Back to Journals » OncoTargets and Therapy » Volume 15
Tumor Heterogeneity and Drug Resistance Mutations Using ctDNA in Metastatic EGFR Mutation-Positive Lung Adenocarcinoma: A Case Report
Authors Sun J, Sun G, Lu K, Xu L, Qu X, Cheng Y, Pan E, Yang P, Wu T, Zhang Y, He H
Received 29 May 2022
Accepted for publication 1 August 2022
Published 30 August 2022 Volume 2022:15 Pages 919—923
DOI https://doi.org/10.2147/OTT.S376647
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Jinghua Sun,1,* Ge Sun,1,* KeMou Lu,1 Lingling Xu,1 XiaoNa Qu,1 Ye Cheng,1 Evenki Pan,2 Peng Yang,2 Tingting Wu,2 Yang Zhang,1 HongMei He1
1The Second Affiliated Hospital of Dalian Medical University, Dalian, People’s Republic of China; 2Nanjing Geneseeq Technology Inc., Nanjing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yang Zhang; HongMei He, The Second Affiliated Hospital of Dalian Medical University, Dalian, No. 467, Zhongshan Road, Shahekou District, Dalian, People’s Republic of China, Email [email protected]; [email protected]
Abstract: For advanced non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) mutations, EGFR tyrosine kinase inhibitors (TKIs) have been approved as the standard therapy and shown clinical benefits. However, the emergence of drug resistance is inevitable. Tumor heterogeneity was often observed by imaging method to evaluate the progression of primary and metastatic lesions. Tissue biopsy was also unlikely to accurately capture the complete genomic landscape from a single tissue sample. Recently, genomic characterization of circulating tumor DNA (ctDNA) offer an opportunity to reveal the clonal dynamics throughout the course of a patient’s illness and provide comprehensive genomic landscape of tumors to assess tumor heterogeneity. Here, we reported a lung adenocarcinoma (LADC) with EGFR mutations who was treated with sequential EGFR TKIs. The CT image of the patient’s different lesions suggested that dynamic change of tumor heterogeneity had occurred. Targeted next-generation sequencing (NGS) analysis of ctDNA revealed dynamic changes of mutational profiles between the primary and metastatic tumors to discover tumor evolution to guide treatment decision-making.
Keywords: tumor heterogeneity, circulating tumor DNA, ctDNA, resistance mutation, next-generation sequencing, NGS
Introduction
In Asian non-small cell lung cancer (NSCLC) patients, somatic activating mutations in the epidermal growth factor receptor (EGFR) such as point mutation L858R within exon 21 and short in-frame deletions within exon 19 are the most common oncogenic driver mutation.1 With the development of targeted therapy and next-generation sequencing (NGS) technology, the therapeutic strategies for lung cancer have evolved into a new era of genomics-guided precision medicine. These EGFR-targeted tyrosine kinase inhibitors (TKIs) have shown improved tumor response and progression-free survival (PFS) outcome in EGFR-mutated NSCLC.2 Although EGFR T790M mutation have been confirmed as first- and second-generation EGFR-TKIs resistance mutations, a third-generation EGFR-TKI of osimertinib has shown efficacy in counteracting the growth of EGFR T790M mutant tumors.3 Although osimertinib has demonstrated high clinical efficacy, developing resistance is also inevitable.
The biggest hurdle for the successful treatment of cancer is the tumor heterogeneity, which can take different forms of intratumor, intermetastatic or intrametastatic heterogeneity within an individual patient.4 Computed tomography (CT) remains the initial imaging method for clinical staging of lung cancer, evaluating the change of primary and metastatic lesions. Recently, gene sequencing of circulating tumor DNA (ctDNA) from a liquid biopsy or blood sample can provide comprehensive genetic information of all cancerous lesions (primary and metastases), which overcomes spatial and temporal heterogeneity of a single-tumor biopsy sample, as well as facilitates dynamic tracking of genomic evolution for formatting a treatment strategy.5
We herein report a case of lung adenocarcinoma (LADC) with EGFR mutations, whose different tumor lesions exhibited different response to the EGFR-TKIs. The NGS analysis of ctDNA revealed dynamic changes of mutational profiles between the primary and metastatic tumors.
Case Report
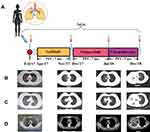
A 44-year-old female was diagnosed with stage IV LADC (cT2bN2M1c) in the right lung center with lymph nodes metastases in February 2017. Positron emission tomography-computed tomography (PET-CT) scan indicated a 14×9 mm density mass in the upper lobe of right lung and a 23×7 mm density mass in the right hilum, respectively (Figure 1). Immunohistochemistry (IHC) staining of tumor biopsy showed positive for LADC markers thyroid transcription factor-1 (TTF-1) and Napsin-A, but negative for lung squamous cell carcinoma (LSCC) markers P40 and P63. NGS demonstrated EGFR L858R with a mutation allelic frequency (MAF) of 0.5%, in plasma and 19.7% in tumor tissue of the upper lobe of right lung, and TP53 R282W with a MAF of 29.9% in plasma and 43.5% in tumor tissue of the upper lobe of right lung (Table 1). Then, the patient was treated with gefitinib (250 mg QD) in April 2017, which is a first-generation EGFR-TKI. In November 2017, the lesions in the upper lobe of right lung deceased to 14×4 mm, the size of the right hilum lesion increased to 27×14 mm and a new 11×8 mm density mass in the mediastinal lymph nodes occurred (Figure 1). The patient reached a progression-free survival (PFS) of 7 months. In order to find more efficient therapeutic strategy, targeted NGS was applied in the plasma sample and it revealed EGFR T790M (MAF = 1.2%), EGFR L858R (MAF = 2%) and TP53 R282W (MAF = 31.7%) (Table 1). The patient was then switched to osimertinib, a third-generation EGFR-TKI (80 mg QD) that is selective for T790M resistance mutations,6 and achieved an initial partial response (PR) with sustained response ongoing for 7 months. In July 2018, the size of the upper lobe of right lung and right hilum lesion increased to 18×13 mm and 65×36 mm, respectively, which indicated a PD (Figure 1). However, the size of lesions in the mediastinal lymph nodes decreased to 8×4 mm. The follow-up genomic testing indicated EGFR L718Q (MAF =1.6%), EGFR T790M (MAF = 1.0%), EGFR L858R (MAF = 5.8%) and TP53 R282W (MAF = 42.1%) in the plasma sample (Table 1). Due to the occurrence of EGFR L718Q, as a mechanism of acquired resistance to osimertinib, the therapy was switched to GC treatment (gemcitabine 1000 mg/m2 and carboplatin AUC 5 on Day 1, and gemcitabine 1000 mg/m2 on Day 8 of a 21-day cycle). However, the size of the upper lobe of right lung, mediastinal lymph nodes and right hilum lesion increased to 18×17 mm, 15×12 mm and 110×80 mm, respectively, which indicated a PD (Figure 1) and targeted NGS revealed EGFR L718Q (MAF =1.6%), EGFR T790M (MAF = 0.8%), EGFR L858R (MAF = 3.4%) and TP53 R282W (MAF = 37.7%) in the plasma sample in December 2018 (Table 1). Unfortunately, the patient died in January 2019.
 |
Table 1 Genetic Alterations by Targeted NGS in the Primary Tumor of the Upper Lobe of Right Lung and Serial Plasma ctDNA |
Discussion
Tumor heterogeneity has been one of the major huddles for the successful treatment of cancer, which is largely attribute to differences in somatic mutations in the tumor.7 Heterogeneity is also often seen in an individual patient and can take different forms including intratumor, intermetastatic or intrametastatic heterogeneity.4 Heterogeneity between different malignant lesions is common in patients with advanced metastatic cancer.8 In this case, the response to the treatments differed in different lesions suggested intermetastatic heterogeneity. Multiple factors may lead to tumor heterogeneity and tumor evolution, including mutational burden, copy number variation and genome doubling.9,10 Some therapies can also contribute to genomic diversity. In this study, the patient was diagnosed with LADC with EGFR L858R. Sequential administration of EGFR TKIs (gefitinib and osimertinib) was performed. For acquired mutations detection, such as EGFR T790M, the usefulness of liquid biopsy has already been reported and used in clinical practice.11 However, such treatment also accelerated the accumulation of EGFR-resistance mutations, including the occurrence of EGFR-resistance mutations, such as EGFR T790M and EGFR L718Q. The emergence of acquired drug resistance limited the duration of tumor response and gave rise to tumor progress as new metastatic lesions or as proliferation of previous tumor lesions, which could be detected by clinical imaging studied. Genomic instability is the most prominent factor that drive tumor heterogeneity since different cells acquire unique mutations that lead to genetically distinct subpopulation.12 Tumor heterogeneity is also acts as a potentiator of acquired resistance regardless of the therapy.13
The patient’s tumor sample was insufficient for genetic testing for the dynamic monitoring. Moreover, molecular characterization of a sample from a primary or metastatic lesion, which often obtained from a needle biopsy or surgical excision, was unlikely to accurately capture the complete genomic landscape. The analysis of ctDNA has been used as a biomarker to assess clonal dynamic progression throughout the course of a patient’s treatment, identify drivers of acquired resistance and assess tumor heterogeneity.14,15 The CT image of the patient’s different lesions suggested that dynamic change of tumor heterogeneity had occurred. Genomic characterization of ctDNA revealed the mutational pattern to help recognize whole genetic alterations occurring in the whole body, which had a potential to demonstrate this heterogeneity (Figure 2), but not in all tumors. The genetic testing based on ctDNA may also offer an opportunity to predict the efficacy of therapeutic approach to improve clinical outcomes.
There are still several limitations of our study which should be further studied in the future. We did not have tumor samples for the dynamic monitoring to validate the tumor heterogeneity. Additionally, further research is warranted to study more effective therapy.
Conclusion
Our report presented LADC patient with EGFR L858R who was treated with sequential EGFR-TKIs and developed resistance mutations, which were detected by dynamic NGS monitoring. The CT image of different lesion during the course of treatment revealed tumor heterogeneity. We highlight the importance of dynamic monitoring of ctDNA using NGS to assess tumor heterogeneity and guide treatment decision-making.
Date Availability
All data sets generated for this study are included in the manuscript.
Ethics Statement
This study involving human participants were reviewed and approved by Ethics Committee of the Second Affiliated Hospital of Dalian Medical University. The patient provided written informed consent to participate in this study.
Acknowledgments
We would like to thank the patient for providing written informed consent for publication.
Author Contributions
All authors performed data analysis, drafted or revised the manuscript, agreed on the journal to which the article was submitted, approved the final version of the manuscript, and agreed to be accountable for all aspects of the work.
Funding
This research did not receive any specific funding.
Disclosure
Authors Evenki Pan, Peng Yang, and Tingting Wu are employed by Nanjing Geneseeq Technology Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res. 2015;5(9):2892–2911.
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654
3. Gristina V, Malapelle U, Galvano A, et al. The significance of epidermal growth factor receptor uncommon mutations in non-small cell lung cancer: a systematic review and critical appraisal. Cancer Treat Rev. 2020;85:101994. doi:10.1016/j.ctrv.2020.101994
4. El-Sayes N, Vito A, Mossman K. Tumor heterogeneity: a great barrier in the age of cancer immunotherapy. Cancers. 2021;13(4):806. doi:10.3390/cancers13040806
5. Sorber L, Zwaenepoel K, Deschoolmeester V, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer. 2017;107:100–107. doi:10.1016/j.lungcan.2016.04.026
6. Cross DAE, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4(9):1046–1061. doi:10.1158/2159-8290.CD-14-0337
7. Lim Z-F, Ma PC. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol. 2019;12(1):134. doi:10.1186/s13045-019-0818-2
8. Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479–485. doi:10.1038/bjc.2012.581
9. Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non–small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. doi:10.1056/NEJMoa1616288
10. Liu Y, Zhang J, Li L, et al. Genomic heterogeneity of multiple synchronous lung cancer. Nat Commun. 2016;7(1):13200. doi:10.1038/ncomms13200
11. Del Re M, Crucitta S, Gianfilippo G, et al. Understanding the mechanisms of resistance in EGFR-positive NSCLC: from tissue to liquid biopsy to guide treatment strategy. Int J Mol Sci. 2019;20(16):3951. doi:10.3390/ijms20163951
12. Hanahan D, Weinberg Robert A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
13. Ramón y Cajal S, Sesé M, Capdevila C, et al. Clinical implications of intratumor heterogeneity: challenges and opportunities. J Mol Med. 2020;98(2):161–177. doi:10.1007/s00109-020-01874-2
14. Ma F, Guan Y, Yi Z, et al. Assessing tumor heterogeneity using ctDNA to predict and monitor therapeutic response in metastatic breast cancer. Int J Cancer. 2020;146(5):1359–1368. doi:10.1002/ijc.32536
15. Nakada H, Nakagomi H, Hirotsu Y, et al. A study of tumor heterogeneity in a case with breast cancer. Breast Cancer. 2017;24(3):483–489. doi:10.1007/s12282-016-0733-0
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.