Back to Journals » OncoTargets and Therapy » Volume 12
TRIM44 Promotes Colorectal Cancer Proliferation, Migration, and Invasion Through the Akt/mTOR Signaling Pathway
Authors Li C, Hu H, Yang X, Huang C, Yu X
Received 25 August 2019
Accepted for publication 14 November 2019
Published 9 December 2019 Volume 2019:12 Pages 10693—10701
DOI https://doi.org/10.2147/OTT.S228637
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Chun-guang Li,1 Hang Hu,1 Xiao-jun Yang,2 Chao-qun Huang,2 Xue-qiao Yu1
1Department of Colorectal & Anal Surgery, Zhongnan Hospital of Wuhan University, Wuhan 430071, Hubei Province, People’s Republic of China; 2Department of Gastrointestinal Surgery, Zhongnan Hospital of Wuhan University, Wuhan 430071, Hubei Province, People’s Republic of China
Correspondence: Chun-guang Li
Zhongnan Hospital of Wuhan University, No. 169 of Donghu Road, Wuchang District, Wuhan 430071, Hubei Province, People’s Republic of China
Tel +8627-67812888
Fax +8627-67812892
Email [email protected]
Purpose: The tripartite motif protein 44 (TRIM44) participates in a variety of biological processes of malignant tumors. However, the expression and molecular mechanism of TRIM44 in colorectal cancer (CRC) remain unclear.
Patients and methods: 123 CRC tissues were used for immunohistochemical assay and survival analysis. Small interfering RNA (siRNA) technology was used to silence the expression of TRIM44 in CRC cell lines. Then, we explored the effect of TRIM44 on the biological behavior of CRC cells. Finally, we studied the underlying mechanisms by Western blot.
Results: We found that TRIM44 is up-regulated in CRC tissues and cells. TRIM44 is a risk factor for poor prognosis in patients with CRC. In vitro, we effectively silenced the expression of TRIM44 in CRC cell lines. Silencing of TRIM44 inhibits the proliferation, migration and invasion of CRC cells. In terms of mechanistic studies, we found that high TRIM44 expression activates the Akt/mTOR signaling pathway.
Conclusion: Our research showed that TRIM44 may serve as a biomarker for CRC patients.
Keywords: TRIM44, CRC, prognosis, Akt/mTOR
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors of the digestive tract and is the leading cause of cancer-related mortality. The number of global cancer-related deaths reached 9.6 million in 2018, of which CRC-related deaths accounted for 10.2%.1 While the development and refinements of comprehensive anti-tumor treatment and precision medicine have greatly improved the prognosis of patients with CRC, the prognosis of patients with advanced CRC remains dismal.2 Knowledge of the molecular mechanism underlying the CRC progression process is necessary. The identification of critical genes and novel therapeutic targets will help improve the prognosis of patients with CRC.
The tripartite motif protein (TRIM) family includes E3 ubiquitin ligase. Members of the TRIM family participate in a variety of biological processes, which include mitosis, apoptosis and proliferation, cell cycle progression, migration, and invasion.3 The TRIM family includes important regulators of multiple human diseases, including cancer.4 The TRIM protein contains the RING domain, B-box structure, and coiled-coil region. The RING domain has E3 ubiquitin ligase activity, which can mediate the ubiquitination of target proteins.5 The B-box structure, comprising conserved cysteine and histidine residues, is a unique domain of the TRIM protein, which might play a decisive role.6 TRIM44 is an important member of the TRIM family. TRIM44 is abnormally expressed and plays a role in promoting malignant solid tumors, including melanoma, cervical cancer, ovarian cancer, esophageal cancer, and liver cancer.7–11 However, the expression and molecular mechanism of TRIM44 in CRC remain unclear.
In this study, we aimed to analyze the expression levels of TRIM44 in human CRC, assess its clinical significance, and reveal the role and mechanism of TRIM44 in cell proliferation, invasion, and migration of CRC.
Materials and Methods
Bioinformatics Analysis
Differential expression of TRIM44 in CRC samples and paracancerous samples was analyzed using the online Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) database. The GEPIA survival analysis tool was used to analyze the relationship between TRIM44 mRNA expression and CRC prognosis. In addition, the expression level of TRIM44 was classified as low and high. Gene Set Enrichment Analysis (GSEA) on signal pathways was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. P-values <0.05 and false discovery rates <0.25 were considered significant.
Tissue Samples and Cell Culture
Overall, 120 paraffin embedded CRC tissues were collected from Wuhan University Zhongnan Hospital. Fresh CRC tissues and paracancerous tissues were also collected from three patients. These 123 CRC patients underwent surgical resection at Zhongnan Hospital from January 2010 to January 2012, and were pathologically diagnosed with CRC. Complete clinical data and follow-up information of 120 patients were obtained. Of them, 72 were men and 48 were women, with a median age of 57 years (range, 39–78 years). The study was performed according to the Helsinki Declaration and was approved by the Ethics Committee of Zhongnan Hospital. All patients signed a written informed consent.
Intestinal mucosal epithelial cells (NCM460) and three CRC cell lines (SW620, LOVO, and HCT116) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured by RPMI-1640 (Gibco, Grand Island, NY, USA) medium with the addition of 10% of fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in an atmosphere of 5% CO2.
Immunohistochemistry and Evaluation
The paraffin-embedded tissues were cut into 4 μm-thick sections for immunohistochemistry (IHC). Xylene and ethanol were used for dewaxing and dehydration, respectively, followed by citrate buffer (pH=6.0) for antigen retrieval. Then, 3% H2O2 was used to block endogenous peroxidase activity, and 5% FBS (Solarbio, Beijing, China) was utilized to assay any non-specific antigen binding of the conjugates. The tissue slices were subsequently incubated with a 1:100 dilution of anti-TRIM44 antibody (Proteintech Group, Inc., Rosemont, IL, USA) at 4°C overnight. The next day, the slices were reacted with horseradish peroxidase-labeled secondary antibody (Beyotime Biotechnology, Shanghai, China) for 1 h at room temperature. The tissue slices were stained with hematoxylin (Sigma-Aldrich, St. Louis, MO, USA), dehydrated using an ethanol gradient, and sealed with neutral gel. In the negative control, the primary antibody was replaced with PBS.
The tissue slices were independently assessed by two pathologists. TRIM44 was primarily expressed in cellular cytoplasm. The IHC staining reaction was evaluated using a semi-quantitative scoring system involving staining intensity and staining degree. Staining intensity was ranked as follows: 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). Staining degree was ranked as follows: 0 (≤5%), 1 (6–25%), 2 (26–50%), 3 (51–75%), or 4 (76–100%). Staining intensity score and staining degree score were multiplied to obtain the final score: ≤1 indicating low expression and >1 indicating high expression.
Western Blotting
Total protein content was measured using a BCA Protein Assay Kit (Thermo Fisher Scientific). Proteins comprising the total protein sample were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). Each membrane was blocked 5% skimmed milk for 1 h and then incubated with primary antibody at 4°C overnight. The following day, the membranes were rinsed three times using Tris buffered saline containing Tween and then incubated with secondary antibody for 1 h at room temperature. Finally, the ECL kit (EMD Millipore, Billerica, MA, USA) was used to visualize the protein bands. The primary antibodies and dilutions used in this study were anti-TRIM44 antibody (Proteintech Group, Inc., 1:1000); anti-mTOR antibody (1:2000), anti-p-mTOR antibody (1:1500), anti-Akt antibody (1:1000), anti-p-Akt antibody (1:1000), anti-P70 antibody (1:1000), and anti-p-P70 antibody (1:2000) (all from Cell Signal Technology, Beverly, MA, USA).
Small Interfering RNA (siRNA)
The expression of TRIM44 in CRC cell lines was silenced by siRNA (si-TRIM44). The target sequence was GGGACCAAATGAAGATGTT. Lipofectamine 2000 reagent (Thermo Fisher Scientific) was used to add si-TRIM44 to CRC cells as described by the manufacturer. Non-specific siRNA (si-NC) and scrambled siRNA (scr-RNA) served as negative controls. Cells were incubated with 5% CO2 at 37°C for 72 h and collected for use in later experiments. The inhibition efficiency was determined by Western blot.
Cell Proliferation
Cell proliferation was evaluated using a Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology, Jiangsu, China). Briefly, CRC cells (3× 103) were seeded into 96-well plates (Corning, Corning, NY, USA) and incubated for indicated time periods. After incubation for 24, 48, and 72 h, 10 μL of CCK-8 solution was added to each well and the samples were incubated for an additional 2 h at 37°C. A microplate reader was used to measure the optical density at 450 nm. The cell growth curve was drawn.
Transwell Assay
The Transwell assay was used to investigate the invasion and migration of CRC cells. For the invasion assay, the upper Transwell chamber (8 μm, BD Bioscience, San Jose, CA, USA) was coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) beforehand. Matrigel was later employed in the migration assay. Cells (5×104) were starved by culture in serum-free medium and then inoculated in the upper chamber. The lower chamber contained culture medium with 10% FBS. After 24 h incubation, the cells that pervaded the surface or migrated into the membrane were fixed, stained using crystal violet, and counted. For the invasion assay, the upper Transwell chamber was also coated with Matrigel. Briefly, 5×104 cells were starved by culture in serum-free medium and inoculated in the upper chamber. The lower chamber contained culture medium with 10% FBS. After incubation for 24 h, the cells that migrated into or invaded the surface of the membrane were fixed, stained with crystal violet, and counted.
Wound Healing Assay
The wound healing assay was used to assess the migration ability of CRC cells. Cells were seeded into six-well plates. When growth was 100% confluent, the monolayers were scraped with a 1000-μL pipette tip and washed with PBS. After being incubated at 37°C for 24 h, the cells were photographed and analyzed using an optical microscope.
Statistical Analyses
Student’s t-test was used to compare the data between the two groups. The relationship between clinicopathological characteristics of CRC patients and TRIM44 expression was analyzed using the Chi-square test. Kaplan-Meier survival curves of CRC patients were drawn. Log rank test was employed to compare the survival curves. Disease-free survival (DFS) and overall survival (OS) were the end points. The Cox proportional hazard regression model was used to analyze whether TRIM44 was a prognostic factor for patients with CRC. GraphPad Prism 6.0 (GraphPad, La Jolla, CA, USA) and SPSS 21.0 (SPSS Inc., Chicago, IL, USA) were used for data analysis. A p-value <0.05 was considered significant.
Results
Upregulation of TRIM44 in CRC
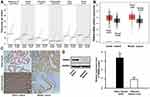
Analysis using the GEPIA online tool indicated an upregulated trend of TRIM44 expression in digestive tract solid tumors compared with that in normal samples (Figure 1A). TRIM44 was also highly expressed in CRC samples (Figure 1B). To verify these predictions, the expression status of TRIM44 protein in CRC tissues was assessed by IHC and Western blot. IHC was first performed to determine the cellular location of TRIM44 in CRC, and the expression of TRIM44 in 120 CRC tissues and paracancerous tissues. The TRIM44 protein was mainly located in the cytoplasm of CRC cells (Figure 1C). TRIM44 was expressed in 54.2% (65/120) CRC tissues, which was significantly higher than that in adjacent tissues (36.7%, 44/120; p=0.006). TRIM44 protein expression was quantitatively determined in three pairs of fresh CRC and paracancerous tissues using Western blot. TRIM44 protein expression was significantly higher in CRC tissues than in paracancerous tissues (p< 0.01; Figure 1D). The data revealed the upregulated expression of TRIM44 in CRC tissues.
Correlation Between TRIM44 and Prognosis of Patients with CRC
The prognostic role of TRIM44 in patients with CRC was assessed. The GEPIA survival tool revealed that CRC patients with high mRNA levels of TRIM44 had poorer prognosis (Figure 2A and B). The 120 patients with CRC were subsequently divided into TRIM44 low expression group (n= 55) and TRIM44 high expression group (n= 65) according to the IHC staining results. The correlation between TRIM44 expression status and clinicopathological characteristics of patients with CRC was determined. High TRIM44 expression was closely associated with lymph node status, distant metastasis, lymphatic and vascular invasion, and carcinoembryonic antigen levels in patients with CRC (Table 1). In addition, long-term follow-up was performed in these patients with CRC, with mean follow-up of 43.6± 38.4 months (range, 4–132 months). Patient survival during follow-up was similar to that of the GEPIA online prediction. CRC patients with higher TRIM44 expression had poorer prognosis (Figure 2C and D). In addition to TRIM44 expression status, univariate analysis indicated that T stage, N stage, M stage, and lymphovascular invasion could affect the survival time of patients with CRC (Table 2). Multivariate Cox analysis showed that T stage, N stage, M stage, and TRIM44 expression status were independent risk factors for prognosis in patients with CRC. These data indicated a key role for TRIM44 in the CRC process.
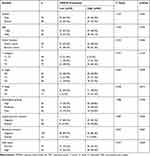 |
Table 1 Association Between TRIM44 Expression and Clinicpathological of CRC Patients |
 |
Table 2 Univariate and Multivariate Cox Proportional Hazards Analysis of TRIM44 Expression and OS for CRC Patients |
Effects of TRIM44 on Biological Behavior of CRC Cells
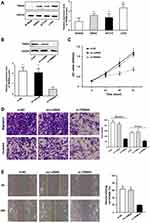
To investigate the effects of TRIM44 on the biological behavior of CRC cells, we examined the abundance of TRIM44 protein expression in CRC cell lines. TRIM44 protein was upregulated in SW620, LOVO, and HCT116 CRC cells compared with the expression in NCM460 normal intestinal mucosal epithelial cells (Figure 3A). The highest level of TRIM44 expression was detected in LOVO cells. These cells were included in the subsequent loss-of-function studies. After effective knockdown of TRIM44 expression in the LOVO cells (Figure 3B), we further examined the effects of TRIM44 on CRC cell proliferation, migration, and invasion. The CCK-8 assay showed that the proliferation of LOVO cells in the si-TRIM44 group was significantly inhibited compared with that in the control group (Figure 3C). The Transwell chamber was used to assess the migration and invasion abilities of CRC cells. TRIM44 silencing effectively inhibited the migration and invasion capacities of LOVO cells (Figure 3D). The wound healing assay was used to further confirm the effects of TRIM44 on the migration ability of CRC cells. The assay demonstrated that knockdown of TRIM44 suppressed the migration of LOVO cells (Figure 3E).
Involvement of TRIM44 in Activation of the Akt/mTOR Signaling Pathway
To explore the potential mechanism of TRIM44 in the CRC process, we performed a GSEA analysis using KEGG data. TRIM44 was closely associated with the Akt/mTOR signaling pathway. To validate the above bioinformatics predictions, the levels of Akt/mTOR signaling pathway-related molecules were determined in the control and si-TRIM44 groups. Western blot revealed that the expression of total mTOR, Akt, and P70 remained unchanged, while the expressions of phosphorylated mTOR, Akt, and P70 were inhibited in si-TRIM44 cells, in comparison with those in control group cells (Figure 4).
Discussion
CRC is a common malignant tumor of the digestive tract. Lifestyle changes have resulted in significantly increasing trends in the incidence and mortality rates of CRC in China.12 Distant metastasis is common in patients with early-stage CRC, especially those with liver metastasis. However, the molecular mechanism of CRC is not yet defined. To improve patient’s prognosis, knowledge of the molecular regulatory mechanism in the CRC process and novel therapeutic targets for CRC are needed. In the present study, the level of TRIM44 expression in CRC tissues increased, and CRC patients with high levels of TRIM44 expression had poorer prognosis. Combined with the predictive results of biological information, we speculate that TRIM44 might play a tumor-promoting role in the CRC process.
Tan et al indicated that through the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway, TRIM44 was upregulated in prostate cancer cell lines and displayed enhanced invasion and proliferation of tumor cells.13 Liu et al reported that expression of TRIM44 was markedly upregulated in tissues of cervical cancer than in nearby normal tissues, and that TRIM44 expression was closely related to FIGO stage, lymph node metastasis, and histological grade; and high levels of expression of TRIM44 was correlated to a poor prognosis in cervical cancer patients.8 TRIM44 also plays an important role in the malignant progression of papillary thyroid carcinoma (PTC). TRIM44 is highly expressed in thyroid PTC tissues and cell lines, and silencing of TRIM44 significantly inhibits PTC cell proliferation, migration/invasion, and epithelial-mesenchymal transition (EMT).14 In other previous studies, the abnormal expression of TRIM44 was closely associated with the progression of malignant tumors, such as gastric cancer, esophageal cancer, and intrahepatic cholangiocarcinoma, thus indicating the poor prognosis of these cancer patients.11,15,16 TRIM44 is highly expressed in gliomas and promotes tumor cell proliferation and cell cycle progression through AKT/p21/p27 signaling pathway.17 Nevertheless, no study has evaluated the effects of TRIM44 on the biological behavior of CRC cells. In this study, siRNA technology was used for effective silencing of TRIM44 expression in LOVO CRC cells. A series of in vitro assays demonstrated that knockdown of TRIM44 significantly inhibited the proliferation, migration, and invasion abilities of LOVO cells. Combined with the outcomes of clinical samples, we preliminarily determined that TRIM44 played a role in the metastatic process of CRC.
GSEA analysis of KEGG data was performed to determine the pathway of TRIM44 in promoting the CRC process. TRIM44 was mainly enriched in the Akt/mTOR signaling pathway. TRIM44 is overexpressed and promotes EMT in esophageal cancer cells via the Akt/mTOR signaling pathway.10 In another study on melanoma, Wei et al found that overexpressed TRIM44 directly binds to TLR4 and activates the Akt/mTOR pathway to ultimately induce tumor cell EMT using Co-IP and mass spectrometry.7 In non-small cell lung cancer, overexpression of TRIM44 induces EMT and enhances the metastatic potential of cancer cells; additionally, the administration of mTOR inhibitors in tumor cells can reverse TRIM44-induced mTOR signaling transmit.18 Based on previous studies and our present bioinformatics predictions, we examined the changes in Akt/mTOR signaling pathway-related molecules after knockdown of TRIM44. TRIM44 was confirmed to exert a tumor-promoting effect in the CRC process by activating the Akt/mTOR signaling pathway.
Conclusion
Collectively, the results demonstrate that TRIM44 is upregulated in CRC and promotes tumor cell proliferation, migration, and invasion by activating the Akt/mTOR signaling pathway. Our findings shed new light on the mechanisms of CRC. TRIM44 may be a useful prognostic indicator and a novel therapeutic target for CRC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.v68.6
2. Schmoll HJ, Van Cutsem E, Stein A, et al. ESMO consensus guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23(10):2479–2516. doi:10.1093/annonc/mds236
3. Venuto S, Merla G. E3 ubiquitin ligase TRIM proteins, cell cycle and mitosis. Cells. 2019;8(5):E510. doi:10.3390/cells8050510
4. Tsai WW, Wang Z, Yiu TT, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468(7326):927–932. doi:10.1038/nature09542
5. Chu Y, Yang X. SUMO E3 ligase activity of TRIM proteins. Oncogene. 2011;30(9):1108–1116. doi:10.1038/onc.2010.462
6. Massiah MA, Simmons BN, Short KM, Cox TC. Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING. J Mol Biol. 2006;358(2):532–545. doi:10.1016/j.jmb.2006.02.009
7. Wei CY, Wang L, Zhu MX, et al. TRIM44 activates the AKT/mTOR signal pathway to induce melanoma progression by stabilizing TLR4. J Exp Clin Cancer Res. 2019;38(1):137–150. doi:10.1186/s13046-019-1138-7
8. Liu S, Meng F, Ding J, et al. High TRIM44 expression as a valuable biomarker for diagnosis and prognosis in cervical cancer. Biosci Rep. 2019;39(3):PMC6400662.
9. Liu S, Yin H, Ji H, Zhu J, Ma R. Overexpression of TRIM44 is an independent marker for predicting poor prognosis in epithelial ovarian cancer. Exp Ther Med. 2018;16(4):3034–3040. doi:10.3892/etm.2018.6541
10. Xiong D, Jin C, Ye X, et al. TRIM44 promotes human esophageal cancer progression via the AKT/mTOR pathway. Cancer Sci. 2018;109(10):3080–3092. doi:10.1111/cas.13762
11. Peng R, Zhang PF, Zhang C, et al. Elevated TRIM44 promotes intrahepatic cholangiocarcinoma progression by inducing cell EMT via MAPK signaling. Cancer Med. 2018;7(3):796–808. doi:10.1002/cam4.1313
12. Chen H, Li N, Ren J, et al. Participation and yield of a population-based colorectal cancer screening programme in China. Gut. 2019;68(8):1450–1457. doi:10.1136/gutjnl-2018-317124
13. Tan Y, Yao H, Hu J, Liu L. Knockdown of TRIM44 inhibits the proliferation and invasion in prostate cancer cells. Oncol Res. 2017;25(8):1253–1259. doi:10.3727/096504017X14854310794561
14. Zhou Z, Liu Y, Ma M, Chang L. Knockdown of TRIM44 inhibits the proliferation and invasion in papillary thyroid cancer cells through suppressing the Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;96:98–103. doi:10.1016/j.biopha.2017.09.132
15. Kashimoto K, Komatsu S, Ichikawa D, et al. Overexpression of TRIM44 contributes to malignant outcome in gastric carcinoma. Cancer Sci. 2012;103(11):2021–2026. doi:10.1111/cas.2012.103.issue-11
16. Ong CA, Shapiro J, Nason KS, et al. Three-gene immunohistochemical panel adds to clinical staging algorithms to predict prognosis for patients with esophageal adenocarcinoma. J Clin Oncol. 2013;31(12):1576–1582.
17. Zhou X, Yang Y, Ma P, et al. TRIM44 is indispensable for glioma cell proliferation and cell cycle progression through AKT/p21/p27 signaling pathway. J Neurooncol. 2019. doi:10.1007/s11060-019-03301-0
18. Xing Y, Meng Q, Chen X, et al. TRIM44 promotes proliferation and metastasis in non‑small cell lung cancer via mTOR signaling pathway. Oncotarget. 2016;7(21):30479–30491. doi:10.18632/oncotarget.8586
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.