Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 16
Untargeted Plasma Lipidomics Reveal Perturbed Metabolites of Glycerophospholipids, and Sphingolipids in Moderate-to-Severe Acne
Authors Zhang D, Yu S, Ou Yang X , Wang X , Zhu Y, Xiao Z, Tan Y, Wu L , Li C
Received 17 June 2023
Accepted for publication 5 August 2023
Published 11 August 2023 Volume 2023:16 Pages 2189—2200
DOI https://doi.org/10.2147/CCID.S426451
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Deng Zhang,1 Simin Yu,1 Xiaoliang Ou Yang,1 Xiuping Wang,1 Yunxia Zhu,1 Zhen Xiao,2 Yanping Tan,3 Liang Wu,1 Chunming Li1
1Department of Dermatology, The Second Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, People’s Republic of China; 2Department of Dermatology, Taiyuan Central Hospital, Taiyuan, Shanxi, People’s Republic of China; 3Department of Dermatology, Jiangxi Provincial Maternal and Child Health Hospital, Nanchang, Jiangxi, People’s Republic of China
Correspondence: Chunming Li, Department of Dermatology, The Second Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, People’s Republic of China, Email [email protected]
Background: Acne vulgaris (AV) is a common inflammatory disorder involving the pilosebaceous unit. The study aimed to explore the plasma lipidome signatures and identify specific lipid biomarkers in moderate-to-severe acne patients.
Patients and Methods: Untargeted plasma lipidomic analysis using ultra-high performance liquid chromatography system (UHPLC) coupled to q-extraction plus was employed on 30 moderate-to-severe acne patients aged between 16– 25 years and 30 healthy controls. Multivariate data analyses were used to identify the distinguishing lipid metabolites.
Results: All 1449 species of 37 lipid subclasses were identified from the MS data. There were apparent differences in plasma lipid profiles between acne groups and control groups. With variable influence on projection (VIP) > 1.0 and P-value < 0.05, 26 significantly different lipid metabolites were identified. These metabolites consisted mainly of glycerophospholipids (GPs), sphingolipids (SPs), and glycerolipids (GLs). Combining with AUC≥ 0.800 as the elected criteria, we obtained five differential lipids with good diagnostic performance for acne severity, including 2 sphingomyelins (SM), 1 phosphatidylglycerol (PG), 1 trihexosylceramide (Hex3Cer), and 1 Phosphatidylcholine (PC). Among them, PG (44:0) had the highest AUC values.
Conclusion: Our study revealed the plasma lipidome signature of patients with moderate-to-severe acne. The results will provide a novel light into the perturbed lipid metabolism leading to the development of acne.
Keywords: moderate-to-severe acne, lipidomics, sphingolipids, glycerophospholipids
Introduction
Acne vulgaris (AV) is a common inflammatory disorder involving the pilosebaceous unit and affects skin sites with a high density of sebaceous glands, such as face, back, and chest.1,2 It affects approximately 85% of world’s population aged 12 to 25 years.3 Typical acne lesions include open and closed comedones, inflammatory papules, pustules, and nodules.4 AV is a multifactorial disease, and its precise pathogenesis remains unknown. Four factors are believed to play a pivotal role in the formation of acne lesions, including excessive sebum, disturbed keratinization within the follicle, altered bacterial colonization with Cutibacterium acnes, and inflammation.5–7
Metabolomics is a newly developing omics after genomics, transcriptomics, and proteomics analysis.8,9 It uses advanced analytical chemistry techniques and complicated statistical methods to comprehensively measure large numbers of small molecule metabolites found in biofluids, tissues, and cells. As a subfield of metabolomics, lipidomics aims to comprehensively and systematically reflect the alterations in lipid profiles and related metabolic pathways within organisms under various physiological or pathological conditions.10 To date, lipidomics has been employed to study some dermatological diseases such as psoriasis,11 atopic dermatitis,12 urticaria,13 and hidradenitis suppurativa.14
Several lipidomics studies have revealed an association between lipid metabolite alterations and acne. Through skin surface lipidomics, Zhou et al15 revealed that the variation of triacylglycerols (TGs), phytosphingosine, and sphinganine was closely related to the severity of acne in adolescents. In addition, skin surface lipids sampled from young males with acne had significantly higher phosphatidylserine levels.16 In a rat acne model, 96 different lipid species were identified by a skin lipidomics study.17 Targeted lipidomics of serum sphingolipids in patients with acne has also been performed.18 However, no untargeted lipidomics studies have been conducted focusing on the plasma of moderate-to-severe acne.
In this study, an untargeted lipidomics analysis using an ultra-high performance liquid chromatography system (UHPLC) coupled with q-extraction plus was adopted to examine of the total plasma lipids in moderate-to-severe acne patients. Our aim is to reveal the discrepancies in lipid metabolites of moderate-to-severe acne patients and screen for potential biomarkers.
Materials and Methods
Patient Population
The patient group consisted of 30 patients aged between 16–25 years who visited our department from 10 January 2016 to 5 September 2021. The Global Acne Grading System (GAGS) criteria was used to estimate the disease severity of acne.19 The control group consisted of 30 gender- and age-matched healthy subjects. Exclusionary criteria for all participants were recent (within one month) treatment with topical and systemic medications, history of smoking, history of alcohol abuse, presence of systemic diseases, and psychiatric diseases. The study was conducted in accordance with the principles in the Declaration of Helsinki and was approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University. Each participant was fully informed of the purpose of the study and signed an informed consent form.
Main Reagents and Instruments
Reagents
Methanol (Thermo Fisher, USA), acetonitrile (Thermo Fisher, USA), propanol (Thermo Fisher, USA), formic acid (Fluka, German), ammonium formate (Sigma-Aldrich, USA), Internal Standards (SPLASH® LIPIDOMIX MASS SPRC STANDARD, AVANTI, 330707-1EA), and Methyl Tert-Butyl Ether (MTBE) (Sigma-Aldrich, USA).
Instruments
UHPLC Nexera LC-30A ultra performance liquid chromatography system (SHIMADZU, Japan), Q Exactive plus mass spectrometer (Thermo Scientific, USA), the chromatographic column: Waters, ACQUITY UPLC CSH C18, 1.7 µm, 2.1 mm × 100 mm column and Low temperature high-speed centrifuge (Eppendorf 5430R, German).
Sample Collection and Preparation
Plasma sample from each participant was collected in vacuum tubes (5 mL) containing EDTA anticoagulant, then centrifuged and stored at −80°C. Each sample (50 μL) was spiked with 20 μL internal standards and then homogenized with 240 μL methanol and 200 μL water. Following the addition of 800 μL of MTBE, the mixture was subjected to ultrasound followed by sitting still for 30 min. The solution was centrifuged (14,000 g, 15 min, 10°C) to extract the top layer, which was then dried under nitrogen. For Liquid Chromatograph-Mass Spectrometer (LC-MS) analysis, the lipid extracts were redissolved and vortexed in 200 μL of an isopropanol solution. Thereafter, 90 µL of the re-solution was collected and centrifuged (14,000 g, 15 min, 10°C). The supernatants were collected for further analysis. By pooling equal amounts of lipid extracts from all samples, quality control (QC) samples were prepared.
Liquid Chromatograph-Mass Spectrometer/Mass Spectrometer (LC-MS/MS) Analysis
Lipidomic analysis was conducted by Shanghai Applied Protein Technology Company. Reversed phase chromatography was selected for LC separation using the CSH C18 column. Solvent A was acetonitrile–water (6:4, vol/vol) with 0.1 mM ammonium formate and 0.1% formic acid, and solvent B was isopropanol–acetonitrile (9:1, vol/vol) with 0.1 mM ammonium formate and 0.1% formic acid. The initial mobile phase consisted of 30% solvent B, which was kept for 2 min, and then linearly increased to 100% solvent B for 23 min, followed by equilibrating in 5% solvent B for 10 min. The autosampler was kept at 10°C. Signal fluctuations were detected randomly to avoid bias due to instrumentation errors. To evaluate and monitor the reliability and stability of the experimental data, sampling of the queue was performed every eight samples by using one QC sample.
Following ultra-HPLC separation, samples were analyzed using a Q Exactive™ plus mass spectrometer. The chromatographic conditions were in accordance with a previous report.20
Data Processing and Statistical Analysis
To process the raw data, the LipidSearch software version 4.2 (Thermo Scientific™, USA) was used. Adducts of +H, +NH4 were chosen for positive mode searches, and -H, +HCOO were chosen for negative mode searches. For the data extracted from LipidSearch, the ion peak with a value of > 50% missing from the group was deleted. After normalization and integration using the Perato scaling method, the processed data were imported into SIMPCA-P 16.1 (Umetrics, Umea, Sweden) for orthogonal partial least squares discriminant analysis (OPLS-DA). Significantly different lipids were identified based on a combination of statistically significant thresholds of variable influence on projection (VIP) values obtained from the OPLS-DA model and two-tailed Student’s t-test.
Results
Clinical Characteristics of the Subjects
A total of 30 moderate-to-severe acne patients (Group P) and 30 healthy subjects (Group C) were enrolled in the study. Detailed clinical characteristics are described in Table 1. No statistically significant differences between acne and healthy controls in age, sex distribution, or body mass index were found.
 |
Table 1 Characteristics of Group P and Group C |
Different Lipidomic Profiles Between the Moderate-to-Severe Acne and Healthy Groups
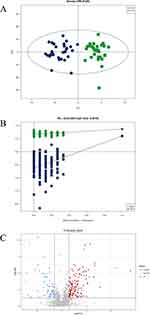
All 1449 species of 37 lipid subclasses were identified from the Mass spectrometry data (Figure 1). The main lipid metabolite subclasses in the plasma samples included phosphatidylcholine (PC), TG, and sphingomyelin (SM). OPLS-DA, a supervised multivariate model, was constructed to analyze the lipidomic data and search for features that may discriminate between Group P and Group C. An apparent distinction between them was observed in the OPLS-DA model, which indicated significant alterations in the plasma lipidomic profile of Group P (Figure 2A). The model was evaluated by monitoring the model R2 (goodness of fit) and Q2 (predictive ability) values and permutation tests (n = 200) were performed on R2 and Q2 as shown in Figure 2B (R2 = 0.6647, Q2 = −0.6375). The permutation test indicated the OPLS-DA model is reliable. The volcano plot displayed significant changes in the plasma metabolite profiles for Group P. Overall, 113 lipid metabolites were upregulated, and 78 were downregulated compared to those in Group C (Figure 2C).
 |
Figure 1 Classification of the identified lipids. |
Screening and Identification of Significantly Altered Lipid Metabolites for Acne
VIP reflected the significance of those variables in the OPLS-DA model and was used to select the significant variables. With VIP > 1.0 and P-value < 0.05, 26 significantly different lipid metabolites were identified (15 up-regulated and 11 down-regulated lipids) (Table 2). These lipid species mainly belonged to Glycerophospholipids (GPs) (n=9), Sphingolipids (SPs) (n=12), and Glycerolipids (GLs) (n=2). Their relationships were revealed by correlation analysis (Figure 3). Furthermore, to visualize the distribution of relative levels of 26 differential lipids in each group, the identified lipid data were analyzed using a clustering heatmap (Figure 4).
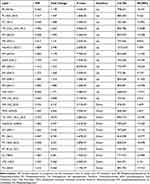 |
Table 2 Identification of Significantly Altered Plasma Lipids Between Group P and Group C |
 |
Figure 3 Correlation clustering-heatmap of selected lipid between Group P and Group C. |
 |
Figure 4 Clustering of differential lipids in Group P and Group C by heatmaps. |
ROC Curve Analysis
To further evaluate the diagnostic performance of the identified lipid species, OPLS-DA-based ROC analysis was performed. The area under the ROC curves (AUC) was calculated to evaluate the reliability and discriminatory power of the potential biomarkers. With AUC≥0.800 as the elected criteria, five differential lipids were identified with good diagnostic performance for acne, including 2 SMs, 1 PG, 1 Hex3Cer, and 1 PC. Among them, PG (44:0) had the highest AUC values (0.941 (95% CI: 0.886–0.996)), indicating outstanding diagnostic ability for acne, whereas Hex3Cer (d32:1) had relatively lower AUC values (AUC = 0.807, 95% CI: 0.697–0.916) (Figure 5).
 |
Figure 5 ROC curves and boxplots for the top 5 differential lipids of highest diagnostic value. (A): SM (d42:1); (B): SM (d36:2); (C): Hex3Cer (d32:1); (D): PG (44:0); (E): PC (16:0_20:3). |
Discussion
Lipids provide the structural basis of the cell membrane bilayer. Moreover, they were involved in various biological processes, such as cell proliferation, apoptosis, angiogenesis, immunity, and inflammation.21–23 Based on “Comprehensive Classification System for Lipids”, eight categories of lipids can be obtained: fatty acids (FA), sphingolipids (SP), glycerolipids (GL), glycerophospholipids (GP), saccharolipids (SL), sterol lipids (ST), polyketides (PK), and prenol lipids (PR).24,25 Currently, lipid dysregulation has been identified in many diseases, including diabetes, respiratory diseases, and Alzheimer’s disease,26–28 suggesting that dysregulated lipids may serve as an important biomarker and be associated with the development of these diseases. In this study, we demonstrated that plasma lipidomic profiles are substantially changed in moderate-to-severe acne patients compared to healthy controls. Besides, 26 differential lipid metabolites were identified between the two groups. They consisted mainly of GPs, SPs, and GLs.
GPs, also known as phospholipids, are critical components of the lipid bilayer of cellular membranes and play a central role in cell signaling and metabolism.29 Previous studies have shown that in skin lipids, GPs were increased in young male acne patients and decreased in infantile acne patients.16,30 Besides, Chen et al found elevated GPs in the skin sample of a rat acne model.17 However, there are no studies on blood GPs of acne. In our study, plasma lipid profiling identified a series of 9 GPs with significant alterations. Three GPs were elevated (PC) (34:1), phosphatidylethanolamine (PE) (40:7e), phosphatidylinositol (PI) (18:0_20:4), and six GPs were decreased (lysophosphatidylcholine (LPC) (18:2), PC (36:1), PC (8:1e_10:1), PG (44:0), PC (16:0_20:3), cardiolipin (CL) (78:2)). The lipid metabolites obtained were mainly from the PC, PG, and PE subgroups. PC constitutes the major component of biological membranes in eukaryotic cells, and its degradation and biosynthesis are essential to regulate cell cycle processes.31 A previous study demonstrated that PC species can regulate the differentiation of keratinocytes. Therefore, the alteration in PC levels suggests a defect in the process, resulting in disturbed keratinization. Notably, the levels of PG (44:0) were decreased. Previous studies have shown that PG can inhibit keratinocyte inflammatory mediator expression.32,33 We postulate that decreased PG (44:0) level may be related to acne inflammation.
SPs occur in the plasma membranes of all eukaryotic cells and play an essential role in maintaining the skin barrier.34 Through untargeted metabolomics, our previous study revealed sphingolipid metabolic pathway was significantly perturbed in a rabbit acne model.35 Similarly, Chen et al conducted a skin lipidomics study of a rat acne model, and metabolic pathway analysis showed that the sphingolipid metabolism pathway had the highest impact factors.17 However, studies on blood sphingolipids in acne patients remain limited. In this study, 12 SPs were markedly altered in moderate-to-severe acne patients. Of note, seven SMs increased, including SM (d34:1), SM (d36:1), SM (d36:2), SM (d38:1), SM (d42:1), SM (d42:2), and SM (d42:3). This result was in agreement with our previous untargeted metabolomics study showed that sphingolipid metabolic pathway was significantly perturbed in moderate-to-severe acne patients.36 Besides, Kaya et al revealed that circulating levels of C16 SM were increased in patients with acne compared to healthy controls.18 SMs are the most abundant circulating lipids. In vitro, SM can stimulate the proliferation of keratinocytes.37 Therefore, we hypothesize that the elevated plasma SMs may be involved in the pathogenesis of acne by regulating abnormal keratinization processes.
GLs are the principal constituent of cell membranes and extracellular protective layers such as the stratum corneum. They are classified as monoacylglycerol (MG), diacylglycerol (DG), or triacylglycerol.38 TGs are the most abundant lipids in the body synthesized from glycerol and fatty acids (FAs), which can be hydrolyzed into DG and FAs by lipase reaction. Most previous studies have mainly measured total TG levels in patients with acne.39–41 However, no studies have examined the association between plasma TGs species levels and acne. In the present study, we found elevated levels of TG (16:0_16:0_18:1) and decreased levels of TG (18:1_18:2_18:3) in moderate-to-severe acne patients.
According to the ROC analysis results, five lipid species had an AUC value of more than 0.800. They were defined as the potential lipid biomarkers of moderate-to-severe acne and can be regarded as the complement of the existing protein or gene biomarkers. Among them, SM (d42:1), SM (d36:2), and Hex3Cer (d32:1) were significantly increasing in moderate-to-severe acne patients, while PG (44:0) and PC (16:0_20:3) were significantly decreasing compared with healthy controls. However, further investigation in larger sizes, even involving cell biology, is required to confirm these potential lipid biomarkers.
Conclusion
Based on plasma lipidomics, we demonstrated the difference in plasma lipid profiles between moderate-to-severe acne patients and healthy controls. 26 significantly different lipid metabolites, mainly belonging to GP, SP, and GL, were identified. However, this study had some potential limitations. The sample size was relatively small. In addition, traditional blood lipid profiles, including total cholesterol (TC), low-density lipoprotein cholesterol (LDL), high-density lipoprotein cholesterol (HDL), and TG levels, were not measured. Overall, our research provides a novel light into the perturbed lipid metabolism leading to the development of acne.
Abbreviations
AV, Acne vulgaris; GAGS, Global Acne Grading System; TLR, Toll-like receptors; UHPLC, Ultra-high performance liquid chromatography system; PCA, Principal component analysis; OPLS-DA, Orthogonal partial least squares discriminant analysis; AUC, Area under the curve; ROC, Receiver operating characteristic; VIP, Variable influence on projection; SP, Sphingolipid; GP, Glycerophospholipids; GL, Glycerolipid; PC, Phosphatidylcholine; PE, Phosphatidylethanolamine; PG, Phosphatidylglycerol; PI, Phosphatidylinositol; LPC, Lysophosphatidylcholine; CL, Cardiolipin; SM, Sphingomyelin; Hex3Cer, Trihexosylceramide; DG, Diacylglycerol; TG, Triacylglycerol; FA, Fatty acids.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The studies involving human participants were reviewed and approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University. Informed written consent was obtained from all study patients and/or from their parents if the study participant was under 18 years of age.
Consent for Publication
All patients signed the informed consent.
Acknowledgments
We thank all the patients for participating in this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The project was supported by the National Natural Science Foundation of China (Project No. 81960569) and the Natural Science Foundation of Jiangxi Province (Project No. 20202BAB206034).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gollnick HP. From new findings in acne pathogenesis to new approaches in treatment. J Eur Acad Dermatol Venereol. 2015;29(Suppl 5):1–7. doi:10.1111/jdv.13186
2. Williams HC, Dellavalle RP, Garner S. Acne vulgaris. Lancet. 2012;379(9813):361–372. doi:10.1016/S0140-6736(11)60321-8
3. Eichenfield DZ, Sprague J, Eichenfield LF. Management of acne vulgaris: a review. JAMA. 2021;326(20):2055–2067. doi:10.1001/jama.2021.17633
4. Mosaico G, Artuso G, Pinna M, Denotti G, Orru G, Casu C. Host microbiota balance in teenagers with gum hypertrophy concomitant with acne vulgaris: role of oral hygiene associated with topical probiotics. Microorganisms. 2022;10(7):1344. doi:10.3390/microorganisms10071344
5. Hazarika N. Acne vulgaris: new evidence in pathogenesis and future modalities of treatment. J Dermatolog Treat. 2021;32(3):277–285. doi:10.1080/09546634.2019.1654075
6. Knutsen-Larson S, Dawson AL, Dunnick CA, Dellavalle RP. Acne vulgaris: pathogenesis, treatment, and needs assessment. Dermatol Clin. 2012;30(1):
7. Degitz K, Ochsendorf F. Acne. J Dtsch Dermatol Ges. 2017;15(7):709–722. doi:10.1111/ddg.13278
8. Wishart DS. Metabolomics for investigating physiological and pathophysiological processes. Physiol Rev. 2019;99(4):1819–1875. doi:10.1152/physrev.00035.2018
9. Huang F, Yu J, Lai T, Luo L, Zhang W. The combination of bioinformatics analysis and untargeted metabolomics reveals potential biomarkers and key metabolic pathways in asthma. Metabolites. 2022;13(1):25. doi:10.3390/metabo13010025
10. Wenk MR. Lipidomics: new tools and applications. Cell. 2010;143(6):888–895. doi:10.1016/j.cell.2010.11.033
11. Zeng C, Wen B, Hou G, et al. Lipidomics profiling reveals the role of glycerophospholipid metabolism in psoriasis. GigaScience. 2017;6(10):1–11. doi:10.1093/gigascience/gix087
12. Janssens M, van Smeden J, Gooris GS, et al. Increase in short-chain ceramides correlates with an altered lipid organization and decreased barrier function in atopic eczema patients. J Lipid Res. 2012;53(12):2755–2766. doi:10.1194/jlr.P030338
13. Li J, Li L, Liu R, et al. Integrative lipidomic features identify plasma lipid signatures in chronic urticaria. Front Immunol. 2022;13:933312. doi:10.3389/fimmu.2022.933312
14. Choi E, Mir SA, Ji S, et al. Understanding the systemic burden of disease in hidradenitis suppurativa from plasma lipidomic analysis. J Dermatol Sci. 2022;107(3):133–141. doi:10.1016/j.jdermsci.2022.08.005
15. Zhou M, Yang M, Zheng Y, et al. Skin surface lipidomics revealed the correlation between lipidomic profile and grade in adolescent acne. J Cosmet Dermatol. 2020;19(12):3349–3356. doi:10.1111/jocd.13374
16. Zhou M, Gan Y, He C, Chen Z, Jia Y. Lipidomics reveals skin surface lipid abnormity in acne in young men. Br J Dermatol. 2018;179(3):732–740. doi:10.1111/bjd.16655
17. Chen T, Zhu Z, Du Q, et al. A skin lipidomics study reveals the therapeutic effects of tanshinones in a rat model of acne. Front Pharmacol. 2021;12:675659. doi:10.3389/fphar.2021.675659
18. Kaya S, Aslan İ, Kıraç E, Karaarslan T, Aslan M. Serum Sphingolipidomic Analysis in Acne Vulgaris Patients. Ann Clin Lab Sci. 2019;49(2):242–248.
19. Doshi A, Zaheer A, Stiller MJ. A comparison of current acne grading systems and proposal of a novel system. Int J Dermatol. 1997;36(6):416–418. doi:10.1046/j.1365-4362.1997.00099.x
20. Xue SS, Zhou CH, Xue F, et al. The impact of repetitive transcranial magnetic stimulation and fluoxetine on the brain lipidome in a rat model of chronic unpredictable stress. Prog Neuropsychopharmacol Biol Psychiatry. 2020;102:109946. doi:10.1016/j.pnpbp.2020.109946
21. Huse M, Le Floc’h A, Liu X. From lipid second messengers to molecular motors: microtubule-organizing center reorientation in T cells. Immunol Rev. 2013;256(1):95–106. doi:10.1111/imr.12116
22. Huse M. Lipid-based patterning of the immunological synapse. Biochem Soc Trans. 2014;42(6):1506–1511. doi:10.1042/BST20140191
23. Höglinger D, Nadler A, Schultz C. Caged lipids as tools for investigating cellular signaling. Biochim Biophys Acta. 2014;1841(8):1085–1096. doi:10.1016/j.bbalip.2014.03.012
24. Fahy E, Subramaniam S, Murphy RC, et al. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50(Suppl):S9–14. doi:10.1194/jlr.R800095-JLR200
25. Fahy E, Subramaniam S, Brown HA, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46(5):839–861. doi:10.1194/jlr.E400004-JLR200
26. González-Domínguez R, García-Barrera T, Gómez-Ariza JL. Metabolomic study of lipids in serum for biomarker discovery in Alzheimer’s disease using direct infusion mass spectrometry. J Pharm Biomed Anal. 2014;98:321–326. doi:10.1016/j.jpba.2014.05.023
27. Angelakopoulou A, Shah T, Sofat R, et al. Comparative analysis of genome-wide association studies signals for lipids, diabetes, and coronary heart disease: Cardiovascular Biomarker Genetics Collaboration. Eur Heart J. 2012;33(3):393–407.
28. Lykkesfeldt J. Malondialdehyde as biomarker of oxidative damage to lipids caused by smoking. Clin Chim Acta. 2007;380(1–2):50–58. doi:10.1016/j.cca.2007.01.028
29. Ivanova PT, Milne SB, Byrne MO, Xiang Y, Brown HA. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:21–57. doi:10.1016/S0076-6879(07)32002-8
30. Zhou M, Wang H, Yang M, et al. Lipidomic analysis of facial skin surface lipids reveals an altered lipid profile in infant acne. Br J Dermatol. 2020;182(3):817–818. doi:10.1111/bjd.18474
31. Huang J, Wang Q, Qi Z, Zhou S, Zhou M, Wang Z. Lipidomic profiling for serum biomarkers in mice exposed to ionizing radiation. Dose Response. 2020;18(2):1559325820914209. doi:10.1177/1559325820914209
32. Xie D, Choudhary V, Seremwe M, et al. Soy phosphatidylglycerol reduces inflammation in a contact irritant ear edema mouse model in vivo. J Pharmacol Exp Ther. 2018;366(1):1–8. doi:10.1124/jpet.117.244756
33. Choudhary V, Uaratanawong R, Patel RR, et al. Phosphatidylglycerol inhibits toll-like receptor-mediated inflammation by danger-associated molecular patterns. J Invest Dermatol. 2019;139(4):868–877. doi:10.1016/j.jid.2018.10.021
34. Gomez-Larrauri A, Presa N, Dominguez-Herrera A, Ouro A, Trueba M, Gomez-Munoz A. Role of bioactive sphingolipids in physiology and pathology. Essays Biochem. 2020;64(3):579–589. doi:10.1042/EBC20190091
35. Ou-Yang X, Zhang D, Wang X, et al. Nontargeted metabolomics to characterize the effects of isotretinoin on skin metabolism in rabbit with acne. Front Pharmacol. 2022;13:963472. doi:10.3389/fphar.2022.963472
36. Yu S, Xiao Z, Ou Yang X, Wang X, Zhang D, Li C. Untargeted metabolomics analysis of the plasma metabolic signature of moderate-to-severe acne. Clin Chim Acta. 2022;533:79–84. doi:10.1016/j.cca.2022.06.012
37. Pillai S, Mahajan M, Carlomusto M. Ceramide potentiates, but sphingomyelin inhibits, vitamin D-induced keratinocyte differentiation: comparison between keratinocytes and HL-60 cells. Arch Dermatol Res. 1999;291(5):284–289. doi:10.1007/s004030050409
38. Park J, Choi J, Kim DD, et al. Bioactive lipids and their derivatives in biomedical applications. Biomol Ther. 2021;29(5):465–482. doi:10.4062/biomolther.2021.107
39. Sobhan M, Seif Rabiei MA, Amerifar M. Correlation between lipid profile and acne vulgaris. Clin Cosmet Investig Dermatol. 2020;13:67–71. doi:10.2147/CCID.S230617
40. Jiang H, Li CY, Zhou L, et al. Acne patients frequently associated with abnormal plasma lipid profile. J Dermatol. 2015;42(3):296–299. doi:10.1111/1346-8138.12761
41. Abulnaja KO. Changes in the hormone and lipid profile of obese adolescent Saudi females with acne vulgaris. Braz J Med Biol Res. 2009;42(6):501–505. doi:10.1590/S0100-879X2009000600005
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.