Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 16
Up-Regulated Expression of ICAM1, MT1A, PTGS2, LCE3D, PPARD, and GM-CSF2 Following Solar Skincare Protection and Repair Strategies in a 3-Dimensional Reconstructed Human Skin Model
Authors Tanaka Y , Parker R, Aganahi A
Received 2 August 2023
Accepted for publication 3 October 2023
Published 12 October 2023 Volume 2023:16 Pages 2829—2839
DOI https://doi.org/10.2147/CCID.S428170
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Video abstract presented by Richard Parker.
Views: 712
Yohei Tanaka,1 Richard Parker,2 Amaryllis Aganahi2
1Clinica Tanaka Plastic, Reconstructive Surgery and Anti-Aging Center, Matsumoto, Nagano, Japan; 2RATIONALE, Kyneton, VIC, Australia
Correspondence: Yohei Tanaka, Clinica Tanaka Plastic, Reconstructive Surgery and Anti-aging Center, M-1 Bld 1F, 3-4-3, Ote, Matsumoto, Nagano, 390-0874, Japan, Tel +81-263-36-0016, Fax +81-263-36-0016, Email [email protected]
Background: Clinical, optical and histological research confirms that solar skin damage continues to pose a threat to human skin health globally despite widespread sunscreen usage and sun awareness campaigns. Despite this, very few studies examine the critical changes in gene expression and DNA repair activity following recommended topical solar protection and repair strategies to ameliorate the harmful effects of ultraviolet, visible light and near-infrared radiation.
Purpose: To investigate alterations in gene expression following topical solar protection and solar repair strategies.
Methods: Using epidermal keratinocytes and dermal fibroblasts derived from a 3-dimensional reconstructed human skin model, gene expression was assessed via the Genemarkers Standard Skin Panel using 112 genes deploying two analytical techniques: DNA microarray and quantitative real-time PCR exploration. Tissues were inoculated with products then collected after 24 hours following application of solar protection formulations and 16 hours following solar repair formulations (The Essential Six, RATIONALE, Victoria, Australia).
Results: A DNA microarray revealed 67 genes that were significantly up-regulated or down-regulated following the treatment. The quantitative real-time PCR revealed that, in comparison to the control, the genes encoding Intercellular Adhesion Molecule 1 (ICAM1), Metallothionein 1A (MT1A), Prostaglandin-Endoperoxide Synthase 1 (PTGS2), Late Cornified Envelope 3D (LCE3D), Peroxisome Proliferator Activated Receptor (PPARD), and Granulocyte/Macrophage Colony Stimulating Factor 2 (GM-CSF2) have been up-regulated following usage of the solar protection regime, 1.87, 861.16, 4.34, 1.91, 1.06, and 3.6, respectively. ICAM1, MT1A, PTGS2, LCE3D, PPARD, and GM-CSF2 were up-regulated following use of the solar repair regime, 3.78, 2.98, 14.89, 5.09, 2.42, and 13.51, respectively.
Conclusion: This study demonstrates that a specific solar protection and repair regime upregulated genes involved in photoprotection and repair mechanisms in a 3-dimensional (3D) reconstructed human-like skin model.
Keywords: gene expression, anti-photoageing, anti-photoimmunosuppression, antioxidant, photoprotection, DNA repair
Introduction
The composition of the sun’s incident solar energy is approximately 50% near-infrared (NIR), 40% visible light (VL) and 10% ultraviolet (UV) radiation.1,2 In recent decades, world-wide educational campaigns have been launched with the aim of preventing solar skin damage and photoaging, leading to the widespread adoption of sunscreen use. Yet despite the universal desire to avoid skin cancer, photodermatoses, photoimmunosuppression and photoageing, solar skin damage continues to pose a threat to human health worldwide.2–6
Accumulating evidence elucidates the combined deleterious effects of VL and NIR solar energy when combined with UV radiation. Daily and sustained solar exposure to the sun’s total energy output is increasingly being implicated in the widespread detection of skin cancer and cutaneous photodamage.2–7
It has previously been reported that NIR emitted from the sun can be considered as a catalyst for skin ageing. When innate NIR protection is inadequate or impaired, solar NIR will induce harmful effects including photodermatoses, skin ptosis, vasodilation, sagging, muscle thinning and possibly photocarcinogenesis following sustained, accumulative exposure.2–7
In previous studies, the authors reported the lack of global sunscreens (SPF50+, PA+++ or ++++) capable of protecting skin against the entire solar spectrum (UV, VL and NIR). Daily skin health regimes that go beyond daily sunscreen use to include immune-enhancing, antioxidant, barrier restoration, pH recalibration and DNA repair strategies are now widely recommended by dermatologists worldwide.2,3
Interestingly, the solar protection and repair regime investigated in previous studies also provided significant improvements in terms of skin texture, appearance, clarity, and firmness as demonstrated in multidimensional assessments (2D and 3D). Most patients participating to the self assessment studies reported high levels of satisfaction.2,3 As the effects of solar energy (UV, VL, NIR) on the skin are now understood to be the major cause of skin ageing, it is reasonable to hypothesize that this skincare approach is able to provide optimal protection and repair of solar damage including photoageing and immunosuppression.2,3
Antiageing strategies using skincare topicals with active agents demand scientific proof of efficacy, and one investigation in this realm involves gene expression testing.8–10 Despite this, there remain few studies that examine gene expression changes and DNA-mitigated skin protection and repair activity following solar protection and repair strategies ranging from UV through to NIR radiation.
To investigate alternations in gene expression following solar protection and solar repair strategies, we assessed changes in gene expression activation or inhibition using various DNA assay and analysis methods on a 3D human skin model containing epidermal keratinocytes and dermal fibroblasts.
Methods
Skin Model
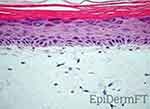
The selected human skin in-vitro replica of full thickness skin (Figure 1) is a reconstructed multilayered 3D human skin model. It contains normal, human-derived epidermal keratinocytes (NHEK) and normal, human-derived dermal fibroblasts (NHFB) (EpiDermFT, MatTek, MA, USA). The EpiDermFT system used in this study consists of NHEK and NHFB. The cultures of NHEK and NHFB are configured to replicate a human multilayer and highly differentiated epidermis and dermis. The skin model is composed of epidermally organized basal, spinous, granular, and cornified epidermal layers similar to those in human (in-vivo) skin anatomy. The dermal compartment contains a collagen matrix incorporating viable normal human dermal fibroblasts (NHDF). The EpiDermFT system is active on a mitotic and metabolic level.
The model presents various markers inherent to mature in vivo epidermal differentiation (pro-filaggrin, the K1/K10 cytokeratin pair, involucrin, and type I epidermal transglutaminase) as well as structural markers (keratohyalin granules, tonofilament bundles, desmosomes), specifically the intercellular lamellar lipids layered in a similar pattern to in situ epidermis.
Additionally, the dermal/epidermal junction (DEJ) is supported by a well-defined basement membrane. Other markers demonstrating the similarity of the EpiDermFT system with the in-vivo skin model include the presence of hemidesmosomes, lamina lucida, lamina densa, anchoring fibril structures, structural and signalling proteins (collagen IV, laminin, collagen VII and integrin α6). EpiDermFT is most commonly used for anti-aging, collagen synthesis, wound healing, and photoprotection studies. Clear and straightforward protocols utilize modern-day techniques to study gene and protein expression, cytokine release, histological changes, and other skin-specific markers. In order to obtain high reproducibility levels, the EpiDermFT tissues have been produced following Good Manufacturing Practice (GMP) procedures.
Structurally and morphologically, the NHEK and NHFB arrays are remarkably accurate in replicating consistent and reliable levels of differentiation and intercellular activity analogous to human skin.
Topical Skincare Formulations
The skincare formulations (The Essential Six from RATIONALE, Victoria, Australia) used to conduct this study represent a comprehensive topical regimen to protect skin from solar assault during the day and repair environmental damage at night. Each formulation is a specific combination of 50 to 100 compounds grouped and listed by function or nature below (Table 1).
 |
Table 1 Solar Protection and Repair Formulation Compositions by Function or Nature |
This approach involves comprehensively shielding the skin against the broader solar spectrum (approximately 290 nm to 3200 nm). The Essential Six daily regime is composed of three solar protection formulations for day (#1 The Serum, #2 The Serum, and #3 The Tinted Serum SPF50+) and three nightly solar repair formulations (#4 The Crème, #5 The Serum, and #6 The Night Crème).
Four cultures were used with each treatment group (Solar Protection and Solar Repair Formulations). At approximately 9:30 am, four tissues were inoculated with 5uL of each of the Solar Repair Formulations at the centre of each EFT-400 culture, one after the other using a calibrated positive displacement pipette and a sterile glass spreader to distribute the topical materials across the surface in between each application.
Post applications, the cultures were returned to the incubator at 37°C with 5% CO2 and ~95% relative humidity. Around 5:20pm the tissues were rinsed off to clear tissues from excess of Formulations and returned to the incubator at 37°C with 5% CO2 and ~95% relative humidity until the next day. At approximately 5:30 pm, four other tissues were inoculated with 5uL of each of the Solar Repair Formulations at the centre of each EFT-400 culture, one after the other using a calibrated positive displacement pipette and a sterile glass spreader to distribute the topical materials across the surface in between each application. The cultures were then returned to the incubator at 37°C with 5% CO2 and ~95% relative humidity and collected at approximately 9:30 am the next morning to be washed and remove any excess from the surface of the culture with sterile DPBS. The Solar Protection samples were also collected on that morning.
Solar Protective Formulations
#1 The Serum
Vitamin B complex and Australian botanical extracts enhance skin immune responses to protect from solar induced photoimmunosuppression.
#2 The Serum
An extensive complex of skin identical vitamins, minerals and enzymatic antioxidants helps prevent the formation of Reactive Oxygen Species (ROS) and free radicals induced from solar energy and environmental pollution.
#3 The Tinted Serum SPF50+
A daily solar spectrum protection formulation (UV + VL and NIR radiation) containing zinc oxide, iron oxides, provitamin D, melanin and heat shock proteins.2
Solar Repair Formulations
#4 The Crème
A skin identical composition of stratum corneum lipids including specialised ceramides, triglycerides and cholesterol assists in restoring and augmenting skin barrier function.
#5 The Serum
This complex of alpha and beta hydroxy acids at low pH is designed to maintain stratum corneum pH at optimal acidic levels.
#6 The Night Crème
A synergistic combination of vitamin A and DNA repair enzymes assists in promoting skin cellular DNA repair processes in response to solar and environmental damage.
RNA Extraction
After each of the tissue samples were collected from the incubator at 37°C with 5% CO2 and ~95% relative humidity, each culture was placed into a RNAlater preservative solution tube and incubated for 1–2 hours at room temperature. Post incubation, each tube was transferred to a 4°C refrigerator until RNA isolation.
The extraction and isolation of the total RNA content was performed out of 12 (4 per test material and the negative control) highly differentiated cultures of 3D reconstructed human dermal fibroblasts and epidermal keratinocyte models (Mattek EFT-400) using the Maxwell RSC Simply RNA kit (Promega). Using UV absorbance, quantitative and qualitative measurements of RNA samples were determined.
cDNA Synthesis
cDNA samples were produced using a High Capacity cDNA Synthesis Kit (Applied Biosystems). About 2000ng total of RNA per sample were required in order to generate first-strand cDNA.
Openarray Processing and Analysis
The method deployed to perform the qPCR reactions required the use of validated gene expression assays in an OpenArray and 384-well format. Assays were run in a Life Technologies QuantStudio 12K Flex instrument. To ensure accurate results, every single gene was assayed in duplicate.
The quality of the raw data files obtained from qPCR method as well as the statistical analysis were assessed and performed using the ThermoFisher Connect Software (Life Technologies). Using the relative quantitation (RQ) method to determine the statistical analysis, the difference (delta) of quantification Cycle (dCq) values was calculated by normalizing the quantification Cycle (Cq) value of the target genes to the Cq value of a control gene (endogenous). This method highlights potential variability happening during the experimental process between several samples.
Endogenous Control Gene Selection
The endogenous control gene selection is critical in order to isolate a consistently expressed control gene. Five candidate control genes, Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH), Peptidylprolyl Isomerase A (PPIA), Hypoxanthine Phosphoribosyltransferase (HPRT1), Polyubiquitin-C (UBC) and β-Glucuronidase (GUSB) were analysed for the OpenArray and two candidate control genes (GAPDH and PPIA) for 384-well plate format. Statistical analysis (unpaired t-tests) was performed for each comparison using normalized dCq values. Two genes (GAPDH and PPIA) were analyzed with the OpenArray method and were selected as candidates to be used as control genes for 384-well plate method. The most consistent endogenous control gene was chosen based on the stability score and range scores calculated by the ThermoFisher Data Connect RQ software. The lower the stability scores, the more consistent will be the gene expression between the different samples. PPIA was selected as the endogenous control for both the OpenArray and 384-well plate formats (Figure 2).
qPCR Data Quality and Statistical Data Analysis
The quality of qPCR data was assessed using a variety of reporting techniques such as visual assessment of the qPCR curve and the Cq value. Cq values are an indication of the transcript total present in the sample and can impact the quality of the qPCR data. qPCR amplification takes place over a total of 40 cycles, and typically occurs before cycle 30 in a 384-well plate format. The quality and relative quantity of transcript is then linked to the Cq values:
- Cq values below 30 correspond to high transcript levels and robust, high-quality PCR data.
- Cq values above 30 correspond to lower level transcript, less robust qPCR data; data should be reviewed cautiously.
Results
A DNA Openarray identified 67 genes that were statically significant up-regulated or down-regulated (Table 2) and 37 genes that were drastically up-regulated at least twofold following solar protection and repair regimes. When compared to the control cells, quantitative real-time PCR identified that the genes encoding for Intercellular Adhesion Molecule 1 (ICAM1), Metallothionein 1A (MT1A), Prostaglandin-Endoperoxide Synthase 1 (PTGS2), Late Cornified Envelope 3D (LCE3D), Peroxisome Proliferator Activated Receptor (PPARD), and Granulocyte/Macrophage Colony Stimulating Factor 2 (GM-CSF2) were up-regulated following usage of the solar protection regime; 1.87, 861.16, 4.34, 1.91, 2.42, and 3.6, respectively. ICAM1, MT1A, PTGS2, LCE3D, PPARD, and GM-CSF2 were up-regulated following the solar repair regime; 3.78, 2.98, 14.89, 5.09, 2.42, and 13.51, respectively (Table 3 and Figure 3).
 |
Table 2 A DNA Microarray Showed 67 Genes That Were Statically Significant Up-Regulated or Down-Regulated |
 |
Table 3 Gene Function and Fold Expression Change of ICAM1, MT1A, PTGS2, LCE3D, PPARD, and GM-CSF2 |
Discussion
Improvement in skin clarity, tone, firmness and texture are the most desired benefits of patients seeking medical procedures including facelifting, cosmetic injectables and phototherapy.2 Aggressive surgical procedures and ablative treatments have not met with widespread acceptance due to the risks they present as well as the inconsistent outcomes and considerable effects on patient lives. Laser or light therapies may provide the expected results as long as the treatment is continuous at clinics. Nevertheless, these results often come with significant inflammation and downtime. Medical procedures such as, facelifting, threadlifting and fillers provide some level of improvement as skin and subtissues are lifted but would not provide the expected visual skin rejuvenation.2
The heightened physical and mental health implications caused by solar damage and skin ageing highlight the imperative for home-based, non-invasive treatments.2,3
In our prior studies, the results induced by this specific topical solar protection and repair regime were significant, as demonstrated and documented via objective digital facial surface analysis and 3-D volumetric assessments.2,3 Through this objective method, outcomes in terms of skin improvements highlighted above were visible and impressive. Patients reported a high level of satisfaction and comfort with low to no downtime, discomfort or side effects.
Post treatment, most patients reported that their skin and facial profile felt and looked rejuvenated, tightened and lifted.2,3
To our knowledge, this study represents the first body of scientific work to examine, measure, assess and report alterations in gene expression following a specific solar protection and repair skincare regime. Gene regulation was investigated using DNA microarray and real-time PCR analysis on a 3-dimensional reconstructed human skin model containing epidermal keratinocytes and dermal fibroblasts. The results induced by this specific topical solar protection and repair regime revealed that 67 genes were significantly up-regulated or down-regulated with accompanying improvements in skin function and appearance (Table 2). ICAM1, MT1A, PTGS2, LCE3D, PPARD, and GM-CSF2 were up-regulated following both of the solar protection and solar repair protocol (Table 3 and Figure 3).
ICAM1 was up-regulated following the solar protection and repair regime; 1.87, and 3.78, respectively. This gene encodes for the production of cell surface glycoproteins which are typically expressed on endothelial and immune cells.11,12 In human skin, ICAM1 is involved in regulation of immunologic responses induced by environmentally stimulated proinflammatory cytokines.12,13 Photoimmunosuppression is increasingly cited as a major cause of photoageing and solar induced skin diseases, including skin cancer.14 Up-regulation of ICAM1 may enhance prevention of photoimmunosuppression.
MT1A was up-regulated following the solar protection and repair regime, 861.16 and 2.98, respectively. Low molecular weight and cysteine-rich proteins binding divalent heavy metal ions, acting as anti-oxidants and protecting against the formation of hydroxyl-free radicals are encoded by this gene.15–17 These proteins are important in intercellular homeostatic control of metals, in particular the detoxification of heavy metals.15,16 In human skin, MT1A is involved with anti-oxidant detoxification of cells and tissues.15,16 Increased metallothionein leads to a universal reduction in inflammation, free radical formation and damage caused by oxidative stress.17,18 The dramatically increased activity of MT1A seen in this study may indicate potent anti-oxidant ability and may be potentially beneficial for skin anti-ageing, as metallothionein is related to anti-apoptotic properties and the ability to scavenge ageing free radicals.17
PTGS2 was up-regulated following the solar protection, and repair regime; 4.34, and 14.89, respectively. PTGS2 encodes for the inducible isozyme in the biosynthesis of prostaglandin, and takes on a double action of dioxygenase and peroxidase.19 It is suggested that PTGS2 plays a role in the pathway of the prostanoid synthesis involved in both mitogenesis and inflammation due to its regulation through specific stimulatory processes.19 In human skin, its expression is induced in fibroblasts, keratinocytes and melanocytes through a series of mitogenic and inflammatory stimuli which include UV radiation, cytokines, hypoxia and growth factors.20,21 Significant up-regulation of PTGS2 following the skin regime may indicate enhanced photoprotection and antiphotoageing potential.
LCE3D was up-regulated following the solar protection, and repair regime; 1.91, and 5.09, respectively. The Late Cornified Envelope gene encodes for the proteins involved in keratinization.22,23 These proteins are the precursors of the stratum corneum cornified envelope.22,24 In human skin, the genes that encode cornified envelope proteins are clustered in an epidermal differentiation complex, of which LCE3D is a key component, regulating epidermal barrier formation and integrity.24 LCE aberrations are associated with multiple skin diseases, including psoriasis and atopic dermatitis.23 LCE gene expression is altered by exposure to UV radiation. LCE3D directly modulates barrier quality.23 Up-regulation of LCE3D is involved in maintaining healthy skin homeostasis and possibly photoprotection.
PPARD was up-regulated following the solar protection, and repair regime; 1.06, and 2.42, respectively. PPARD regulates epidermal metabolism and lipid synthesis, and the encoded protein is expected to act as a transcriptional repression integrator and nuclear receptor signalling target.25–27 In human skin, PPARD activation involved in the regulation of genes encoding for epidermal differentiation, skin barrier homeostasis, lipid biosynthesis and inflammation.26–28 Up-regulation of PPARD is thought to be helpful in maintaining skin barrier and cellular repair processes.
GM-CSF2 was up-regulated following the solar protection, and repair regime; 3.6, and 13.51, respectively. This gene is responsible for encoding a cytokine engaged in the optimum functioning (production, differentiation, and function) of granulocyte and macrophage colonies.29,30 In human skin, GM-CSF2 is involved in epidermal and dermal immune responses in addition to the control of proliferation and differentiation.31 Significant up-regulation of GM-CSF2 appears to be beneficial for skin repair and rejuvenation.
For this study, an in vitro skin model was deployed specifically epidermal keratinocytes and dermal fibroblasts from a multilayered 3-dimensional cultured human skin model. This reconstructed skin model is highly comparable to that of living human skin. This was established through histological analysis which revealed a fully stratified epidermis containing all major epidermal layers and component cells as well as a dermal compartment and its collagen matrix (Figure 1). This highly analogous human skin model proved highly predictive and accurate in understanding the biological impacts of this specific topical solar protection and repair regime.
Our findings that specific genes involved in enhanced photoprotection and repair of solar skin damage, namely, ICAM1, MT1A, PTGS2, LCE3D, PPARD, and GM-CSF2, warrant further investigation, particularly in vivo studies. Although significant upregulation of the gene expressions occurred following this skincare regime, further research is needed to determine whether other skincare ingredients, treatments or medical procedures could promote enhance further changes in gene expression. Furthermore, this study was a preliminary assessment, suggesting that a larger pool of samples as well as a protein expression study could follow.
Conclusion
This study demonstrates that a specific solar protection and repair regime is capable of upregulating genes that are significantly active in endogenous solar skin protection and repair processes involving the UV, VL and NIR spectra.
Funding
The authors disclose that this study was entirely funded by RATIONALE Skincare Pty Ltd, Victoria, Australia.
Disclosure
Professor Yohei Tanaka is a paid consultant plastic surgeon for RATIONALE. Richard Parker and Amaryllis Aganahi are paid employees of RATIONALE. The authors report no other conflicts of interest in this work.
References
1. Kochevar IE, Pathak MA, Parrish JA, et al. Photophysics, photochemistry and photobiology. In: Freedberg IM, Eisen AZ, Wolff K, editors. Fitzpatrick’s Dermatology in General Medicine. New York: McGraw-Hill; 1999:220–2299.
2. Tanaka Y. Long-term objective assessments of skin rejuvenation using solar protection and solar repair shown through digital facial surface analysis and three-dimensional volumetric assessment. Clin Cosmet Investig Dermatol. 2019;12:553–561. doi:10.2147/CCID.S218176
3. Tanaka Y. Three-dimensional quantification of skin surface displacement following skin rejuvenation using solar protection and solar repair. J Clin Aesthet Dermatol. 2020;13(7):47–50.
4. Tanaka Y, Gale L. Beneficial applications and deleterious effects of near-infrared from biological and medical perspectives. Opt Photonics J. 2013;3:31–39. doi:10.4236/opj.2013.34A006
5. Tanaka Y, Gale L. Protection from near-infrared to prevent skin damage. Opt Photonics J. 2015;5:113–118. doi:10.4236/opj.2015.54010
6. Tanaka Y, Nakayama J. Up-regulated epidermal growth factor receptor expression following near-infrared irradiation simulating solar radiation in a 3-dimensional reconstructed human corneal epithelial tissue culture model. Clin Interv Aging. 2016;11:1027–1033. doi:10.2147/CIA.S111530
7. Tanaka Y. The impact of near-infrared radiation in dermatology. World J Dermatol. 2012;1:30–37. doi:10.5314/wjd.v1.i3.30
8. Widgerow AD, Garruto JA. Gene Expression Studies Pertaining to Extracellular Matrix Integrity and Remodeling: nuances and Pitfalls of In Vitro Investigations. J Drugs Dermatol. 2019;18(12):1255–1259.
9. Smith K, Nido P, Maitra P, Cheng T, Kadoya K. Rationale and Preclinical Evaluation of a Multimodal Topical Body Skincare Product for Toning and Tightening. J Drugs Dermatol. 2021;20(10):1041–1044. doi:10.36849/JDD.6401
10. McDaniel DH, Dover JS, Wortzman M, Nelson DB. In vitro and in vivo evaluation of a moisture treatment cream containing three critical elements of natural skin moisturization. J Cosmet Dermatol. 2020;19(5):1121–1128. doi:10.1111/jocd.13359
11. Available from: https://www.ncbi.nlm.nih.gov/gene/3383.
12. Xiong H, Xu Y, Tan G, et al. Glycyrrhizin ameliorates imiquimod-induced psoriasis-like skin lesions in BALB/c mice and inhibits TNF-alpha-induced ICAM-1 expression via NF-kB/MAPK in HaCaT Cells. Cell Physiol Biochem. 2015;35(4):1335–1346. doi:10.1159/000373955
13. Bito T, Roy A, Sen CK, et al. Flavonoids differentially regulate IFN gamma-induced ICAM-1 expression in human keratinocytes: molecular mechanisms of action. FEBS Lett. 2002;520(1–3):145–152. doi:10.1016/S0014-5793(02)02810-7
14. Salminen A, Kaarniranta K, Kauppinen A. Photoaging: UV radiation-induced inflammation and immunosuppression accelerate the aging process in the skin. Inflammation Res. 2022;71(7–8):817–831. doi:10.1007/s00011-022-01598-8
15. Available from: https://www.ncbi.nlm.nih.gov/gene/4489.
16. Haq F, Mahoney M, Koropatnick J. Signaling events for metallothionein induction. Mutat Res. 2003;533(1–2):211–226. doi:10.1016/j.mrfmmm.2003.07.014
17. Swindell WR. Metallothionein and the biology of aging. Ageing Res Rev. 2011;10(1):132–145. doi:10.1016/j.arr.2010.09.007
18. Qu W, Waalkes MP. Metallothionein blocks oxidative DNA damage induced by acute inorganic arsenic exposure. Toxicol Appl Pharmacol. 2015;282(3):267–274. doi:10.1016/j.taap.2014.11.014
19. Available from: https://www.ncbi.nlm.nih.gov/gene/5743.
20. Kim JY, Shin JY, Kim MR, et al. siRNA-mediated knock-down of COX-2 in melanocytes suppresses melanogenesis. Exp Dermatol. 2012;21(6):420–425. doi:10.1111/j.1600-0625.2012.01483.x
21. Arai KY, Fujioka A, Okamura R, et al. Stimulatory effect of fibroblast-derived prostaglandin E2 on keratinocyte stratification in the skin equivalent. Wound Repair Regen. 2014;22(6):701–711. doi:10.1111/wrr.12228
22. Available from: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=84648.
23. Niehues H, van Vlijmen-Willems IMJJ, Bergboer JGM, et al. Late cornified envelope (LCE) proteins: distinct expression patterns of LCE2 and LCE3 members suggest nonredundant roles in human epidermis and other epithelia. Br J Dermatol. 2016;174(4):795–802. doi:10.1111/bjd.14284
24. Marshall D, Hardman MJ, Nield KM, et al. Differentially expressed late constituents of the epidermal cornified envelope. Proc Natl Acad Sci. 2001;98(23):13031–13036. doi:10.1073/pnas.231489198
25. Available from: https://www.ncbi.nlm.nih.gov/gene/5467.
26. Blunder S, Krimbacher T, Moosbrugger-Martinz V, et al. Keratinocyte-derived IL-1β induces PPARG downregulation and PPARD upregulation in human reconstructed epidermis following barrier impairment. Exp Dermatol. 2021;30(9):1298–1308. doi:10.1111/exd.14323
27. Michalik L, Wahli W. Peroxisome proliferator-activated receptors (PPARs) in skin health, repair and disease. Biochim Biophys Acta. 2007;1771(8):991–998. doi:10.1016/j.bbalip.2007.02.004
28. Mao-Qiang M, Fowler AJ, Schmuth M, et al. Peroxisome-proliferator-activated receptor (PPAR) gamma activation stimulates keratinocyte differentiation. J Invest Dermatol. 2004;123(2):305–312. doi:10.1111/j.0022-202X.2004.23235.x
29. Available from: https://www.ncbi.nlm.nih.gov/gene/1437.
30. Samavedam UKSRL, Iwata H, Schulze FS, et al. GM-CSF modulates autoantibody production and skin blistering in experimental epidermolysis bullosa acquisita. J Immunol. 2014;192(2):559–571. doi:10.4049/jimmunol.1301556
31. Shiomi A, Usui T, Mimori T. GM-CSF as a therapeutic target in autoimmune diseases. Inflamm Regen. 2016;536:8. doi:10.1186/s41232-016-0014-5
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.