Back to Journals » Cancer Management and Research » Volume 10
Type-specific high-risk human papillomavirus viral load as a viable triage indicator for high-grade squamous intraepithelial lesion: a nested case–control study
Authors Dong B, Sun P , Ruan G , Huang W , Mao X , Kang Y , Pan D , Lin F
Received 10 July 2018
Accepted for publication 24 August 2018
Published 23 October 2018 Volume 2018:10 Pages 4839—4851
DOI https://doi.org/10.2147/CMAR.S179724
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Binhua Dong,1,* Pengming Sun,1,2,* Guanyu Ruan,1 Weiyi Huang,1 Xiaodan Mao,1 Yafang Kang,1 Diling Pan,3 Fen Lin1
1Laboratory of Gynecologic Oncology, Fujian Provincial Maternity and Children’s Hospital, Affiliated Hospital of Fujian Medical University, Fuzhou 350001, Fujian, P.R. China; 2Department of Gynecology, Fujian Provincial Maternity and Children’s Hospital, Affiliated Hospital of Fujian Medical University, Fuzhou 350001, Fujian, P.R. China; 3Department of Pathology, Fujian Provincial Maternity and Children’s Hospital, Affiliated Hospital of Fujian Medical University, Fuzhou 350001, Fujian, P.R. China
*These authors contributed equally to this work
Purpose: Currently, the associations between type-specific high-risk human papillomavirus (HR-HPV) viral loads and cervical lesions are still inconsistent. We aimed to assess the type-specific HR-HPV viral load as a risk triage indicator for development of high-grade squamous intraepithelial lesion or worse (≥HSIL).
Patients and methods: A total of 19,446 women who underwent primary screening for cervical cancer using Cervista® HR-HPV and cytology assays were enrolled. The viral loads of 1,396 HR-HPV-positive specimens confirmed by Cervista® assay were detected by BioPerfectus Multiplex Real-Time PCR assay. The correlation between viral loads and cervical lesions was analyzed. The optimal cutoffs of individual HR-HPV viral loads used to predict ≥HSIL were determined from the receiver operating characteristic curve. A logistic regression model was used to analyze the relationship between covariates and the probability of ≥HSIL.
Results: The viral loads of HPV-16, -31, -33, -52, and -58 were positively correlated with the severity of the cervical lesion, which was significantly elevated in patients with ≥HSIL, whereas those of HPV-18, -45, -56, -59, and other types were not. The optimal cutoffs of the log10-transformed viral loads for HPV-16, -31, -33, -52, and -58 in identifying ≥HSIL were 4.26, 4.46, 4.48, 4.36, and 4.26 copies per 10,000 cells, respectively. Furthermore, multivariate analysis indicated that type-specific viral loads of HPV-16, -31, -33, -52, and -58 exceeding the cutoffs could be independent risk factors for the incidence of ≥HSIL.
Conclusion: The BioPerfectus Multiplex Real-Time PCR viral load assay provides viable triage for ≥HSIL when using appropriate cutoff levels.
Keywords: high risk, human papillomavirus, viral load, high-grade squamous intraepithelial lesion, BioPerfectus Multiplex Real-Time PCR
Introduction
Cervical cancer is the fourth most frequent malignancy in women, and an estimated 530,000 new cases are diagnosed annually.1 The incidence and mortality rate2,3 of cervical cancer in P.R. China are 7.5 and 3.4 per 100,000 women, respectively. The development of cervical cancer is usually related to persistent infections with human papillomavirus (HPV).4–6 To date, >200 HPV genotypes have been characterized based on their specific sequence information.4 Among them, HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -66, and -68 are classified as high-risk types and HPV-26, -53, -73, and -82 are classified as probable high-risk types5,6 by the WHO.
The most common HPV types identified in cervical cancer worldwide are HPV-16, -18, -45, -31, -33, -52, -58, and -35, with the proportion of cervical cancers associated with HPV-16 and/or HPV-18 (HPV-16/18) being >70%.6–8 Type-specific, high-risk HPV (HR-HPV) infection rates in cervical cancer patients vary by geographic region.7,8 In P.R. China, our previous work9 and other studies10 have shown that HPV-16, -18, -58, -59, -33, -52, -56, and -31 are the most common infection genotypes in cervical cancer.
The relationship between HR-HPV viral loads and cervical lesions remains unclear. The earliest measurements of HR-HPV viral load were performed semi-quantitatively using Hybrid Capture 211,12 and PCR-ELISA,13 but the data generated regarding HR-HPV viral load were inaccurate and inconsistent. The use of reverse transcription-PCR (RT-PCR)14 for HR-HPV viral load determination has become common. Although RT-PCR can absolutely quantify the viral load of HR-HPV, the results14 are not accurate because the viral load is not normalized relative to the viral copies per cell or to the amount of an internal control gene. In recent years, some studies15–17 have normalized the absolute viral load of HPV, but studies15–19 of the relationship between HR-HPV viral load and cervical lesions have focused mainly on HPV-16 and -18 without considering non-HPV-16/18 types. Most studies have reported that the HPV-16 viral load varies directly with the grade of cervical lesions.14–17 However, some studies have suggested that a single measurement of HPV-16 viral load is not a clinically useful biomarker.19 HPV-18 viral load measurement is also questionable as a diagnostic tool for cervical lesions. It has been reported that high HPV-18 viral loads are correlated with the severity of cervical lesions,15 but other studies have stated that HPV-18 viral load is not significant12,16,19 and may, in fact, be lower in women with cervical intraepithelial neoplasia grade 2/3 (CIN2/3) than in those without it.18 Similarly, the clinical relevance of the HR-HPV viral loads of non-HPV-16/18 types is dubious and inconsistent.20–23 Moreover, no cutoffs have been established for the viral loads of individual types of HPV to distinguish between HR-HPV–positive women with high-grade squamous intraepithelial lesion or worse (≥HSIL) and those without it.
A previous study reported24 the BioPerfectus Multiplex Real-Time PCR (BMRT) assay, which was a newly developed, highly automated approach for quantifying viral loads for 14 HR-HPV types. This method provides an absolute quantitative measure of individual types with normalization according to the amount of human DNA.24,25 In order to confirm that type-specific HPV viral loads correlate with the grade of cervical lesions and determine whether the type-specific HPV viral load is a viable risk identification tool for ≥HSIL in HR-HPV–positive women, the present study analyzed type-specific viral loads of 14 HR-HPV types using the BMRT assay in a large-scale clinical trial.
Patients and methods
Patients and study design
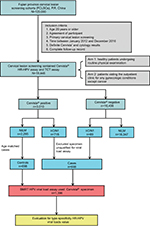
All clinical samples in this study were organized using the Fujian Province Cervical Lesions Screening Cohorts, Fujian, P.R. China. In one of these cohorts, 19,446 women assessed using Cervista® HR-HPV assay and the Thinprep® Cytologic Test at the Fujian Provincial Maternity and Children’s Hospital between January 2012 and December 2016 were enrolled. The population eligible for this study involved two arms, one consisting of healthy patients undergoing routine physical examination and another consisting of patients visiting the outpatient clinic for any gynecologic conditions other than cancer. Qualified participants were those for whom HPV was positively detected by the Cervista® assay, with definite cervical histopathology results, and who had a complete follow-up record (Figure 1). In total, 698 eligible women who were histopathologically abnormal were classified in the case group. Another 698 women who were age matched with the case group and who were histopathologically normal were placed in the control group. All participants were examined, followed, and diagnosed using standardized protocols. All participants in this study provided written informed consent. The protocol was reviewed and approved by the Ethical Committees of the Fujian Provincial Maternity and Children’s Hospital.
Cervical specimen collection
Cervical cells were collected from the cervical canal of all participants with plastic brushes, stored in ThinPrep® Pap Test PreservCyt® Solution (Hologic Inc., Madison, WI, USA), and immediately transported to the laboratory, where they were stored at 4°C. Thinprep® Cytologic Test, Cervista® HR-HPV, and HPV viral load testing were performed on ThinPrep cervical cells.
DNA preparations
DNA from the cervical specimens collected in Thinprep® Pap Test PreservCyt® solution was extracted and purified using a Genfind DNA Extraction Kit (Hologic Inc.) according to the manufacturer’s instructions. The DNA was extracted by magnetic bead adsorption. The kit purified the DNA by removing blood, inflammatory cells, necrotic debris, HPV virions, and mucus. The concentration and purity of the cervical cell DNA were determined using a Q5000 spectrophotometer (Quawell Technology, Inc., San Jose, CA, USA).
Liquid-based cytology
Cytological specimens were blindly evaluated, independent of the results of the other assays, by two experienced cytopathologists. If the diagnosis was different, the cervical samples were reviewed again and a consensus diagnosis was obtained. The results were evaluated using the Bethesda system. Samples were classified as: negative for intraepithelial lesion or malignancy (NILM); atypical squamous cells of undetermined significance; low-grade squamous intraepithelial lesion (LSIL); atypical squamous cells, not possible to exclude HSIL; HSIL; squamous cervical cancer (SCC); atypical glandular cells; and adenocarcinoma (ADC) in situ.
Cervista® HR-HPV DNA test
Cervista® HR-HPV (Hologic Inc.) is a qualitative method for detecting DNA from 14 HR-HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68) in cervical specimens. The test uses invader oligonucleotides that amplify signals to detect specific nucleic acid sequences. This method involves two isothermal reactions: 1) a primary reaction with the targeted DNA sequence and 2) a secondary reaction producing a fluorescent signal. In this test, three oligonucleotide mixtures were created, in which the 14 HR-HPV types were classified into A9, A7, or A5/A6 groups based on phylogenetic relationships. The human histone two gene (HIST2H2BE) served as an internal control for all three oligonucleotide mixtures. The A9 group detected six HPV subtypes (HPV-16, -31, -33, -35, -52, and -58). If at least one of these subtypes was positive, then A9 was taken to be positive. Similarly, A5/A6 included HPV-51, -56, and -66, whereas A7 consisted of HPV-18, -39, -45, -59, and -68. The experimental procedure was conducted according to the kit manufacturer’s instructions.
BMRT HR-HPV viral load assay
Type-specific HPV viral load measurement and genotyping were simultaneously conducted using a BMRT assay. The BMRT assay was performed with the fluorescence-based multiplex HPV DNA genotyping kit (Bioperfectus Ltd, Jiangsu, P.R. China). Specific primers and their corresponding TaqMan probes were designed to detect each of the 14 HR-HPV genotypes (HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -66, and -68), and the viral loads of the 14 high-risk genotypes were simultaneously quantified. A human single-copy gene encoding DNA topoisomerase III (TOP3) was tested to calibrate the standards for absolute quantification of the viral copies in a given sample.24 The experimental procedure was conducted according to the kit manufacturer’s instructions. PCR was performed on an ABI Prism 7500 Fast Dx System (Thermo Fisher Scientific, Waltham, MA, USA). Five-point standard curves for HPV and the cellular log-phase were established for absolute quantification. The standard curves for HPV and TOP3 were Y = –3.85633(log10X)+36.93833 and Y = –3.34656(log10X)+38.51644, respectively. The number of viral copies was normalized according to the cellular input and log10-transformed. In cervical specimens, the distribution of HPV infection is uneven.21 Obtained specimens of cervical cells may, therefore, be partially infected with HPV, whereas other parts may not be infected, so the calculated viral load is the average viral load of all cells entering the test. Furthermore, a previous study has indicated that the viral load per 10,000 cells is considered the applicable unit of HPV load.24 Therefore, the normalization of HPV type-specific viral loads was performed as follows: viral load = log10[Cn HPV/Cn TOP3)×10,000] copies/10,000 cells, where Cn HPV is the quantity of HPV DNA and Cn TOP3 is the number of human cells. Perfectus v1.0 was used for genotyping and quantitative analysis of HPV nucleic acids (Bioperfectus Ltd).
Histology
Women who were HPV positive and/or had an abnormal cytological result were referred for colposcopy and punch biopsy. Women with a punch biopsy diagnosis ≥HSIL underwent a loop electrosurgical excision procedure cone biopsy or conization by cold knife. Specimens were fixed in 10% formalin and routinely processed for paraffin embedding. Subsequently, 4-μm-thick histological sections were cut and stained with H&E using the standard method. Cervical biopsy specimens were then histologically examined and classified according to the CIN system. The cervical lesions were diagnosed by two professional pathologists.
Statistical analysis
The target metric was the viral load of the 14 HR-HPV types at the first visit producing a positive test result. The mean and SD of the log10-transformed absolute virus copy numbers per 10,000 human cells were considered the type-specific HPV viral load. The viral loads of the 14 type-specific HR-HPVs were stratified by cytological and colposcopic grade. Pearson’s correlation coefficient (r) was applied to determine the association between viral load and cervical lesion severity. One-way ANOVA was performed to compare type-specific viral load with cervical cytology or histology. The Scheffé method was used for multiple pairwise comparisons of all HR-HPV types.
Viral loads in single and multiple infections were separately correlated with cervical lesion severity. A paired t-test was used to compare type-specific HR-HPV loads between women with and without diagnoses of ≥HSIL. A receiver operating characteristic (ROC) curve was utilized to identify the optimal cutoff value of the type-specific HR-HPV viral loads for predicting ≥HSIL. A logistic regression model was used to analyze the relationship between covariates and the probability of ≥HSIL in HR-HPV–positive women; the OR was adjusted for covariates. SPSS v17.0 (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses. All tests were run at a two-sided significance level of 0.05.
Results
Patient characteristics and HPV infection status
The present study included 19,446 participants. Among them, 698 women exhibited abnormal cervical histopathology (223 CIN1, 162 CIN2, 168 CIN3, 128 SCC, and 17 ADCs) and were classified as the case group. Moreover, 698 women with positive Cervista® results and normal histopathology were placed in the control group. The BMRT assay revealed that 962 (68.9%) of the women had single infection, 346 (24.8%) had multiple infections, and 88 (6.3%) were negative. Among the single-infection cases, 474 (134 CIN1, 109 CIN2, 119 CIN3, 99 SCC, and 13 ADC) were diagnosed as CIN1 or worse. Among the multiple infection cases, 186 cases (71 CIN1, 44 CIN2, 41 CIN3, 27 SCC, and 3 ADC) were diagnosed as CIN1 or worse. Thirty-eight uninfected cases were diagnosed as CIN1 or worse (18 CIN1, 9 CIN2, 8 CIN3, 2 SCC, and 1 ADC). The mean ages of the women with various histological diagnoses in this study were 46.7±10.47 years (NILM), 43.6±10.69 years (CIN1), 45.3±9.62 years (CIN2), 48.7±9.41 years (CIN3), 49.9±9.69 years (SCC), and 44.7±7.16 years (ADC).
Correlation between type-specific HR-HPV viral load and cervical lesions
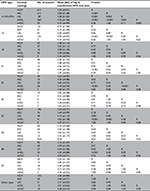
The combined viral loads of the 14 HR-HPVs and the individual viral loads of HPV-16, -31, -33, -52, and -58 increased linearly as the cytological cervical status changed from normal to HSIL (r14 HR-HPV = 0.47, P14 HR-HPV<0.001; rHPV16 = 0.46, PHPV16<0.001; rHPV31 = 0.60, PHPV31<0.001; rHPV33 = 0.51, PHPV33<0.001; rHPV52 = 0.45, PHPV52<0.001; rHPV58 = 0.42, PHPV58<0.001), whereas the individual viral loads of HPV-18 (r = 0.07, P = 0.47), HPV-45 (r = –0.34, P = 0.26), HPV-56 (r = –0.16, P = 0.31), HPV-59 (r = 0.14, P = 0.28), and other high-risk types (OT, r = 0.10, P = 0.24) did not. The combined viral loads of the 14 HR-HPVs and the individual viral loads of HPV-16, -31, -33, -52, and -58 were significantly higher in women with cytological diagnoses of ≥HSIL (all P-values <0.001) than in those with normal cytology. Nevertheless, there were no significant differences between atypical gland cells and normal cytology in terms of viral loads (Table 1).
There was also a linear increase in the viral loads of the 14 HR-HPVs and HPV-16, -31, -33, -52, and -58 as the cervical histopathologic status changed from normal to SCC (r14 HR-HPV = 0.65, P14 HR-HPV<0.001; rHPV16 = 0.53, PHPV16<0.001; rHPV31 = 0.68, PHPV31<0.001; rHPV33 = 0.74, PHPV33<0.001; rHPV52 = 0.49, PHPV52<0.001; rHPV58 = 0.62, PHPV58<0.001), but there was no significant increase in the viral loads of HPV-18 (r = –0.04, P = 0.67), HPV-45 (r = –0.32, P = 0.29), HPV-56 (r = 0.00, P = 1.00), HPV-59 (r = 0.26, P = 0.05), or OT (r = 0.15, P = 0.07). Relative to normal histology, the viral loads among women with a histopathologic diagnosis of HSIL or SCC were significantly higher for the combined 14 HR-HPVs (PHSIL<0.001, PSCC<0.001), HPV-16 (PHSIL<0.001, PSCC<0.001), HPV-31 (PHSIL<0.001, PSCC<0.001), HPV-33 (PHSIL<0.001, PSCC<0.001), HPV-52 (PHSIL<0.001, PSCC = 0.27), HPV-56 (PHSIL = 0.01, PSCC = 0.02), and HPV-58 (PHSIL<0.001, PSCC<0.001) but not for the other types. Compared to LSIL, the viral loads in those with a histopathologic diagnosis of HSIL or SCC were significantly higher for the 14 HR-HPVs (PHSIL<0.001, PSCC<0.001), HPV-16 (PHSIL = 0.003, PSCC<0.001), HPV-31 (PHSIL = 0.002, PSCC = 0.003), HPV-33 (PHSIL = 0.003, PSCC<0.001), and HPV-58 (PHSIL = 0.02, PSCC = 0.03), as shown in Table 2. Conversely, the viral loads of HPV-16, -18, -31, -33, -45, -52, -56, -58, -59, and OT were not significantly higher in women with SCC than in those with HSIL. In fact, the viral loads of HPV-31, -52, -56, and OT were slightly lower. Among the women histologically confirmed to have ADC, the viral loads of the 14 HR-HPVs, HPV-16, -18, -56, -59, and OT were lower than those in women histologically confirmed to have LSIL, HSIL, or SCC. Nevertheless, statistically significant differences were found only for the 14 HR-HPVs (PADC-CIN1 = 0.002, PADC-CIN2/3<0.001, PADC-SCC<0.001) and HPV-16 (PADC-CIN1 = 0.04, PADC-CIN2/3 = 0.02, PADC-SCC = 0.002). The results show that the HR-HPV viral load is not an effective indicator of ADC diagnosis (Table 2).
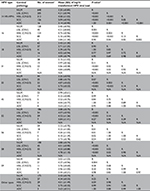
Comparisons of type-specific HR-HPV viral loads in HPV-infected women with and without ≥HSIL
Histopathology confirmed that 475 women had ≥HSIL, and the BMRT assay was positive in 455 women. For both single-infection and multiple infection cases, the viral loads of the 14 HR-HPVs (Psingle<0.001, Pmultiple<0.001), HPV-16 (Psingle<0.001, Pmultiple<0.001), HPV-31 (Psingle<0.001, Pmultiple<0.001), HPV-33 (Psingle<0.001, Pmultiple<0.001), HPV-52 (Psingle<0.001, Pmultiple = 0.04), and HPV-58 (Psingle<0.001, Pmultiple<0.001) were significantly higher in ≥HSIL cases than in NILM/LSIL cases. In contrast, HPV-18 (Psingle = 0.21, Pmultiple = 0.06), HPV-45 (Psingle = 0.13, Pmultiple = 0.56), HPV-56 (Psingle = 0.09, Pmultiple = 0.66), and HPV-59 (Psingle = 0.37, Pmultiple = 0.44) viral loads were not significantly different in cases with or without ≥HSIL (Table 3).
Performance and cutoffs of BMRT viral load assay for the incidence of ≥HSIL in HR-HPV infections
We analyzed the type-specific HPV viral load data using ROC curve models for all HR-HPV–positive women and determined the most useful cutoff for the identification of ≥HSIL (Figure 2). The area under the ROC curve (area under the curve [AUC]) for viral load was calculated for the ≥HSIL endpoints. According to the ROC curve analysis, the 14 HR-HPV viral load had an optimal cutoff of 3.17 copies/10,000 cells (log10-transformed). At this cutoff level, the viral load was able to identify ≥HSIL with a sensitivity (Se) of 84.0%, a specificity (Sp) of 81.0%, and an AUC of 0.90 (95% CI = 0.877–0.911; P<0.001). For the individual HR-HPV viral loads, only the AUC estimates for HPV-16, -31, -33, -52, and -58 were significant. HPV-16 had an AUC of 0.82 (95% CI = 0.766–0.863; P<0.001) and an optimal cutoff of 4.26 copies/10,000 cells (Se = 79.0%, Sp = 81.0%, positive likelihood ratio [PLR] = 4.16, and negative likelihood ratio [NLR] = 0.26). HPV-31 had an AUC of 0.91 (95% CI = 0.827–0.995; P<0.001) and an optimal cutoff of 4.46 copies/10,000 cells (Se = 89.0%, Sp = 89.0%, PLR = 8.09, and NLR = 0.12). HPV-33 had an AUC of 0.91 (95% CI = 0.839–0.978; P<0.001) and an optimal cutoff of 4.48 copies/10,000 cells (Se = 83.0%, Sp = 83.0%, PLR = 4.88, and NLR = 0.20). HPV-52 had an AUC of 0.75 (95% CI = 0.669–0.828; P<0.001) and an optimal cutoff of 4.36 copies/10,000 cells (Se = 62.0%, Sp = 87.0%, PLR = 4.77, and NLR = 0.44). HPV-58 had an AUC of 0.80 (95% CI = 0.726–0.865; P<0.001) and an optimal cutoff of 4.26 copies/10,000 cells (Se = 67.0%, Sp = 79.0%, PLR = 3.19, and NLR = 0.42). The AUC estimate for HPV-18, -45, -56, -59, and OT were not significant.
According to the multivariate logistic regression model analysis (Table 4), the independent risk factors for the incidence of ≥HSIL in HPV infections following log10 transformation were: 14 HR-HPV viral load ≥3.17 copies/10,000 cells (OR = 2.53, 95% CI = 1.73–3.71; P<0.001), HPV-16 viral load ≥4.26 copies/10,000 cells (OR = 21.97, 95% CI = 13.56–35.60; P<0.001), HPV-31 viral load ≥4.46 copies/10,000 cells (OR = 12.17, 95% CI = 2.56–57.81; P = 0.002), HPV-33 viral load ≥4.48 copies/10,000 cells (OR = 6.15, 95% CI = 1.50–25.33; P = 0.012), HPV-52 viral load ≥4.36 copies/10,000 cells (OR = 5.12, 95% CI = 2.77–9.47; P<0.001), and HPV-58 viral load ≥4.26 copies/10,000 cells (OR = 6.35, 95% CI = 3.48–11.58; P<0.001).
Discussion
In recent years, quantitative measurements of HR-HPV load have become common. However, associations between HPV viral loads and cervical lesions are inconsistent. Furthermore, previous studies15,16 focused mainly on HPV-16 and -18, regardless of the absolute load of individual types.12 This study was conducted to evaluate the correlations between the absolute viral loads of the 14 type-specific HR-HPVs, as detected by BMRT HPV viral load assay, and the degree of cervical lesions. In addition, the optimal cutoffs for type-specific HR-HPV viral loads were defined and the ability of the assay to identify ≥HSIL in HR-HPV infections was evaluated according to the cutoffs.
This study showed that the viral loads of HPV-16, -31, -33, -52, and -58, as well as of the 14 HR-HPVs linearly increased as cervical cytological severity also increased. Similarly, for both single infection and multiple infection cases, there were significantly higher viral loads for these types among women with ≥HSIL than among those with NILM/LSIL. We noticed that these five genotypes were associated with a substantially higher risk of ≥HSIL. The present study indicates that the most useful cutoff levels for the log10-transformed viral loads of HPV-16, -31, -33, -52, -58, and the 14 HR-HPVs were 4.26, 4.46, 4.48, 4.36, 4.26, and 3.17 copies per 10,000 cells, respectively, and that these cutoffs provided viable triage for the risk of ≥HSIL in HR-HPV infections (all P<0.05). This finding is consistent with previous studies reporting the effect of HPV-16 viral load on the development of cervical cancer.15,26,27 In our research, the cutoff level for the log10-transformed viral load of HPV-16 was 4.26 copies per 10,000 cells, which is similar to the cutoff level reported by Segondy et al.15 Moreover, Sun et al24 also reported that using 16,600 copies/10,000 cells (log10-transformed: 4.22 copies/10,000 cells) as a cutoff value can predict CIN2+. Among non-16/18 HR-HPV genotypes, special attention should be given to HPV-31 and -33 due to their high carcinogenicity6 and to HPV-52 and -58 due to their high prevalence in Asian women.28 However, earlier studies have seldom or inconsistently reported correlations between HPV-31, -33, -52, and -58 viral loads and cervical lesions.21,29–32 Moreover, HPV-16, -31, -33, -52, and -58 are assigned to species 9, consistent with the ALTS trial,21 and we found that the elevated risk of ≥HSIL for high viral loads appeared to be specific to alpha-9-associated types. In addition, because the HPV-16 viral load predominated, estimates of the viral load of the combined 14 HR-HPVs showed similar trends to those for HPV-16 in terms of cervical cancer disease progression.
In contrast, the viral loads of HPV-18, -45, -56, -59, and other high-risk types did not correlate with cervical lesion severity and were of limited clinical utility in this study. High viral loads of HPV-18, -45, -56, and -59 were not effective at detecting ≥HSIL in HR-HPV–positive women. The data indicated that the viral loads of HPV-56 and -59 are relatively high in ≥HSIL cases with both single and multiple infections, but not significantly so. In contrast, the viral loads of HPV-18 and -45 in ≥HSIL cases were lower than those in NILM/LSIL cases. This result corroborates a previous study indicating that HPV-18 DNA loads were lower in women with HSIL than in those without.18 This finding indicates that cervical lesions induced by HPV-18, -45, -56, and -59 and other high-risk types are more likely to be associated with increased HPV transcript levels rather than with viral copies33 and with more integration rather than episomal status in invasive cancer.34 Additionally, HPV-18, -45, and -59 as representative types of the alpha-7 species were found to be strongly associated with ADC.35 The glandular epithelium of ADC does not support productive HPV infection and may limit intracellular viral replication;36 therefore, the low viral loads in these types of malignancy result in difficulty in detecting HPV and underestimations of the association of HPV-18, -45, and -59 loads with glandular lesions.
The HPV life cycle strictly follows the differentiation program of cellular DNA machinery. The HPV viral load, reflecting cytological changes in exfoliated cells, is closely correlated with the number of infected cells and the viral copies in these cells.37 Therefore, a possible mechanism underlying the association between high viral loads and incremental incidence of cervical neoplasia is an elevated likelihood of cells with transforming infections and viral integration into the cellular genome, concurrent with an increased number of infected cells and/or increased number of viral copies in individual cells.38 Cervical lesions often start from one or more cells and then develop by clonal expansion. The association between ≥HSIL and HPV-16, -31, -33, -52, and -58 viral loads may be explained by the possibility that the number of cells with transforming infections increases with the number of viral copies per cell and/or the number of infected cells.
The higher viral loads of HPV-16, -31, -33, -52, and -58 in cases of ≥HSIL should not be considered to be from ≥HSILs themselves, as cancer cells undergoing clonal expansion may contain fewer viral copes than proliferating cells undergoing productive viral replication. Cervical specimens were obtained by scraping maturing cells from broad areas of the cervical epithelium. Even when they are present, ≥HSILs constitute only a small proportion of the cells removed by scraping.39
There are some limitations of this study. First, the lengths of the infections in the women participating in this study are unknown. Therefore, no conclusion regarding transient infection was drawn. Second, this study had a small number of women diagnosed with ADC. Consequently, no general conclusion could be drawn regarding the difference between control and ADC cases in terms of viral loads. Further studies are needed to confirm our findings and bridge these information gaps. Third, we did not assess the correlation between infection type (episomal, integrated, mixed) and viral load. Evaluation of this relationship will give us a deeper understanding of the biological significance of viral loads. This is the direction in which we plan to continue our efforts in the future. Finally, although HPV viral load testing has potential applications as an alternative to cytology in future clinical practice, the extent to which these findings can be transformed into clinical practice must be rigorously verified in the context of viral load measurement methods and variation in type-specific carcinogenicity among regions.
Conclusion
Our research demonstrates the potential value of type-specific HPV viral load assay. The viral loads of HPV-16, -31, -33, -52, and -58 were positively correlated with cervical disease progression. Optimal cutoffs of type-specific HR-HPV viral loads were established to screen for ≥HSIL. The type-specific viral load assay provides a viable triage tool for ≥HSIL in HR-HPV infections.
Abbreviations
AGC, atypical gland cells; ASC, atypical squamous cells; CIN1+, cervical intraepithelial neoplasia grade one or worse; TCT, Thinprep® Cytologic Test
Acknowledgments
The authors would like to thank the Fujian Province Cervical Lesions Screening Cohorts Investigators for their planning and conducting the trial and for providing the biological specimens and data to the present study. Above all, they are grateful to the patients who made this study possible. This work was supported by grants from the Fujian Provincial Natural Science Foundation of China (grant no. 2017J01232), Fujian Provincial Maternity and Children’s Hospital Natural Science Foundation (grant no. 16–24) and the Fujian Provincial Health and Family Planning Commission Innovation Project (grant no. 2009-CXB-33).
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Centers for Disease Control and Prevention. Global cancer statistics. Available from: http://www.cdc.gov/cancer/international/statistics.htm. Accessed May 13, 2016. | ||
Vaccarella S, Laversanne M, Ferlay J, Bray F. Cervical cancer in Africa, Latin America and the Caribbean and Asia: regional inequalities and changing trends. Int J Cancer. 2017;141(10):1997–2001. | ||
Pan XF, Zhao ZM, Sun J, et al. Acceptability and correlates of primary and secondary prevention of cervical cancer among medical students in southwest China: implications for cancer education. PLoS One. 2014;9(10):e110353. | ||
Brancaccio RN, Robitaille A, Dutta S, et al. Generation of a novel next-generation sequencing-based method for the isolation of new human papillomavirus types. Virology. 2018;520:1–10. | ||
Bouvard V, Baan R, Straif K, et al. WHO International Agency for Research on Cancer Monograph Working Group. A review of human carcinogens – part B: biological agents. Lancet Oncol. 2009;10(4):321–322. | ||
Muñoz N, Bosch FX, de Sanjosé S, et al. International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–527. | ||
Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: variation by geographical region, histological type and year of publication. Int J Cancer. 2011;128(4):927–935. | ||
de Sanjose S, Quint WG, Alemany L, et al. Retrospective International Survey and HPV Time Trends Study Group. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11(11):1048–1056. | ||
Sun P, Song Y, Ruan G, et al. Clinical validation of the PCR-reverse dot blot human papillomavirus genotyping test in cervical lesions from Chinese women in the Fujian province: a hospital-based population study. J Gynecol Oncol. 2017;28(5):e50. | ||
Wang H, Cheng X, Ye J, et al. Distribution of human papilloma virus genotype prevalence in invasive cervical carcinomas and precancerous lesions in the Yangtze River Delta area, China. BMC Cancer. 2018;18(1):487. | ||
Hesselink AT, van den Brule AJC, Brink AA, et al. Comparison of hybrid capture 2 with in situ hybridization for the detection of high-risk human papillomavirus in liquid-based cervical samples. Cancer. 2003;102(1):11–18. | ||
Thomsen LT, Frederiksen K, Munk C, Junge J, Iftner T, Kjaer SK. Long-term risk of cervical intraepithelial neoplasia grade 3 or worse according to high-risk human papillomavirus genotype and semi-quantitative viral load among 33,288 women with normal cervical cytology. Int J Cancer. 2015;137(1):193–203. | ||
Zerbini M, Venturoli S, Cricca M, et al. Distribution and viral load of type specific HPVs in different cervical lesions as detected by PCR-ELISA. J Clin Pathol. 2001;54(5):377–380. | ||
Ylitalo N, Sørensen P, Josefsson AM, et al. Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case–control study. Lancet. 2000;355(9222):2194–2198. | ||
Segondy M, Ngou J, Kelly H, et al. Diagnostic value of human papillomavirus (HPV) 16 and HPV18 viral loads for the detection of high-grade cervical intraepithelial neoplasia (CIN2+) in a cohort of African women living with HIV. J Clin Virol. 2018;100:9979–83. | ||
Wu Z, Qin Y, Yu L, et al. Association between human papillomavirus (HPV) 16, HPV18, and other HR-HPV viral load and the histological classification of cervical lesions: results from a large-scale cross-sectional study. J Med Virol. 2017;89(3):535–541. | ||
Sundstrom K, Ploner A, Dahlstrom LA, et al. Prospective study of HPV16 viral load and risk of in situ and invasive squamous cervical cancer. Cancer Epidemiol Biomarkers Prev. 2012;22(1):150–158. | ||
Xi LF, Koutsky LA, Castle PE, et al. Human papillomavirus type 18 DNA load and 2-year cumulative diagnoses of cervical intraepithelial neoplasia grades 2–3. J Natl Cancer Inst. 2009;101(3):153–161. | ||
Constandinou-Williams C, Collins SI, Roberts S, Young LS, Woodman CB, Murray PG. Is human papillomavirus viral load a clinically useful predictive marker? A longitudinal study. Cancer Epidemiol Biomarkers Prev. 2010;19(3):832–837. | ||
del Río-Ospina L, Soto-de León SC, Camargo M, et al. The DNA load of six high-risk human papillomavirus types and its association with cervical lesions. BMC Cancer. 2015;15:100. | ||
Fu Xi L, Schiffman M, Ke Y, et al. Type-dependent association between risk of cervical intraepithelial neoplasia and viral load of oncogenic human papillomavirus types other than types 16 and 18. Int J Cancer. 2017;140(8):1747–1756. | ||
Liu J, Lu Z, Wang G, et al. Viral load and integration status of HPV58 associated with cervical lesion severity in women of Northeast China. Jpn J Clin Oncol. 2017;47(2):123–129. | ||
Depuydt CE, Criel AM, Benoy IH, Arbyn M, Vereecken AJ, Bogers JJ. Changes in type-specific human papillomavirus load predict progression to cervical cancer. J Cell Mol Med. 2012;16(12):3096–3104. | ||
Sun Z, Zhang R, Liu Z, et al. Development of a fluorescence-based multiplex genotyping method for simultaneous determination of human papillomavirus infections and viral loads. BMC Cancer. 2015;15:860. | ||
Moberg M, Gustavsson I, Gyllensten U. Real-time PCR-based system for simultaneous quantification of human papillomavirus types associated with high risk of cervical cancer. J Clin Microbiol. 2003;41(7):3221–3228. | ||
Xi LF, Kiviat NB, Galloway DA, Zhou XH, Ho J, Koutsky LA. Effect of cervical cytologic status on the association between human papillomavirus type 16 DNA load and the risk of cervical intraepithelial neoplasia grade 3. J Infect Dis. 2008;198(3):324–331. | ||
van Duin M, Snijders PJ, Schrijnemakers HF, et al. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int J Cancer. 2002;98(4):590–595. | ||
Wang R, Guo XL, Wisman GB, et al. Nationwide prevalence of human papillomavirus infection and viral genotype distribution in 37 cities in China. BMC Infect Dis. 2015;15:257. | ||
Liu X, Schiffman M, Hulbert A, et al. Association of human papillomavirus 31 DNA load with risk of cervical intraepithelial neoplasia grades 2 and 3. J Clin Microbiol. 2015;53(11):3451–3457. | ||
Ho CM, Chien TY, Huang SH, Lee BH, Chang SF. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol Oncol. 2006;102(1):54–60. | ||
Cheung JL, Cheung TH, Tang JW, Chan PK. Increase of integration events and infection loads of human papillomavirus type 52 with lesion severity from low-grade cervical lesion to invasive cancer. J Clin Microbiol. 2008;46(4):1356–1362. | ||
Chan PK, Cheung JL, Cheung TH, et al. Profile of viral load, integration, and E2 gene disruption of HPV58 in normal cervix and cervical neoplasia. J Infect Dis. 2007;196(6):868–875. | ||
Ho CM, Lee BH, Chang SF, et al. Type-specific human papillomavirus oncogene messenger RNA levels correlate with the severity of cervical neoplasia. Int J Cancer. 2010;127(3):622–632. | ||
Kurman RJ, Schiffman MH, Lancaster WD, et al. Analysis of individual human papillomavirus types in cervical neoplasia: a possible role for type 18 in rapid progression. Am J Obstet Gynecol. 1988;159(2):293–296. | ||
Hadzisejdć I, Krasević M, Haller H, Grahovac B. Distribution of human papillomavirus types in different histological subtypes of cervical adenocarcinoma. Coll Antropol. 2007;31(Suppl 2):97–102. | ||
Zielinski GD, Snijders PJ, Rozendaal L, et al. The presence of high-risk HPV combined with specific p53 and p16INK4a expression patterns points to high-risk HPV as the main causative agent for adenocarcinoma in situ and adenocarcinoma of the cervix. J Pathol. 2003;201(4):535–543. | ||
Peitsaro P, Johansson B, Syrjänen S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J Clin Microbiol. 2002;40(3):886–891. | ||
Brotherton JML, Tabrizi SN, Phillips S, et al. Looking beyond human papillomavirus (HPV) genotype 16 and 18: defining HPV genotype distribution in cervical cancers in Australia prior to vaccination. Int J Cancer. 2017;141(8):1576–1584. | ||
Sherman ME, Wang SS, Tarone R, Rich L, Schiffman M. Histopathologic extent of cervical intraepithelial neoplasia 3 lesions in the atypical squamous cells of undetermined significance low-grade squamous intraepithelial lesion triage study: implications for subject safety and lead-time bias. Cancer Epidemiol Biomarkers Prev. 2003;12(4):372–379. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.