Back to Journals » Cancer Management and Research » Volume 9
Unintended target effect of anti-BCL-2 DNAi
Authors Ebrahim AS, Kandouz M, Emara N, Sugalski AB, Lipovich L, Al-Katib AM
Received 7 April 2017
Accepted for publication 15 July 2017
Published 22 September 2017 Volume 2017:9 Pages 427—432
DOI https://doi.org/10.2147/CMAR.S139105
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Abdul Shukkur Ebrahim,1 Mustapha Kandouz,2 Nada Emara,1 Amara B Sugalski,3 Leonard Lipovich,3 Ayad M Al-Katib1
1Lymphoma Research Laboratory, 2Department of Pathology, School of Medicine, 3Center for Molecular Medicine and Genetics, Wayne State University, Detroit, MI, USA
Introduction: Previous research suggested that a novel compound PNT2258 inhibits B-cell lymphoma 2 (BCL-2) transcription by DNA interference (DNAi) and demonstrated its activity in preclinical xenograft models and in a pilot Phase II clinical trial in non-Hodgkin’s lymphoma (NHL). While the drug downregulates BCL-2 at the promoter, mRNA, and protein levels, there is a significant homology (13–16 bases) between PNT100 and a number of promoters of genes involved in cell cycle regulation and survival. In this study, we identify cyclin-dependent kinase-4 (CDK4) as an unintended target gene of PNT2258 and examine its relevance to NHL.
Methods: We performed a Basic Local Alignment Search Tool (BLAST) homology search using PNT100 DNAi sequences. Also, we conducted CDK4 promoter assay in K562 cells and studied the protein expression of CDK4 in Wayne State University (WSU)-follicular small cleaved cell lymphoma (FSCCL), WSU-diffuse large cell lymphoma, and WSU-Waldenström’s macroglobulinemia (WM) lymphoma cells.
Results: BLAST homology search showed that PNT100 completely binds to BCL-2 gene as expected. However, there was 100% homology in a stretch of 14 bases (8–21) between PNT100 and CDK4. PNT2258 strongly inhibited CDK4 promoter activity in K562 cells. Moreover, CDK4 protein expression was significantly downregulated by PNT2258 in WSU-FSCCL and WSU-WM cell lines.
Discussion: DNAi may work not only through knocking down the intended gene but also by knocking down other genes. PNT2258 affects CDK4 expression and promoter activity. Results of the present study suggest a broader mechanism of action for DNAi targeting both intended (BCL-2) and unintended (CDK4) genes.
Keywords: non-Hodgkin’s lymphoma, BCL-2, PNT2258, BLAST, CDK4
Introduction
Non-Hodgkin’s lymphoma is the most common blood cancer in USA with >72,000 people estimated to be diagnosed in 2016; >20,000 will die of this disease.1 Our group has had a long-standing interest and track record in developing and evaluating different anti-B-cell lymphoma 2 (anti-BCL-2) strategies including small molecule inhibitors2–5 and antisense oligonucleotides.6,7 More recently, we have reported DNA interference (DNAi) as a new strategy for cancer therapy.8,9 DNAi represents nucleic acid based drugs that block transcription of specific genes involved in complex diseases such as cancer. PNT2258 is a BCL-2-targeting DNAi molecule which contains a single stranded sequence of 24 bases of unmodified phosphodiester DNA (known as PNT100) with a specific sequence, 5′-CACGCACGCGCATCCCCGCCCGTG-3′. PNT100 was designed to hybridize with genomic sequences that reside within the 5′-untranscribed regulatory region of the BCL-2 gene to block its transcription via DNAi. The drug was evaluated in preclinical models of lymphoma and prostate cancer8 and has undergone a Phase I clinical trial.10
We recently demonstrated that PNT2258 significantly and dose-dependently inhibited BCL-2 at the promoter, RNA, and protein expression levels thus causing apoptosis.9 Expression of other BCL-2 family members, BCL-xL, BAX, BAK, and BID, was unaffected. Interestingly, PNT2258 also induced S-phase arrest although BCL-2 is not involved in cell cycle regulation. This observation suggested that PNT2258 might silence other unintended genes (off-target effects). In fact, a Basic Local Alignment Search Tool (BLAST) search revealed that promoter sequences for a number of genes were partially homologous (13–16 out of 24 bases) with PNT100 sequences. In this report, we focus on cyclin-dependent kinase-4 (CDK4) gene as a primary unintended target given its roles in cell cycle regulation as a key regulator of the G1–S transition. We used three lymphoma cell lines with distinct genetic characteristics to investigate the unintended gene effect of PNT2258 and its role as DNAi. Our results show significant downregulation of CDK4 promoter activity and protein expression following PNT2258 exposure in Wayne State University (WSU)-follicular small cleaved cell lymphoma (FSCCL) and WSU-Waldenström’s macroglobulinemia (WM) cells.
Methods and materials
Cell culture
The cell lines used in this study were WSU-FSCCL, WSU-diffuse large cell lymphoma (DLCL2), and WSU-WM, which were established in our laboratory at Wayne State University (WSU).11–13 WSU-FSCCL (t(14;18) translocation for BCL-2 and t(8;11) c-MYC rearrangements), WSU-DLCL2 (t(14;18) BCL-2 rearrangement), and WSU-Waldenstrom’s macroglobulinemia (WM; t(8;14) c-MYC rearrangement). All cell lines are Epstein–Barr virus negative and grow in liquid culture as previously described.14 The molecular characterization, translocations, and breakpoints of each cell line have been published.11 Human K562 cell line was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cells were maintained according to a standard protocol.
Algorithm
“BLAST 2 SEQUENCES” is an interactive tool that utilizes the BLAST engine for pairwise DNA–DNA or protein–protein sequence comparison and is based on the same algorithm and statistics of local alignments that have been described earlier.15 The BLAST 2.0 algorithm generates a gapped alignment by using dynamic programming to extend the central pair of aligned residues. The heuristic methods confine the alignments to a predefined region of the path graph. A performance evaluation of the new gapped BLAST algorithm and its comparison to that of the original ungapped BLAST has been presented.16
Luciferase assay
The 404-bp fragment, containing the CDK4 promoter subcloned into pGL3-basic luciferase expression vector (Promega, Madison, WI, USA), was a kind gift of Dr Gary L Firestone (Department of Molecular and Cell Biology, University of California at Berkeley, Berkeley, CA, USA). All transfections were carried out in 24-well plates. Briefly, plasmids (1 μg CDK4 promoter construct, 0.1 μg Renilla luciferase expressing reporter vector pRL-Null [Promega]), were introduced into <50% confluent K562 cells with Lipofectamine® LTX transfection reagent (Life Technologies, Grand Island, NY, USA). The CDK4 and control empty vectors were transfected into K562 cells at a density of 0.2×106 viable cells/mL per well in a 24-well plate. Twenty-four hours after transfection, cells were treated with 2.5 μM PNT2258 for 48 hours. Cells were lysed 48 hours later, and promoter activity analyzed in a MicroLumat Plus LB96V reader (Berthold Technologies, Bad Wildbad, Germany) using the Dual Luciferase® Reporter Assay System (Promega). The firefly luciferase values were normalized to those of Renilla luciferase; all transfections were repeated at least three times.
Antibodies and reagents
CDK4 antibody (DCS-35: sc-23896, 1:500) was purchased from Santa Cruz Biotechnology (Dallas, TX, USA) and the actin antibody (ACTN05, 1:2,000) from Thermo Fisher Scientific (Waltham, MA, USA). Protein concentrations were determined using the micro bicinchoninic acid protein assay (Pierce Chemical Company, Rockford, IL, USA). PNT2258 and associated reagents were provided by ProNAi Therapeutics (Plymouth, MI, USA). The drug PNT2258 consists of a 24 base unmodified DNA phosphodiester oligonucleotide encapsulated in amphoteric proprietary liposomes as previously described.8
Immunoblotting
PNT2258-treated and untreated cells were harvested, washed in phosphate-buffered saline, and lysed in M-PER lysis buffer containing a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific).9 Equal amounts of protein lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by blotting with the indicated antibodies and detection by Western SuperSignal West Pico Chemiluminescent Substrate reagents (Thermo Fisher Scientific). Select images were quantified using the ImageJ densitometry software (version 1.45; US National Institutes of Health, Bethesda, MD, USA) and normalized to the actin signal. Data are presented as relative band signal intensity compared to control.
Data analysis and statistical significance
Statistics were conducted using GraphPad Prism 5.0 for Windows (GraphPad Software, Inc, La Jolla, CA, USA) and tests were done with one-way analysis of variance (*P<0.05, **P<0.01, ***P<0.001). P-values <0.05 were considered to be statistically significant. Results are displayed as averages with error bars indicating standard deviations. ImageJ (National Institute of Mental Health, Bethesda, MD, USA) densitometry software was used for quantification of Western blot bands. Selected bands were quantified based on their relative integrated intensities, calculated as the product of the selected pixel area and the mean gray value for those pixels normalized to the internal control.
Results
BLAST search identifies multiple targets for the PNT2258 DNAi oligonucleotides
The BLAST homology search showed that PNT100 does fully bind to the BCL-2 gene as predicted. However, portions of PNT100 sequence averaging 13–14 bases showed 100% identity with four different genes: CDK4, insulin-like growth factor-1 receptor (IGF-1R), p21-activated kinase 3, and transferrin receptor (TFRC/CD71). As expected, CDK4, IGF-1R, and TFRC are involved in cell cycle regulation. CDK4, IGF-1R, and TFRC may also modulate cell cycle/cell death via effects on cell metabolism. Specifically, there was 100% homology in 14 bases (8–21) between PNT100 and CDK4 (Figure 1) which is the subject of this report.
PNT2258 represses the CDK4 promoter
We have previously reported that the 24 bases PNT100 sequence included within PNT2258 is complementary to a segment of the consensus sequence recognized by the Sp1 transcription factor in the BCL-2 P1 promoter.9 To examine whether a putative region in the CDK4 promoter confers PNT2258 responsiveness, we transiently transfected K562 cells with a plasmid containing a 404-bp long CDK4 promoter construct before exposure to PNT2258 for 48 hours. As shown in Figure 2, PNT2258 strongly downregulated CDK4 promoter activity.
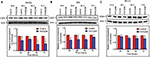
PNT2258 downregulates CDK4 protein in lymphoma cells
We examined whether PNT2258 inhibits CDK4 expression in lymphoma cells. Western blot analysis revealed that CDK4 protein levels were significantly decreased 24 through 72 hours following exposure to 2.5 μM PNT2258 in WSU-FSCCL and WSU-WM cells (Figure 3A and B). However, the CDK4 protein levels in WSU-DLCL2 cells were lower at 48–72 hours, but were not statistically significant (Figure 3C).
Discussion
WSU-FSCCL, WSU-DLCL2, and WSU-WM cells were used in a previous study as tools to investigate the effect of PNT2258 in vitro at different concentrations (2.5, 5.0, and 10 μM) on cell growth and survival. PNT2258 induced apoptosis which can be explained by its effect on BCL-2 expression.9 A significant decrease in cell viability was observed after 48 hours in all of the cell lines exposed to PNT2258 at the lowest concentration (2.5 μM).9,17 Nevertheless, PNT2258 has additional effects on WSU-FSCCL cells, such as cell cycle arrest, that cannot be explained by BCL-2 targeting.18 Results of the present study suggest a broader mechanism of action for DNAi targeting both intended (BCL-2) and unintended (CDK4) genes. Such an effect can enhance the DNAi anticancer effectiveness.
Our findings in this study have two implications: 1) PNT2258 inhibits the expression of CDK4 (which is one of the mechanisms of action of PNT2258 not previously recognized or predicted), and 2) decreased expression of CDK4 explains the previously observed effect of PNT2258 on cell cycle, a finding that cannot be explained by the effect of PNT2258 on the target gene (BCL-2).
Our previous findings suggest that BCL-2 family member MCL-1 protein levels were lower in PNT2258-treated cells.9 However, we believe that this decrease might be due to knocking down the unintended target gene CDK4, which may lead to a cell cycle arrest induced by PNT2258. MCL-1 downregulation was recently shown to be associated with G1/S transition.19 Moreover, it is not clear if complete 24 bases of DNAi against CDK4 will have a more significant knockdown of CDK4 expression compared with partial homology of PNT100 sequence. CDK4 has been a target for cancer therapy with recent US Food and Drug Administration approval of CDK4/6 inhibitors palbociclib (PD-0332991) and ribociclib (LEE011) for breast cancer.20,21
Inhibition of CDK4, IGF-1R, and TFRC leads to decreased proliferation and G1 cycle arrest.22–24 IGF-1R can regulate cell cycle progression at several steps by facilitating G0–G1 transition through p70 S6K and promoting G1–S transition by increasing cyclin D1 and CDK4 gene expression. Since activation of IGF-1R and TFRC enhances reactive oxygen species, their inhibition improves mitochondrial function and lowers intrinsic apoptosis, reducing cytochrome c release and increasing caspase-3 and poly(ADP-ribose) polymerase cleavage.22–24 These proapoptotic effects are consistent with similar effects of PNT2258 and, although awaiting experimental evidence, suggest that the latter’s unintended effects on CDK4, IGF-1R, and TFRC are unlikely to result in harmful “off-target” effects.
Additional investigations are required to explore CDK4 targeting by DNAi as an alternative anti-CDK4 strategy. There remains the possibility of additional unintended target genes for PNT2258 that may also play a significant role as part of its broad mechanism of action.
Conclusion
In our study, we demonstrated that DNAi may work not only through knocking down their intended gene, but also by knocking down other genes. These possibilities should be kept in mind as new DNAi agents are developed.
Abbreviations
WSU: Wayne State University
NHL: non-Hodgkin’s lymphoma
FSCCL: follicular small cleaved cell lymphoma
WM: Waldenström’s macroglobulinemia
DLCL2: diffuse large cell lymphoma
DNAi: DNA interference
Acknowledgments
The authors wish to thank ProNAi Therapeutics for providing PNT2258 and related agents used in this study. This study was funded by a grant from ProNAi Therapeutics, by St John Hospital and Medical Center Foundation, and by Michigan Corporate Relations Network’s Small Company Innovation Program. The data sets supporting the conclusions of this article are included within the article.
Author contributions
ASE designed and carried out all experiments, correlated and analyzed the resulting data, and organized and prepared the manuscript for publication. MK designed the promoter assay. NE provided technical assistance to ASE in carrying out experiments. LL and SBA provided guidance and analysis of BLAST search. AMA conceived and coordinated the overall project, participated in the design of experiments, and manuscript writing. All authors read and approved the final manuscript. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66(1):7–30. | ||
Mohammad RM, Wang S, Aboukameel A, et al. Preclinical studies of a nonpeptidic small-molecule inhibitor of Bcl-2 and Bcl-X(L) [(-)-gossypol] against diffuse large cell lymphoma. Mol Cancer Ther. 2005;4(1):13–21. | ||
Mohammad RM, Goustin AS, Aboukameel A, et al. Preclinical studies of TW-37, a new nonpeptidic small-molecule inhibitor of Bcl-2, in diffuse large cell lymphoma xenograft model reveal drug action on both Bcl-2 and Mcl-1. Clin Cancer Res. 2007;13(7):2226–2235. | ||
Arnold AA, Aboukameel A, Chen J, et al. Preclinical studies of Apogossypolone: a new nonpeptidic pan small-molecule inhibitor of Bcl-2, Bcl-XL and Mcl-1 proteins in follicular small cleaved cell lymphoma model. Mol Cancer. 2008;7:20. | ||
Al-Katib AM, Sun Y, Goustin AS, et al. SMI of Bcl-2 TW-37 is active across a spectrum of B-cell tumors irrespective of their proliferative and differentiation status. J Hematol Oncol. 2009;2:8. | ||
Smith MR, Abubakr Y, Mohammad R, et al. Antisense oligodeoxyribonucleotide down-regulation of bcl-2 gene expression inhibits growth of the low-grade non-Hodgkin’s lymphoma cell line WSU-FSCCL. Cancer Gene Ther. 1995;2(3):207–212. | ||
Mohammad R, Abubakr Y, Dan M, et al. Bcl-2 antisense oligonucleotides are effective against systemic but not central nervous system disease in severe combined immunodeficient mice bearing human t(14;18) follicular lymphoma. Clin Cancer Res. 2002;8(4):1277–1283. | ||
Rodrigueza WV, Woolliscroft MJ, Ebrahim AS, et al. Development and antitumor activity of a BCL-2 targeted single-stranded DNA oligonucleotide. Cancer Chemother Pharmacol. 2014;74(1):151–166. | ||
Ebrahim AS, Kandouz M, Liddane A, et al. PNT2258, a novel deoxyribonucleic acid inhibitor, induces cell cycle arrest and apoptosis via a distinct mechanism of action: a new class of drug for non-Hodgkin’s lymphoma. Oncotarget. 2016;7(27):42374–42384. | ||
Tolcher AW, Rodrigueza WV, Rasco DW, et al. A phase 1 study of the BCL2-targeted deoxyribonucleic acid inhibitor (DNAi) PNT2258 in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2014;73(2):363–371. | ||
Mohammad RM, Mohamed AN, Smith MR, Jawadi NS, al-Katib A. A unique EBV-negative low-grade lymphoma line (WSU-FSCCL) exhibiting both t(14;18) and t(8;11). Cancer Genet Cytogenet. 1993;70(1):62–67. | ||
Al-Katib AM, Smith MR, Kamanda WS, et al. Bryostatin 1 down-regulates mdr1 and potentiates vincristine cytotoxicity in diffuse large cell lymphoma xenografts. Clin Cancer Res. 1998;4(5):1305–1314. | ||
al-Katib A, Mohammad R, Hamdan M, Mohamed AN, Dan M, Smith MR. Propagation of Waldenstrom’s macroglobulinemia cells in vitro and in severe combined immune deficient mice: utility as a preclinical drug screening model. Blood. 1993;81(11):3034–3042. | ||
Al-Katib AM, Aboukameel A, Mohammad R, Bissery MC, Zuany-Amorim C. Superior antitumor activity of SAR3419 to rituximab in xenograft models for non-Hodgkin’s lymphoma. Clin Cancer Res. 2009;15(12):4038–4045. | ||
Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. | ||
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. | ||
Ebrahim AS, Sabbagh H, Liddane A, Raufi A, Kandouz M, Al-Katib A. Hematologic malignancies: newer strategies to counter the BCL-2 protein. J Cancer Res Clin Oncol. 2016;142(9):2013–2022. | ||
Moldoveanu T, Follis AV, Kriwacki RW, Green DR. Many players in BCL-2 family affairs. Trends Biochem Sci. 2014;39(3):101–111. | ||
Morciano G, Giorgi C, Balestra D, et al. Mcl-1 involvement in mitochondrial dynamics is associated with apoptotic cell death. Mol Biol Cell. 2016;27(1):20–34. | ||
O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13(7):417–430. | ||
Finn RS, Aleshin A, Slamon DJ. Targeting the cyclin-dependent kinases (CDK) 4/6 in estrogen receptor-positive breast cancers. Breast Cancer Res. 2016;18(1):17. | ||
Hamilton E, Infante JR. Targeting CDK4/6 in patients with cancer. Cancer Treat Rev. 2016;45:129–138. | ||
Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28(1):20–47. | ||
O’Donnell KA, Yu D, Zeller KI, et al. Activation of transferrin receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis. Mol Cell Biol. 2006;26(6):2373–2386. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.