Back to Journals » OncoTargets and Therapy » Volume 12
TRIM31 promotes glioma proliferation and invasion through activating NF-κB pathway
Authors Zhou L, Deng ZZ, Li HY, Jiang N, Wei ZS, Hong MF , Chen XD, Wang JH, Zhang MX, Shi YH, Lu ZQ, Huang XM
Received 12 August 2018
Accepted for publication 10 December 2018
Published 27 March 2019 Volume 2019:12 Pages 2289—2297
DOI https://doi.org/10.2147/OTT.S183625
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Li Zhou,1 Zhe-Zhi Deng,2 Hai-Yan Li,2 Nan Jiang,3 Zhi-Sheng Wei,4 Ming-Fan Hong,4 Xiao-Dang Chen,2 Ji-Hui Wang,2 Ming-Xing Zhang,1 Yi-Hua Shi,1 Zheng-Qi Lu,1,2 Xu-Ming Huang1
1Department of Rehabilitation, The First Affiliated Hospital of Guangdong Pharmaceutical University, Guangzhou 510080, China; 2Department of Neurology, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou 510000, China; 3Department of Hepatic Surgery, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou 510000, China; 4Department of Neurology, The First Affiliated Hospital of Clinical Medicine of Guangdong Pharmaceutical University, Guangzhou 510080, China
Background: Glioma is the most lethal primary brain tumor, the survival rate still isn’t improved in the past decades. It’s essential to study the regulatory mechanism of glioma progression, hoping to find new therapy targets or methods. The family of tripartite motif (TRIM) containing proteins are E3 ubiquitination ligases, which play critical role in various tumor progression.
Methods: Cell proliferation and invasion were analyzed by colony formation assay, soft agar growth assay, BrdU incorporation assay and transwell invasion assay. Luciferase reporter analysis was used to analyze NF-κB pathway activity.
Results: We found TRIM31 was upregulated in glioma cells and tissues, its overexpression significantly promoted glioma cell proliferation and invasion, while its knockdown significantly inhibited glioma cell proliferation and invasion. Mechanism analysis found TRIM31 promoted NF-κB pathway activity and increased its targets expression. NF-κB inhibition reversed the phenotype caused by TRIM31, confirming TRIM31 promoted glioma progression through activating NF-κB pathway. Using clinical specimens found TRIM31 expression was positively correlative with NF-κB activity.
Conclusion: This study found TRIM31 promoted glioma proliferation and invasion through activating NF-κB activity.
Keywords: TRIM31, glioma, NF-κB, proliferation, invasion
Introduction
Protein ubiquitination is critical for proteasome system and participates in many biological processes, such as posttranslational regulation, DNA repair, and tumorigenesis.1,2 The canonical ubiquitination cascade is mediated by E1-activating enzymes, E2 conjugation enzymes, and E3 ligase. E3-ligases mainly control substrate specificity and E2 recruitment selection and are divided into two subgroups: HECT domain ligases, which have catalytic activity,3,4 and noncatalytic E3 ligases, E3-RINGs and cullin E3 multisubunit complexes.5–7 The number of E3-RING proteins has 300 members at last; the family of tripartite motif (TRIM) containing proteins belongs to RING-type E3 ubiquitin ligase. Many TRIMs have been shown to play critical role in various tumor progression.8 For example, TRIM24 is a liver-specific tumor suppressor and inhibits cell proliferation and anchorage-independent colony formation.9 TRIM32 functions as a tumor suppressor in leukemias through activating RARα-mediated transcription,10 TRIM32 also binds to and degrades tumor suppressor ABI2 and promotes cell proliferation, transformation, and metastasis in squamous cell carcinoma, papilloma, and head and neck carcinoma.11 TRIM31 is downregulated in non-small cell lung cancer and inhibits cell growth through regulating cyclin D1 and cyclin E expression.12 TRIM31 is upregulated in early stage gastric cancer tissues and is a potential biomarker for gastric cancer.13,14 TRIM31 can upregulate inflammatory factors in colorectal cancer, such as IL6, TNF, and IL-1β, in turn activating NF-κB pathway to promote colorectal cancer cell invasion and metastasis.15 TRIM31 promotes gallbladder cancer cell proliferation and invasion through activating PI3K/Akt pathway.16 miR-551b promotes ovarian cancer cell proliferation, invasion, and drug resistance through targeting FOXO3 and TRIM31,17 suggesting TRIM31 plays different role in different kinds of tumors. The role of TRIM31 in glioma has not been studied; in this study, we main studied the role of TRMI31 in the progression of glioma and found TRIM31 promoted glioma cell proliferation and invasion.
Materials and methods
Cell culture
Primary normal human astrocytes (NHAs) were obtained from ScienCell Research Laboratories. Glioma cells including U373MG, LN-Z308, LN319, LN444, LNN382T, SNB19, U87MG, and LN464 were purchased from ATCC or Cell Bank of Chinese Academy of Sciences and maintained in high-glucose DMEM (Hyclone, Logan, UT, USA) supplemented with 10% FBS (Excell, Shanghai, China).
Clinical specimens
Ten fresh glioma and their adjacent normal brain tissues were obtained from the First Affiliated Hospital of Clinical Medicine of Guangdong Pharmaceutical University. The donor consent was written informed consent, this procedure was conducted in accordance with the Declaration of Helsinki, and the approval of the Institutional Research Ethics Committee was obtained.
RNA isolation and qPCR
Total RNA was isolated from cells and tissues using TRIzol (Thermo, Waltham, MA, USA), cDNA was reverse transcribed using TranScript First-Strand cDNA Synthesis SuperMix (Transgen Biotech, Beijing, China), and qPCR was performed using TransScript® II Green One-Step qRT-PCR SuperMix (Transgen Biotech) and CFX-96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, China). The expression of various genes was defined based on threshold cycle (Ct) and calculated using 2−ΔΔCt method. GAPDH was used as the endogenous control. Several primers were shown as follows: TRIM31, forward: 5′-CCAGAGTCAAACCGTGAGCG-3′ and reverse: 5′-GGCAACTTGGAGCCCGAA-3′; BCL2L1, forward: 5′-TCCCCATGGCAGCAGTAAAG-3′ and reverse: 5′-GTGATGTGGAGCTGGGATGT-3′; Snail, forward: 5′-GAC CAC TAT GCC GCG CTC TT-3′ and reverse: 5′-TCG CTG TAG TTA GGC TTC CGA TT3-′; MMP3, forward: 5′-GAGGACACCAGCATGAACCT-3′ and reverse: 5′-CACCTCCAGAGTGTCGGAGT-3′; IL8, forward: 5′-TCTGCAGCTCTGTGTGAAGG-3′ and reverse: 5′-TGGGGTGGAAAGGTTTGGAG-3′; GAPDH, forward: 5′-AAGAAGGTGGTGAAGCAGGC-3′ and reverse: 5′-TCCACCACCCAGTTGCTGTA-3′.
Western blot
Total proteins form cells and tissues were isolated using RIPA buffer (Millipore, Boston, MA, USA) supplemental with protease inhibitor cocktail (Roche, Basel, Switzerland). Nuclear proteins were isolated using nuclear protein isolation kit (KGP1100, KeyGEN BioTECH, Nanjing, China). Twenty micrograms of protein suspended in SDS loading buffer were run on 10% SDS-PAGE gels and electrotransferred to PVDF membranes (Millipore). Anti-TRIM31 (1:1,000, an67785, Abcam, Cambridge, UK), p65 (1:1,000, 8242, Cell Signaling, Danvers, CO, USA), p84 (1:1,000, ab487, Abcam), IkBα (1:1,000, 4812, Cell Signaling), phospho-IkBα (1:1,000, 2859, Cell Signaling), and α-Tubulin (1:1,000, 3873, Cell Signaling) antibodies were used.
Vectors, siRNA, transfection, and infection
To overexpress TRIM31, the coding sequence was amplified using PCR from cDNA of NHA and subcloned into retroviral vector pBABE vector; the empty vector was used as the negative control. To knock down TRIM31, two shRNAs for TRIM31 were cloned into PLKO.1 lentivirus vector, the empty vector was used as the control, and the targets of shRNAs for TRIM31 were shown as follow: shRNAi1#: CCACAGTTGAACGATCTCAA and shRNAi2#: CGTGAATCCAAGGACCACAAA. The plasmids were transfected into 293 T cells using standard calcium phosphate transfection method; viral supernatants were collected 48 hours after transfection and infected into indicated cells for overnight.
Cell proliferation assay
For colony formation assay, cells (0.5×103) were seeded at 6-well plates, and cultured for 14 days, and then colonies were fixed with 4% paraformaldehyde for 30 minutes, and stained with 1% crystal violet for 1 hour. BrdU incorporation assay was performed using previous method;18 cells (1.5×105) were grown for 3 days before labeling with 3 μg/mL BrdU for 4 hours and stained with 95% ethanol under agitation for overnight, and DNA was denatured with 2N HCl, 0.5% Triton X-100 for 1 hour. Note that, 0.1 M sodium tetraborate was used for neutralization. Cells were incubated with anti-BrdU antibody (ab1893, Abcam) for overnight at 4°C, then cells were washed and incubated with anti-Alexa Fluor 594-conjugated second antibody (Thermo) for 2 hours at room temperature. DAPI (Sigma-Aldrich Co., St Louis, MO, USA) was used to stain nucleus. For anchorage-independent growth assay, a mixture of cells (1×104) and 0.35% low-melt agarose was plated on a base layer of 0.7% low-melt agarose, cells were cultured for 3 weeks, and colonies whose diameter was larger than 0.1 mm were counted. Every experiment was assessed in triplicate.
Transwell invasion assay
Cells (2×105) were seeded on the top side of polycarbonate Transwell filter with Materigel (Corning, Corning, NY, USA) in the upper chamber of the BioCoat invasion chambers (BD, Franklin Lakes, NJ, USA) and incubated for 22 hours at 37°C, cells in the upper chamber were removed with cotton swabs, and invasive cells on the lower membrane surface were fixed in 1% paraformaldehyde and stained with hematoxylin. Cells were counted and expressed as the mean number of cells per field of view. This experiment was assessed in triplicate.
Luciferase reporter assay
Cells were cotransfected with 100 ng pNF-κB reporter luciferase plasmid with 5 ng pRL-TK Renilla plasmid using Lipofectamine 2000 (Thermo). Luciferase and Renilla signals were determined using Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) after 36 hours of transfection. This experiment was assessed in triplicate.
Statistical analysis
Student’s t-test was used to examine statistical differences between two groups. Significant differences were examined using SPSS 19.0. All values are presented as means ± SD. For all tests, a P-value of <0.05 was considered statistically significant. All experiments were performed in triplicate.
Results
TRIM31 is upregulated in glioma cells and tissues
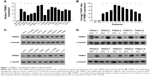
To determine the role of TRIM31 in glioma progression, we first determined the expression of TRIM31 in glioma and primary NHA cell; qPCR and Western blot analysis suggested that TRIM31 was significantly upregulated in glioma cells compared to NHA cell (Figure 1A and C). We also analyzed TRIM31 expression in glioma tissues and adjacent normal brain tissues and found TRIM31 was significantly upregulated in glioma tissues determined by qPCR and Western blotting (Figure 1B and D). These results showed TRIM31 was significantly upregulated in glioma cells and tissues, suggesting TRIM31 might promote glioma development.
Upregulation of TRIM31 promotes glioma proliferation and invasion
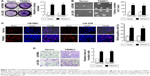
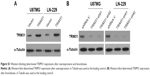
We overexpressed TRIM31 in glioma cell U87MG and LN-229 to determine its function in tumor progression (Figure S1A). TRIM31 overexpression significantly promoted glioma cell proliferation determined by colony formation assay (Figure 2A). Soft agar growth assay suggested colony numbers were significantly increased in cells with TRIM31 overexpression compared to the vector control (Figure 2B). BrdU incorporation analysis suggested that the number of BrdU positive cells was significantly increased compared to the vector control (Figure 2C). Transwell analysis with Matrigel coated analysis showed that TRIM31 overexpression significantly promoted glioma cell invasion (Figure 2D). These results suggested TRIM31 overexpression promoted glioma cell proliferation and invasion.
Downregulation of TRIM31 inhibits glioma proliferation and invasion
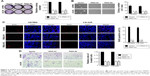
We downregulated TRIM31 in glioma cell U87MG and LN-229 to confirm its function in tumor progression (Figure S1B). TRIM31 knockdown significantly inhibited glioma cell proliferation determined by colony formation assay (Figure 3A). Soft agar growth assay suggested that colony numbers were significantly inhibited in cells with TRIM31 downregulation compared to the vector control (Figure 3B). BrdU incorporation analysis suggested that the number of BrdU positive cells was significantly decreased compared to the vector control (Figure 3C). Transwell analysis with Matrigel-coated analysis showed that TRIM31 downregulation significantly inhibited glioma cell invasion (Figure 3D). These results suggested TRIM31 downregulation inhibited glioma cell proliferation and invasion, confirming TRIM31 promoted glioma cell proliferation and invasion.
TRIM31 promotes glioma cell proliferation and invasion through activating NF-κB pathway
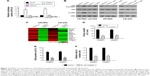
Previous studies find that many TRIM proteins can activate NF-κB pathway through interacting with IKK proteins19,20 and NF-κB pathway plays critical role in tumor progression, such as tumor proliferation, invasion, and metastasis.21 We analyzed the role of TRIM31 in NF-κB pathway activity; TRIM31 overexpression significantly increased NF-κB pathway activity, while TRIM31 knockdown significantly decreased NF-κB pathway activity (Figure 4A). Translocation into nucleus of p65 and the phosphorylation of IκBα are the markers for NF-κB pathway activation, Western blot assay suggested that TRIM31 overexpression increased the nuclear translocation of p65 and the phosphorylation of IκBα, and TRIM31 knockdown inhibited the nuclear translocation of p65 and the phosphorylation of IκBα (Figure 4B). We also analyzed the expression of genes associated with tumor proliferation, invasion, and metastasis, such as BCL2L1, Snail, MMP3, IL8, MYC, MMP9, MMP13, IL1β, XIAP, CXCL5, MMP1, TWIST1, and CCND1. qPCR analysis suggested that TRIM31 overexpression increased their expression and TRIM31 knockdown inhibited their expression, supporting our findings (Figure 4C). To confirm whether TRIM31 promoted glioma proliferation and invasion through regulating NF-κB activity, we inhibited NF-κB activity through adding NF-κB inhibitor JSH-23 (10 μm, Selleck) in TRIM31-overexpressing cells. Colony formation assay and BrdU incorporation assay found that inhibition of NF-κB activity inhibited TRIM31-overexpressing cell proliferation (Figure 4D and E). Transwell assay found inhibition of NF-κB activity in TRIM31 overexpressing inhibited cell invasion (Figure 4F). These results suggested TRIM31 promoted glioma cell proliferation through activating NF-κB activity.
We also confirmed above results using clinical specimens and found that specimens with high TRIM31 expression had high p65 accumulation in nucleus and high Snail, CCND1, and XIAP expression (Figure 5A). TRIM31 expression was positively correlated with p65 accumulation in nucleus (Figure 5B). These findings confirmed that TRIM31 was associated with tumor invasion, proliferation, and NF-κB activity.
Discussion
Glioma is the most frequent malignant primary brain tumor; despite the improvement in surgery, chemotherapy and radiotherapy have been used for glioma therapy. Its 5-year survival rate is still below than 5%,22 so it is important to explore the regulatory mechanism of glioma progression. Many genes have been found to influence glioma, for example, RNA-binding protein LARP4B inhibits glioma growth and induces mitotic arrest and apoptosis.23 Heparanase promotes glioma growth and viability by regulating tumor microenvironment.24 In present study, we found that TRIM31 promoted glioma proliferation and invasion through activating NF-κB pathway. Although TRIM family has been shown to regulatory various tumor progression, TRIM31’s role in glioma progression has not been studied. Our studied enriched the knowledge of TRIM31 in glioma progression.
NF-κB pathway could regulate many biological functions and signaling, such as MAPK, insulin resistance, apoptosis, cell cycle progression, tumor metastasis, keratinocyte differentiation, and inflammation-mediated angiogenesis.25,26 Previous studies find that NF-κB pathway promotes glioma cell survival, resistance to therapy, migration, invasion, metastasis, tumor inflammation microenvironment, and the expansion of glioma stem cells.27,28 We found that TRIM31 could activate NF-κB signaling though luciferase reporter assay; p65 translocating into nucleus is a marker for NF-κB activation. IkBα is NF-κB inhibitor and restricts NF-κB dimer in cytoplasm, IκB kinase IKK1, IKK2 and NEMO induce IκB phosphorylation, releasing NF-κB dimer translocate to nucleus.29,30 Western blotting analyzed further found that TRIM31 promoted the nuclear translocation of p65 and the phosphorylation of IkBα, confirming TRIM31 activates NF-κB.
In summary, we found TRIM31 promoted glioma cell proliferation and invasion through activating NF-κB pathway.
Acknowledgment
This work is supported by Guangdong Provincial Natural Science Foundation (NO. 2018A0303130323).
Disclosure
The authors report no conflicts of interest in this work.
References
Micel LN, Tentler JJ, Smith PG, Eckhardt GS. Role of ubiquitin ligases and the proteasome in oncogenesis: novel targets for anticancer therapies. J Clin Oncol. 2013;31(9):1231–1238. | ||
Lub S, Maes K, Menu E, De Bruyne E, Vanderkerken K, van Valckenborgh E. Novel strategies to target the ubiquitin proteasome system in multiple myeloma. Oncotarget. 2016;7(6):6521–6537. | ||
Scheffner M, Kumar S. Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim Biophys Acta. 1843;2014(1):61–74. | ||
Sluimer J, Distel B. Regulating the human hect E3 ligases. Cell Mol Life Sci. 2018;75(17):3121–3141. | ||
Metzger MB, Pruneda JN, Rere K, Amam W. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta. 1843;2014(1):47–60. | ||
Dubiel W, Dubiel D, Wolf DA, Naumann M. Cullin 3-based ubiquitin ligases as master regulators of mammalian cell differentiation. Trends Biochem Sci. 2018;43(2):95–107. | ||
Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12(4):220. | ||
Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11(11):792–804. | ||
Khetchoumian K, Teletin M, Tisserand J, et al. Loss of TRIM24 (TIF1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet. 2007;39(12):1500–1506. | ||
Sato T, Okumura F, Kano S, Kondo T, Ariga T, Hatakeyama S. TRIM32 promotes neural differentiation through retinoic acid receptor-mediated transcription. J Cell Sci. 2011;124(Pt 20):3492–3502. | ||
Horn EJ, Albor A, Liu Y, et al. Ring protein TRIM32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis. 2004;25(2):157–167. | ||
Li H, Zhang Y, Zhang Y, Bai X, Peng Y, He P. TRIM31 is downregulated in non-small cell lung cancer and serves as a potential tumor suppressor. Tumor Biol. 2014;35(6):5747–5752. | ||
Sugiura T, Miyamoto K. Characterization of TRIM31, upregulated in gastric adenocarcinoma, as a novel RBCC protein. J Cell Biochem. 2008;105(4):1081–1091. | ||
Sugiura T. The cellular level of TRIM31, an RBCC protein overexpressed in gastric cancer, is regulated by multiple mechanisms including the ubiquitin-proteasome system. Cell Biol Int. 2011;35(7):657–661. | ||
Wang H, Yao L, Gong Y, Zhang B. TRIM31 regulates chronic inflammation via NF-κB signal pathway to promote invasion and metastasis in colorectal cancer. Am J Transl Res. 2018;10(4):1247–1259. | ||
Li H, Zhang Y, Hai J, et al. Knockdown of TRIM31 suppresses proliferation and invasion of gallbladder cancer cells by down-regulating MMP2/9 through the PI3K/Akt signaling pathway. Biomed Pharmacother. 2018;103:1272–1278. | ||
Wei Z, Liu Y, Wang Y, et al. Downregulation of FOXO3 and TRIM31 by miR-551b in side population promotes cell proliferation, invasion, and drug resistance of ovarian cancer. Med Oncol. 2016;33(11):126. | ||
Dewaal D, Nogueira V, Terry AR, et al. Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat Commun. 2018;9(1):446. | ||
McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 2011;23(1):46–56. | ||
Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med. 2011;3(9):513–527. | ||
Rajabi H, Kufe D. MUC1-C oncoprotein integrates a program of EMT, epigenetic reprogramming and immune evasion in human carcinomas. Biochim Biophys Acta. 1868;2017(1):117–122. | ||
Jung E, Osswald M, Blaes J, et al. Tweety-Homolog 1 drives brain colonization of gliomas. J Neurosci. 2017;37(29):6837–6850. | ||
Koso H, Yi H, Sheridan P, et al. Identification of RNA-binding protein Larp4B as a tumor suppressor in glioma. Cancer Res. 2016;76(8):2254–2264. | ||
Kundu S, Xiong A, Spyrou A, et al. Heparanase promotes glioma progression and is inversely correlated with patient survival. Mol Cancer Res. 2016;14(12):1243–1253. | ||
Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25(51):6680–6684. | ||
Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-κB signaling pathways. Nat Immunol. 2011;12(8):695–708. | ||
Cahill KE, Morshed RA, Yamini B. Nuclear factor-κB in glioblastoma: insights into regulators and targeted therapy. Neuro Oncol. 2016;18(3):329–339. | ||
Zhou Y, Tan Z, Chen K, et al. Overexpression of SHCBP1 promotes migration and invasion in gliomas by activating the NF-kappaB signaling pathway. Mol Carcinog. 2018;57(9):1181–1190. | ||
Wuerzberger-Davis SM, Chen Y, Yang DT, et al. Nuclear export of the NF-κB inhibitor IκBα is required for proper B cell and secondary lymphoid tissue formation. Immunity. 2011;34(2):188–200. | ||
Mooster JL, Le Bras S, Massaad MJ, et al. Defective lymphoid organogenesis underlies the immune deficiency caused by a heterozygous S32I mutation in IκBα. J Exp Med. 2015;212(2):185–202. |
Supplementary material
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.