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56–66. | ||
Yong W, Yu D, Jun Z, et al. Long noncoding RNA NEAT1, regulated by Lin28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death Dis. 2018;9(9):861. | ||
Lingadahalli S, Jadhao S, Sung YY, et al. Novel lncRNA LINC00844 regulates prostate cancer cell migration and invasion through AR signaling. Mol Cancer Res. 2018;16(12):1865–1878. | ||
Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell. 2017;66(1):e27:9–21. | ||
Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. | ||
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. | ||
Nicolet BP, Engels S, Aglialoro F, van den Akker E, von Lindern M, Wolkers MC. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 2018;46(16):8168–8180. | ||
Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. | ||
Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. | ||
Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23(14):3918–3928. | ||
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. | ||
Shin D, Protano MA, Polydorides AD, et al. Quantitative analysis of high-resolution microendoscopic images for diagnosis of esophageal squamous cell carcinoma. Clin Gastroenterol Hepatol. 2015;13(2):272–279. e272. | ||
Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17(1):140. | ||
Chen F, Feng Z, Zhu J, et al. Emerging roles of circRNA_NEK6 targeting miR-370-3p in the proliferation and invasion of thyroid cancer via Wnt signaling pathway. Cancer Biol Ther. 2018;19(12):1139–1152. | ||
Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. | ||
Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11(1):21. | ||
Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. | ||
Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. | ||
Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. 2018;37(1):172. | ||
Zhang HD, Jiang LH, Hou JC, et al. Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics. 2018;10(9):1229–1242. | ||
Li X, Wang J, Zhang C, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246(2):166–179. | ||
Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. | ||
Dancau AM, Wuth L, Waschow M, et al. PPFIA1 and CCND1 are frequently coamplified in breast cancer. Genes Chromosomes Cancer. 2010;49(1):1–8. | ||
Choi EJ, Yun JA, Jabeen S, et al. Prognostic significance of TMEM16A, PPFIA1, and FADD expression in invasive ductal carcinoma of the breast. World J Surg Oncol. 2014;12:137. | ||
Yang J, Wu NN, Huang DJ, et al. PPFIA1 is upregulated in liver metastasis of breast cancer and is a potential poor prognostic indicator of metastatic relapse. Tumour Biol. 2017;39(7):1010428317713492. | ||
Xiong DD, Dang YW, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220. | ||
Cheng J, Zhuo H, Xu M, et al. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16(1):216. | ||
Shi C, Xu X. miR-670-5p induces cell proliferation in hepatocellular carcinoma by targeting PROX1. Biomed Pharmacother. 2016;77:20–26. | ||
Jiang D, He Z, Wang C, et al. Epigenetic silencing of ZNF132 mediated by methylation-sensitive Sp1 binding promotes cancer progression in esophageal squamous cell carcinoma. Cell Death Dis. 2018; 10(1):1. | ||
Jian H, Zhao Y, Liu B, Lu S. SEMA4B inhibits growth of non-small cell lung cancer in vitro and in vivo. Cell Signal. 2015;27(6):1208–1213. | ||
Zhang B, Ji S, Ma F, Ma Q, Lu X, Chen X. miR-489 acts as a tumor suppressor in human gastric cancer by targeting PROX1. Am J Cancer Res. 2016;6(9):2021–2030. | ||
Wang LQ, Zhang Y, Yan H, Liu KJ, Zhang S. MicroRNA-373 functions as an oncogene and targets YOD1 gene in cervical cancer. Biochem Biophys Res Commun. 2015;459(3):515–520. | ||
Bhardwaj A, Singh S, Srivastava SK, et al. Restoration of PPP2CA expression reverses epithelial-to-mesenchymal transition and suppresses prostate tumour growth and metastasis in an orthotopic mouse model. Br J Cancer. 2014;110(8):2000–2010. | ||
Wang H, Yu J, Wang X, Zhang Y. The RNA helicase DHX33 is required for cancer cell proliferation in human glioblastoma and confers resistance to PI3K/mTOR inhibition. Cell Signal. 2019;54:170–178. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.