Back to Journals » Cancer Management and Research » Volume 11
Tumor-educated platelet miR-34c-3p and miR-18a-5p as potential liquid biopsy biomarkers for nasopharyngeal carcinoma diagnosis
Authors Wang H, Wei X, Wu B, Su J, Tan W, Yang K
Received 12 December 2018
Accepted for publication 27 February 2019
Published 17 April 2019 Volume 2019:11 Pages 3351—3360
DOI https://doi.org/10.2147/CMAR.S195654
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Hui Wang,1 Xiuqi Wei,1 Bian Wu,2 Jingyu Su,1 Wenbin Tan,3 Kunyu Yang2
1Department of Laboratory Medicine, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, People’s Republic of China; 2Cancer Center, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430023, People’s Republic of China; 3Department of Cell Biology and Anatomy, University of South Carolina School of Medicine, Columbia, SC 29208, USA
Background: Nasopharyngeal carcinoma (NPC) is the common malignant tumor of nasopharynx in southern China and other southeastern Asian countries. MicroRNAs (miRNAs) have been shown to play important roles in carcinogenesis. Recently, miR-34c-3p and miR-18a-5p have been found to be involved in carcinogenesis of NPC. Furthermore, platelets in NPC patients may acquire RNAs from NPC cells and turn into “tumor-educated platelet (TEP)”, which may serve as potential biomarkers for a diagnosis of NPC. However, the expression profiles of TEP miR-34c-3p and miR-18a-5p in NPC patients and their diagnostic values are yet to be determined.
Aims: To investigate expression levels of TEP miR-34c-3p and miR-18a-5p and determine their diagnostic values for NPC.
Materials and methods: Relative quantitative real-time PCR was used to determine the expression levels of TEP miR-34c-3p and miR-18a-5p in NPC patients (n=54) as compared to normal subjects (n=36). The receiver operating characteristic (ROC) curve analysis was performed to assess the diagnostic values of TEP miR-34c-3p and miR-18a-5p for NPC.
Results: The expression levels of TEP miR-34c-3p and miR-18a-5p were significantly higher in NPC patients as compared to healthy subjects. The ROC analysis showed that the area under the ROC curve (AUC), sensitivity, specificity and accuracy for TEP miR-34c-3p, miR-18a-5p, or a combination of both miRNAs for NPC diagnostic tests were 0.952, 94.44%, 86.11%, 91.11%, or 0.884, 85.19%, 86.11%, 85.55%, or 0.954, 92.59%, 86.11%, 90.00%, respectively. No correlation was found between expression levels of TEP miR-34c-3p or miR-18a-5p and patients’ demographic variables and their NPC tumor/node/metastasis stages. The positive rates of TEP miR-34c-3p and miR-18a-5p for NPC diagnosis were 93.8% and 87.5%, respectively, which were significantly higher than Epstein-Barr virus DNA with a positive rate of 66.7%.
Conclusion: The expression levels of TEP miR-34c-3p and miR-18a-5p are upregulated in NPC, rendering a significant clinical value for NPC diagnosis. The TEP miRNAs might serve as a novel type of liquid biopsies for NPC diagnosis.
Keywords: tumor-educated platelet, miRNA, liquid biopsy, diagnostic value, nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is the common malignant tumor of nasopharynx with an annual incidence from 15 to 50 cases per 100,000 in southern China and other southeastern Asian countries.1 Many factors contribute to the tumorigenesis of NPC, including viral infection, environmental influences and genetic susceptibility.2 Early diagnosis of NPC is a clinical challenge since patients are usually diagnosed at a late stage due to ambiguous early symptoms.3 The peripheral blood levels of Epstein-Barr virus (EBV) DNA and antibodies against EBV are among the current laboratory tests for diagnosis of NPC, but the sensitivity and specificity of these methods are not satisfied for many patients.4 Therefore, there is an urgent need to define novel biomarkers which can determine the pathological progression of NPC for an early diagnosis.
Platelets are abundant cell fragments in circulation which execute multifunctional actions including forming thrombi, monitoring pathogens in peripheral blood, interacting with immune cells, etc. More importantly, they can be “educated” by cancer cells. They can modulate the splicing of their pre-mRNAs in response to signals from cancer cells, resulting in changes in their transcriptome and molecular profiles which reflect pathological progressions.5 Therefore, platelets can acquire tumor-derived RNAs after interactions with tumor cells. These tumor-educated platelet (TEP) RNAs have been emerging as potential novel serum biomarkers for disease diagnosis, prognosis and prediction.6 There are many advantages of TEP RNAs as biomarkers for cancer diagnosis: 1) platelets contain no nuclei thus interference from genomic DNA is very minimal; 2) platelets are easy to be collected, isolated and analyzed, which are ready to be standardized in general clinical laboratories; and 3) TEP RNA profiles reflect the pathological progression of cancer cells, which potentially render a better sensitivity for an early detection of cancer cells.
MicroRNAs (miRNAs) are small noncoding RNA molecules containing about 18–24 nucleotides. The function of miRNAs includes development, cell differentiation, and regulation of cell cycle and apoptosis.7 miRNAs are mainly located within cells while they can also be found in circulatory system and extracellular environment. The circulating miRNAs can serve as liquid biopsies and have become a growing research hotspot for their potential diagnostic values for an early detection of tumors.8 Particularly, Wan found that miR-34c (including miR-34c-3p, miR-34c-5p) and miR-18a (including miR-18a-3p, miR-18a-5p) were dysregulated in NPC using microarrays.9 The miRNA-34 family contributes to the anti-proliferation and pro-apoptosis of cancer cells functioning as a p53 effector.10,11 The miR-18a-5p belongs to the miRNA 17–92 family, locating at 13q31.3.12 Both miR-34c-3p and miR-18a-5p have been found to play roles in tumorigenesis and metastasis. However, their roles in development, progression and diagnosis of NPC remain to be determined. In this study, we attempted to determine the expression levels of TEP miR-34c-3p and miR-18a-5p in NPC as compared with normal subjects as well as evaluate their diagnostic values for NPC. We found that the expression levels of both TEP miRNAs are significantly upregulated in NPC and they provide a good specificity and sensitivity for NPC diagnosis.
Methods and materials
Sample collection
The study was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (No: IORG0003571). The clinically leftover, de-identified whole blood samples (2 mL) were collected from NPC patients (n=54) and healthy subjects (n=36) from March to April 2018 at the Cancer Center of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology. Written informed consent was obtained by the patients. The inclusion criteria of NPC subjects were as follows: 18–85 years old, newly diagnosed with a NPC, no radiotherapy or chemotherapy being received, and no other primary tumors. The samples were subsequently subject to platelet isolation within 3 hrs after blood collection. The healthy subjects were independent to the NPC group with matching age ranges.
Platelet isolation
The whole blood (in EDTAK2) (2 mL) was centrifuged at 120 g for 20 mins at room temperature. The platelet-enriched top-layer (~600 µL) was collected and subjected to spinning at 360 g for 20 mins to precipitate the platelets. The platelets were washed twice by phosphate buffer saline. The number of platelets was counted by hemocytometer after Crystal Violet Staining. A high purity platelet preparation was determined by a ratio of <5 karyocytes per 107 platelets.13 The quality and quantity of PLT were controlled and counted to be the same for each sample in the subsequent experiments.
RNA extraction and relative real-time quantitative PCR (qPCR) analysis
The total RNA was extracted from isolated platelets or blood plasma using a Liquid Total RNA Isolation Kit (RP4002, BioTeke, Beijing, China). The quantity and quality of RNAs were determined by Nanodrop 2000 spectrophotometer and analyzed by an agarose gel electrophoresis. In total, 2 µg of RNAs were used for cDNA synthesis using a PrimeScript™RT reagent Kit with gDNA Eraser (RR047A, TAKARA, Dalian, China). The qPCR was performed using iQ™ SYBR® Green Supermix (BIO-RAD, CA, USA) on a BIO-RAD CFX96 system. The U6 was used as the internal amplification control. The primers used for amplification of miR-34c-3p, miR-18a-5p and U6 are listed in Table 1. The URP was universal amplification primer, which was used in pairs with hsa-miR-34c-3p-S and hsa-miR-18a-5p-S. Relative quantification of RT-PCR was based upon the amplification efficiency of the target and reference genes, and the cycle number at which fluorescence crossed a prescribed background level cycle threshold (Ct) as we described previously.14 Specifically, the relative expression levels of miRNAs were calculated by 2−△CT method; while △CT = CTmiRNAs - CTU6.
 | Table 1 Primers used for cDNA synthesis and real-time PCR |
EBV DNA levels from lymphocyte and plasma were determined using EB viral nucleic acid quantitative fluorescent probe PCR assay (Shengxiang Biotechnology, Hunan, China) on the Stratagene Mx3000P system (Agilent Technologies, CA, USA) at Department of Laboratory Medicine of Union Hospital.
Data analysis
SPSS19.0 software was used for data analysis. The Mann–Whitney U test was used to compare the differential expression levels of miR-34c-3p and miR-18a-5p among NPC, normal groups and subgroups with different NPC tumor/node/metastasis (TNM) stages. The logistic regression was used to access the joint-probability of TEP miR-34c-3p and TEP miR-18a-5p in diagnosis of NPC. The independent-samples t-test and Fisher test were used to determine the correlations between expression levels of miR-34c-3p or miR-18a-5p and various clinical parameters of subjects. The McNemar and Kappa tests were used to compare the positive rates of miR-34c-3p or miR-18a-5p with EBV DNA for NPC diagnosis. A two-sided test was performed in all statistical analyses. Statistically, significant difference was set at p<0.05.
Results
Upregulation of miR-34c-3p and miR-18a-5p in NPC platelets
The expression levels of TEP miR-34c-3p and miR-18a-5p were significantly higher in NPC patients as compared to healthy control subjects (p<0.001, Figure 1, Figure S1). There was no significant difference in expression levels of both miRNAs in plasma from subjects in both groups (p=0.352 and 0.401, respectively, Figure 1).
Diagnostic value of TEP miR-34c-3p and miR-18a-5p in NPC
The area under the receiver operating characteristic curve (AUC) of TEP miR-34c-3p in the NPC group was 0.952 (Figure 2A). The maximum value of the Youden index was 0.8056 and the criterion was >3.4074. The sensitivity, specificity and accuracy of TEP miR-34c-3p for a NPC diagnostic test were 94.44%, 86.11% and 91.11%, respectively (Table 4). The AUC of TEP miR-18a-5p in NPC was 0.884 (Figure 2B). The maximum value of the Youden index was 0.7130 and the criterion was >0.0602. The sensitivity, specificity and accuracy of TEP miR-18a-5p for a NPC diagnostic test were 85.19%, 86.11% and 85.55%, respectively (Table 4). The AUC of the joint-probability of TEP miR-34c-3p and TEP miR-18a-5p in NPC was 0.954 (Figure 2C). When the joint-probability >0.2783, the corresponding values of TEP miR-34c-3p and TEP miR-18a-5p were >3.4074 and >0.2020, respectively. The sensitivity, specificity and accuracy of the joint-probability of TEP miR-34c-3p and TEP miR-18a-5p for a NPC diagnostic test were 92.59%, 86.11% and 90.00%, respectively (Table 4).
 | Table 2 Correlations between expression levels of target miRNAs and clinical parameters |
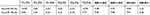 | Table 3 The p-values of comparisons of relative expression levels of TEP hsa-miR-34c-3p or hsa-miR-18a-5p among subgroups with different TNM stages of NPC |
 | Table 4 Statistic data of ROC curves |
Correlations between the expression levels of TEP miR-34c-3p and miR-18a-5p and clinical parameters of NPC subjects
We next attempted to investigate whether expression levels of TEP miR-34c-3p and miR-18a-5p were associated with patients’ demographic variables and their NPC TNM stages. We divided the NPC patients into a high-level group and a low-level group according to the cutoff point in the Youden index analysis, which corresponded to the values of relative expression levels of 3.4074 and 0.0602 for TEP miR-34c-3p and miR-18a-5p as compared to U6, respectively. The subjects in the NPC group with a high level of TEP miR-34c-3p were significantly older than those in the NPC group with a low level of TEP miR-34c-3p (p=0.0492, Table 2). However, the subjects in both NPC groups with a high or low level of TEP miR-18a-5p showed no age difference (p=0.4863, Table 2). There were no significant differences in expression levels of both miRNAs and fasting blood glucose levels in NPC patients (p=0.4178 and 0.5234, respectively, Table 2) or NPC differentiation status (p=0.614and 0.386, respectively, Table 2). The expression levels of TEP miR-34c-3p or miR-18a-5p showed no significant correlation to patients’ gender or NPC TNM stages (p>0.05, Table 2). The NPC diagnostic positive rates of TEP miR-34c-3p and miR-18a-5p were 93.8% and 87.5%, respectively, which were significantly higher as compared to EBV DNA (p=0.004 and 0.041, respectively, Table 2). The expression levels of TEP miR-34c-3p and miR-18a-5p showed no significant differences among different TNM stages of NPC (p>0.05, Figure 3 and Table 3).
Discussion
NPC is a multifactorial disease which takes decades to develop. A majority of patients are misdiagnosed as cervical lymph node metastasis or long distant metastasis during their first clinical visits.15,16 Therefore, it has been a major clinical barrier to determine essential biomarkers for early diagnosis of NPC. In this study, we found that the expression levels of miR-34c-3p and miR-18a-5p in TEP, but not plasma, were significantly higher in NPC patients as compared to normal subjects. Both biomarkers have shown significant clinical values by showing a high sensitivity, specificity and accuracy for NPC diagnosis. Therefore, our study has provided a molecular basis for a prospective clinical application using these miRNAs for NPC diagnosis.
There has been a growing body of evidence to reveal the roles of TEP miRNAs in various pathophysiological processes as well as cancer pathogenesis.13 TEP miRNA signatures along with biomolecular readouts from other liquid biopsies, such as plasma RNA, cell-free DNA and circulating tumor cells, have been drawing great attention for detecting cancer-specific aberrances in genomes and transcriptomes.8 TEP miRNAs have provided many unprecedented characteristics for detecting the status of a disease. Firstly, platelets can react to tumor growth and treatment to become TEP via various biological processes, such as regulation of RNA splicing and tumor RNA sequestration.5 Therefore, the TEP miRNA signatures can reflect molecular profiles of tumorigenesis and cancer metastasis. Secondly, TEP miRNA signatures can pinpoint a specific tumor type with an accuracy of 71%,17 indicating tumor type-specific TEP miRNA processing. Thirdly, TEP miRNAs respond to external stimuli rapidly, providing dynamic miRNA signatures for the progression of a cancer.5 Finally, platelet isolation from blood samples can be easily predefined and standardized for clinical applications.
Many miRNAs have been reported to be associated with the development and progression of NPC, including miR-216b,18 miR-218,15 miR-26a/b,19,20 miR-10b,21 let-7,22 miR-14123 and miR-200a.24 The functional abnormality of p53 has been involved in NPC development. Many miRNA and long non-coding RNAs have participated in modulations of p53 signaling pathways,25 thus regulating NPC development and progression. The miR-34c-3p is one of the P53 effectors.11 Not surprisingly, miRNAs derived from EBV can be oncogenic for NPC as well.9,26 The functions and diagnostic values of miR-18a-5p and miR-34c-3p in NPC are yet to be determined although there are many reports showing their functions in other types of tumors and diseases. For example, upregulation of miR-18a-5p has been reported in triple-negative breast carcinoma (TNBC).16 Inhibition of miR-18a-5p by RNA FENDRR can decrease the invasion and migration ability of prostate cancer.27 Consistent with our data, these evidences together suggest that the upregulation of miR-18a-5p is positively associated with the cancer oncogenic process. Meanwhile, miR-34c-3p appears to have a tumor suppressive function in some cancer types. For example, miR-34c-3p can inhibit proliferation and invasion of non-small cell lung cancer by blockage of eIF4E expression and the PAC1-MAPK pathway.28,29 Ectopic expression of miR-34c-3p inhibits migration, invasion and epithelial–mesenchymal transition in TNBC cells.30 On the contrary, Xiao et al have reported that miR-34c-3p promotes cell proliferation and invasion in hepatocellular carcinoma31 which aligns with our current result showing an upregulation of TEP miR-34c-3p in NPC. Therefore, the functional outputs of miR-34c-3p in tumorigenesis may be of great complexity and diverse and even contradict, depending on the types and progressive stages of cancer cells.
In this study, we have found that TEP miR-34c-3p and miR-18a-5p have significant clinical values for NPC diagnosis due to their high sensitivity, specificity and accuracy in the diagnostic assay. We, however, did not find such altered expression patterns in plasma miR-34c-3p and miR-18a-5p, suggesting that the aberrances of TEP miR-34c-3p and miR-18a-5p are results of an “education” from NPC to platelets. We did not find significant links between TEP miR-34c-3p or miR-18a-5p and patients’ demographic variables and their NPC TNM stages. There are many possible reasons accounting for this result. First, the sample size (n=54) we used for this analysis is not big enough. Second, both miR-34c-3p and miR-18a-5p are associated with tumorigenesis but not the progression of NPC. A functional study of both miRNAs in NPC will be required to answer these questions which will be our next research focus. EBV-DNA detection has become one of the diagnostic tests for NPC in clinics. In this study, we have shown that both TEP miR-34c-3p and miR-18a-5p render significant better positive rates for detection of NPC as compared to EBV-DNA. Our data suggest that TEP miR-34c-3p and miR-18a-5p provide potentially novel diagnostic tools which can be used along with other biomarkers, such as EBV-DNA, to facilitate the diagnosis of NPC in the future.
In our study, we used the U6 as the amplification internal control which may raise a concern of bias in results that are caused by a sole internal control. U6 is one of the most widely used internal standards for monitoring the amplification efficiency during real-time PCR due to its relative stable expression level crossing variety of tissues. More importantly, the Ct values of U6 and our target genes are relatively reasonably close and they showed similar amplification efficiencies crossing samples (Figure S1). Therefore, the potential bias in amplification efficiency by using U6 alone is minimized. Another potential concern is that how the results are not biased by amplification of non-TPE miRNAs. We prepared a high purity platelet with <5 karyocytes per 107 platelets for this study, which ensured to minimize the potential RNA contaminations from any types of cells. It will be interesting to know whether both miRNAs exist in serum exosomes, which will be our next research focus. It is unlikely that exosomal miRNAs have been amplified and biased the results since exosomes (50–200 nm) are too small to be prepared by the low-speed centrifugation method that is used for platelet (2–3 µm) isolation. In this study, we focus on TEP miR-34c-3p and miR-18a-5p. However, we are aware of the variations of TEP RNA profiles. The TEP RNA signatures not only reflect the RNA alterations from NPC tissues but also contain their intrinsic RNA variations corresponding to tumor development in patients. Therefore, a full characterization of the entire TEP RNAs by RNA-seq will be crucial to render a novel strategy for cancer surveillance.
Collectively, in this study we have shown that TEP but not plasma miR-34c-3p and miR-18a-5p are upregulated in NPC patients as compared to healthy subjects. The upregulation of TEP miR-34c-3p or miR-18a-5p is not associated with patients’ demographic variables and their TNM stages of NPC. Our data suggest that TEP miR-34c-3p and miR-18a-5p can potentially serve as essential biomarkers for a novel diagnostic test for NPC.
Acknowledgments
We are very grateful to Dr Fubin Wang at the Department of Laboratory Medicine, Zhongnan Hospital, Wuhan University for his kind assistance in analyzing imaging data. The work was supported by the National Natural Science Foundation of China grant 81374255 to KY.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wu M, Ye X, Wang S, Li Q, Lai Y, Yi Y. MicroRNA-148b suppresses proliferation, migration, and invasion of nasopharyngeal carcinoma cells by targeting metastasis-associated gene 2. Onco Targets Ther. 2017;10:2815–2822. doi:10.2147/OTT.S135664
2. Wang J, Hu JL, Cao RB, et al. Small hairpin RNA-mediated Kruppel-like factor 8 gene knockdown inhibits invasion of nasopharyngeal carcinoma. Oncol Lett. 2015;9(6):2515–2519. doi:10.3892/ol.2015.3099
3. Gao W, Chan JY, Wong TS. Differential expression of long noncoding RNA in primary and recurrent nasopharyngeal carcinoma. Biomed Res Int. 2014;2014:404567. doi:10.1155/2014/404567
4. Twu C-W, Wang W-Y, Liang W-M, et al. Comparison of the prognostic impact of serum anti-EBV antibody and plasma EBV DNA assays in nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 2007;67(1):130–137. doi:10.1016/j.ijrobp.2006.07.012
5. Sol N, Wurdinger T. Platelet RNA signatures for the detection of cancer. Cancer Metastasis Rev. 2017;36(2):263–272. doi:10.1007/s10555-017-9674-0
6. Best MG, Vancura A, Wurdinger T. Platelet RNA as a circulating biomarker trove for cancer diagnostics. J Thromb Haemost. 2017;15(7):1295–1306. doi:10.1111/jth.13720
7. Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi:10.1146/annurev.med.59.053006.104707
8. Bardelli A, Pantel K. Liquid biopsies, what we do not know (Yet). Cancer Cell. 2017;31(2):172–179. doi:10.1016/j.ccell.2017.01.002
9. Wan XX, Yi H, Qu JQ, He QY, Xiao ZQ. Integrated analysis of the differential cellular and EBV miRNA expression profiles in microdissected nasopharyngeal carcinoma and non-cancerous nasopharyngeal tissues. Oncol Rep. 2015;34(5):2585–2601. doi:10.3892/or.2015.4237
10. Lopez JA, Alvarez-Salas LM. Differential effects of miR-34c-3p and miR-34c-5p on SiHa cells proliferation apoptosis, migration and invasion. Biochem Biophys Res Commun. 2011;409(3):513–519. doi:10.1016/j.bbrc.2011.05.036
11. Wu Z, Wu Y, Tian Y, et al. Differential effects of miR-34c-3p and miR-34c-5p on the proliferation, apoptosis and invasion of glioma cells. Oncol Lett. 2013;6(5):1447–1452. doi:10.3892/ol.2013.1579
12. Ichihara A, Wang Z, Jinnin M, et al. Upregulation of miR-18a-5p contributes to epidermal necrolysis in severe drug eruptions. J Allergy Clin Immunol. 2014;133(4):1065–1074. doi:10.1016/j.jaci.2013.09.019
13. Best MG, Sol N, Kooi I, et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28(5):666–676. doi:10.1016/j.ccell.2015.09.018
14. Gao L, Nadora DM, Phan S, et al. Topical axitinib suppresses angiogenesis pathways induced by pulsed dye laser. Br J Dermatol. 2015;172(3):669–676. doi:10.1111/bjd.13439
15. Alajez NM, Lenarduzzi M, Ito E, et al. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011;71(6):2381–2391. doi:10.1158/0008-5472.CAN-10-2754
16. Calvano Filho CM, Calvano-Mendes DC, Carvalho KC, et al. Triple-negative and luminal A breast tumors: differential expression of miR-18a-5p, miR-17-5p, and miR-20a-5p. Tumour Biol. 2014;35(8):7733–7741. doi:10.1007/s13277-014-2025-7
17. Joosse SA, Pantel K. Tumor-educated platelets as liquid biopsy in cancer patients. Cancer Cell. 2015;28(5):552–554. doi:10.1016/j.ccell.2015.10.007
18. Deng M, Tang H, Zhou Y, et al. miR-216b suppresses tumor growth and invasion by targeting KRAS in nasopharyngeal carcinoma. J Cell Sci. 2011;124(Pt 17):2997–3005. doi:10.1242/jcs.085050
19. Ji Y, He Y, Liu L, Chong X. MiRNA-26b regulates the expression of cyclooxygenase-2 in desferrioxamine-treated CNE cells. FEBS Lett. 2010;584(5):961–967. doi:10.1016/j.febslet.2010.01.036
20. Lu J, He ML, Wang L, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71(1):225–233. doi:10.1158/0008-5472.CAN-10-1850
21. Li G, Wu Z, Peng Y, et al. MicroRNA-10b induced by Epstein-Barr virus-encoded latent membrane protein-1 promotes the metastasis of human nasopharyngeal carcinoma cells. Cancer Lett. 2010;299(1):29–36. doi:10.1016/j.canlet.2010.07.021
22. Wong TS, Man OY, Tsang CM, et al. MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression. J Cancer Res Clin Oncol. 2011;137(3):415–422. doi:10.1007/s00432-010-0898-4
23. Zhang L, Deng T, Li X, et al. microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis. 2010;31(4):559–566. doi:10.1093/carcin/bgp335
24. Xia H, Ng SS, Jiang S, et al. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun. 2010;391(1):535–541. doi:10.1016/j.bbrc.2009.11.093
25. Gong Z, Yang Q, Zeng Z, et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 2016;37(3):3683–3695. doi:10.1007/s13277-015-4156-x
26. Zhu LH, Miao XT, Wang NY. Integrated miRNA-mRNA analysis of Epstein-Barr virus-positive nasopharyngeal carcinoma. Genet Mol Res. 2015;14(2):6028–6036. doi:10.4238/2015.June.1.20
27. Zhang G, Han G, Zhang X, et al. Long non-coding RNA FENDRR reduces prostate cancer malignancy by competitively binding miR-18a-5p with RUNX1. Biomarkers. 2018;23(5):435–445. doi:10.1080/1354750X.2018.1443509
28. Liu F, Wang X, Li J, et al. miR-34c-3p functions as a tumour suppressor by inhibiting eIF4E expression in non-small cell lung cancer. Cell Prolif. 2015;48(5):582–592. doi:10.1111/cpr.12201
29. Zhou Y, Xu Y, Qiao C. MiR-34c-3p suppresses the proliferation and invasion of non-small cell lung cancer (NSCLC) by inhibiting PAC1_MAPK pathway. Int J Clin Exp Pathol. 2015;8(6):6312–6322.
30. Wu J, Li WZ, Huang ML, et al. Regulation of cancerous progression and epithelial-mesenchymal transition by miR-34c-3p via modulation of MAP3K2 signaling in triple-negative breast cancer cells. Biochem Biophys Res Commun. 2017;483(1):10–16. doi:10.1016/j.bbrc.2017.01.023
31. Xiao CZ, Wei W, Guo ZX, et al. MicroRNA-34c-3p promotes cell proliferation and invasion in hepatocellular carcinoma by regulation of NCKAP1 expression. J Cancer Res Clin Oncol. 2017;143(2):263–273. doi:10.1007/s00432-016-2280-7
Supplementary material
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




