Back to Journals » Journal of Inflammation Research » Volume 15
Toll-like Interleukin -1 Receptor Regulator (TILRR) Protein, a Major Modulator of Inflammation, is Expressed in Normal Human and Macaque Tissues and PBMCs
Authors Kashem MA, Li L, Yuan XY, Plummer FA, Luo M
Received 8 February 2022
Accepted for publication 27 April 2022
Published 12 May 2022 Volume 2022:15 Pages 2925—2937
DOI https://doi.org/10.2147/JIR.S357866
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Mohammad Abul Kashem,1– 3 Lin Li,2 Xin-Yong Yuan,2 Francis A Plummer1 ,† Ma Luo1,2
1Department of Medical Microbiology and Infectious Diseases, University of Manitoba, Winnipeg, MB, Canada; 2JC Wilt Infectious Diseases Research Center, National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, MB, Canada; 3Department of Microbiology and Veterinary Public Health, Chittagong Veterinary and Animal Sciences University, Chittagong, Bangladesh
†Dr Francis A Plummer passed away on February 04, 2020
Correspondence: Ma Luo, JC Wilt Infectious Diseases Research Center, National Microbiology Laboratory, 745 Logan Avenue, Winnipeg, MB, R3E 3L5, Canada, Tel +1 204-789-5072, Fax +1 204-789-2018, Email [email protected]
Purpose: TILRR is a modulator of genes in the NF-κB inflammation pathway. It regulates inflammation-responsive genes, the secretion of inflammatory mediators, and the migration of immune cells. Because inflammation drives the pathogenesis of many infectious and inflammatory diseases, it is important to know the expression of TILRR protein in tissues and cells. This study examined TILRR protein expression in healthy adult human and macaques’ tissues and PBMCs (peripheral blood mononuclear cells).
Methods and Results: Tissues (trachea, lungs, stomach, small intestine [ileum], cecum, colon, rectum, vagina, cervix, uterus, and penis) and PBMCs from humans and macaques were lysed in RIPA (radioimmunoprecipitation assay) lysis buffer. The TILRR protein was examined by fluorescent Western blot analysis. The relative fluorescence units (rfu) of TILRR protein expression were quantified by Image Studio software (LI-COR). The results showed that adult healthy female (n=1) rectal and cervicovaginal tissues expressed a higher level of TILRR protein than the other tissues (trachea, lungs, stomach, small intestine [ileum], cecum, colon, uterus, and penis) examined. Like humans, the lungs, colon, and rectal tissues of healthy adult female cynomolgus monkeys (Macaca fascicularis) (n=2) expressed the TILRR protein. In addition, PBMCs of healthy adult women (n=4), adult female cynomolgus monkeys (Macaca fascicularis) (n=4), and adult male and female rhesus monkeys (Macaca mulatta) (n=4) showed a similar expression level of TILRR protein (p= 0.2858). TILRR protein was not detected in most of the human cell lines examined, except in Jurkat cells.
Conclusion: Our study for the first time showed that TILRR protein is expressed in healthy adult human and monkey tissues and PBMCs. The TILRR protein in these tissues and PBMCs may play a role in the inflammatory response of these tissues and cells in response to infectious pathogens.
Keywords: TILRR, tissues, PBMCs, human, rhesus monkey (Macaca mulatta), cynomolgus monkey (Macaca fascicularis), inflammation
Introduction
Toll-like interleukin-1 receptor regulator (TILRR) potentiates inflammation by binding with interleukin-1-interleukin-1 receptor type 1 (IL-1-IL-1R1) receptor complex and activation of NF-κB (nuclear factor-kappa-light-chain-enhancer of activated B cells) transcription factor.1,2 The engagement of TILRR with IL-1-IL-1R1 receptor complex influences the recruitment of MYD88 (myeloid differentiation primary response 88) adaptor protein to the cytoplasmic TIR (toll/IL-1 receptor) domain and instigates the elevated level of NF-κB activation, inflammatory responses, and inflammation.1,3 On the other hand, genetic deletion or antibody blocking of TILRR showed a reduced level of NF-κB activation and inflammatory responses.3,4 Our previous studies showed that TILRR modulated the expression of many NF-κB inflammation-responsive genes,5 promoted the secretion of pro-inflammatory mediators,5 and migration of immune cells.6 Recently, we identified TILRR protein in the blood of healthy women,7 and find that the level of plasma TILRR protein is positively correlated with several plasma inflammatory mediators.8 Furthermore, high plasma TILRR protein level is a risk factor in HIV seroconversion.8 Studies showed that inflammation drives the pathogenesis of many infectious diseases, including SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2),9,10 MERS-CoV (middle eastern respiratory syndrome coronavirus),11 H5N1 influenza A virus,12 RSV (respiratory syncytial virus),13 and HIV (human immunodeficiency virus),14–16 common inflammatory diseases, such as COPD (chronic obstructive pulmonary disease),17 and IBD (inflammatory bowel disease),18,19 and the progression of cancer.20 Proinflammatory mediators produced by the epithelial cells and tissues attract immune cells at the site of infections or injuries, leading to cycles of inflammatory reactions and pathogenesis of diseases.13,17,21–27 Because TILRR increases the expression of genes involved in the NF-κB inflammatory pathway and induces secretion of proinflammatory mediators,1,3,4 and promotes immune cell migration,6 the knowledge of TILRR protein expression in different tissues and PBMCs (peripheral blood mononuclear cells) is important for understanding the spectrum of its potential influence in inflammatory responses to infectious pathogens. In this study, we evaluated TILRR protein expression in different human and monkey tissues and PBMCs and showed that TILRR protein is expressed in all tissues and PBMCs examined. Thus, TILRR may influence inflammatory responses in immune cells and these tissues.
Materials and Methods
Study Subjects
Human PBMCs were collected from women (n=4) enrolled in the Pumwani sex worker cohort (PSWC), Nairobi, Kenya between 2004 and 2017 as previously described.8 The PSWC was established in 1985 for the study of the epidemiology and immunobiology of STIs (sexually transmitted infections).8,28
Animal Samples
The cells isolated from the tissues of healthy adult female cynomolgus macaques (Macaca fascicularis) (n=2) were from a previous study that was performed in accordance with Canadian Council on Animal Care guidelines.29–31 The tissues were collected at necropsy. The PBMCs from healthy adult male and female Rhesus monkeys (Macaca mulatta) (n=4) were from a previous contract study carried out at the ABL (Advanced Bioscience Laboratories) facility.
Preparation of Tissue, PBMCs, and Cell Line Lysates
The adult human tissue (healthy) lysates (n=11) were purchased from Novus Biologicals (Bio-Techne Canada) (Table 1). The tissue lysates were from one tissue donor only (n=1). The lysates were supplied in a buffer containing HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) pH 7.9, MgCl2 (magnesium chloride), KCl (potassium chloride), <3% EDTA (ethylenediaminetetraacetic acid), sucrose, glycerol, <0.5% sodium orthovanadate, <5% sodium deoxycholate, <4% NP-40 (nonidet P-40), and a cocktail of protease inhibitors (<0.5%). Tissue lysates were processed by using a total protein extraction kit (Novus Biologicals, Canada; catalog# NBP2-37853) as follows: tissues were weighed, chopped into small pieces, and kept on dry ice. 1X protease inhibitors (2.5 mL per gram of tissue) were added to the tissues and kept on ice for 5 min. After the incubation, tissues were homogenized for 20 sec, put on dry ice for 15 sec, and then again homogenized for 20 sec (a third time 20 sec homogenization was used when the tissues were not well homogenized). The homogenized tissues were rotated at 4° C for 20 min, centrifuged at 18,000 xg at 4° C for 20 min, and the supernatants were collected to determine the total protein concentration. The protein concentration of tissue lysates was 5 mg/mL (as supplied). The lysates were diluted at 1:8 with RIPA (radioimmunoprecipitation assay) lysis buffer (Thermo Fisher Scientific, Toronto, Canada) containing protease inhibitors (10 µL per mL of buffer).
 |
Table 1 Adult Human Normal Tissue Lysates and Monkey Normal Tissue Cell Lysates |
Tissue cells from healthy adult female cynomolgus monkeys (n=2) (Table 1) were isolated and preserved by using a slightly modified protocol as described earlier.32 Briefly, the tissues were cut into 4–5 cm pieces and placed in 1X ice-cold PBS (phosphate-buffered saline) pH 7.2. The tissues were then transferred to a sterile 6-well culture plate and washed with 1X PBS pH 7.2 several times, followed by 70% ethanol for 3 min. After a final wash with 1X PBS pH 7.2, a 3 mL R10 media (RPMI 1640 medium [Sigma-Aldrich, Oakville, Canada] containing 10% fetal bovine serum [FBS], 250 µg/mL pen-strep, and 50 µg/mL gentamicin) was added to the tissues in a new culture plate for 2 min. Washed tissues were further cut into 1 cm pieces and transferred to a new sterile 6-well plate containing 3 mL of working digestion buffer (2 mg/mL collagenase II [Sigma-Aldrich, Oakville, Canada] in R10 media). Using a syringe with a needle, a small volume of working digestion buffer was injected into the tissues and incubated for 3 min at room temperature. Afterward, the tissues were finely cut by scissors to disrupt all small tissue pieces and then incubated for 1 hour at 37° C. After incubation, a 60 µL of EDTA (100 nM) was added to the tissues, and the media containing the small pieces of tissues were slowly taken out until no visible tissue fragment remained. Cells were transferred to a 15 mL centrifuge tube containing 10 mL R10 media and centrifuged at 1800 rpm (no break) for 8 min. The supernatants were carefully removed and 5 mL of freshly made Percoll working solution (31.5 mL of 100% Percoll pH 8.5–9.5 [Sigma-Aldrich, Oakville, Canada] plus 3.5 mL of 10X PBS pH 7.2 and 65 mL of 1X PBS pH 7.2) was added to the cells, vortexed, centrifuged at 2000 rpm for 12 min, and removed the supernatants. After discarding supernatants, 5 mL of ACK lysis buffer (Ammonium-Chloride-Potassium; prepared by adding 8.29 g of 0.15 M NH4Cl, 1g of 10 mM KHCO3, and 41.6 mg of 0.1 mM Tetrasodium salt EDTA to 100 mL distilled water, adjusted pH at 7.2) was added to eliminate the contamination of red blood cells, vortexed, and incubated for 3 min. ACK lysis reaction was stopped by adding 7 mL of R10 media. The tube was centrifuged at 1000 rpm for 10 min (no break) and the supernatants were removed. Finally, 1 mL of freezing media (92.5% FBS and 7.5% DMSO) was added to the cells and preserved in liquid nitrogen.
Peripheral blood mononuclear cells (PBMCs) from healthy women (n=4) (Table 2) were isolated by using a slightly modified method of Ficoll-Hypaque density gradient centrifugation and preserved as previously described.33,34 In brief, heparinized blood was placed into a 50 mL conical centrifuge tube, and an equal volume of room-temperature 1X PBS pH 7.2 (1:1) was added using a sterile serological pipette and mixed well. A 15 mL blood/PBS mixture was then slowly layered over 10 mL Ficoll (Lymphoprep, MJS BioLynx Inc. Ontario, Canada) in a separate tube and the tube was centrifuged at 240 xg for 25 min at 20° C (no brake). Using a sterile pipette, the upper layer containing the plasma and most of the platelets was removed. With a separate sterile pipette, the mononuclear cell layer (white cell layer) was transferred to another centrifuge tube. The cells were then washed once by adding three times the volume of room-temperature PBS pH 7.2 and centrifuging at 240 xg for 10 min. Washing of cells was done once with R10 media (RPMI 1640 media containing 10% FBS and 1% pen-strep). The supernatants were discarded and the cells were resuspended in 10 mL R10 media, counted (3x106 cells/mL) using a hemocytometer and Trypan blue exclusion dye (Thermo Fisher Scientific, Toronto, Canada), and preserved using freezing media (90% FBS and 10% DMSO).
 |
Table 2 Adult Human and Monkey Normal PBMCs Lysates |
Since Ficoll-Hypaque compounds somehow interact with the macaque erythrocytes and lymphocytes causing severe erythrocyte contamination to the lymphocyte layer in the gradient, the PBMCs from healthy adult female cynomolgus monkeys (n=4), and healthy adult male and female rhesus monkeys (n=4) (Table 2) were isolated by a Percoll density gradient centrifugation method as described elsewhere.35 Briefly, the vacutainer tube containing the blood sample was centrifuged at 1200 xg for 10 min (no brake). Plasma was removed from the top of the tube as much as possible without disturbing the buffy coat using a transfer pipette. Afterward, the buffy coat (mostly white blood cells) was removed from the blood and transferred to the dilution tube containing 7.5 mL R0 media (RPMI 1640 medium without added supplements). The cell mixture (8 mL maximum) was then overlaid onto the 4 mL 60% Percoll solution (6 parts of 100X Percoll plus 4 parts of 1X PBS pH 7.2) in a 15 mL Falcon tube and centrifuged at 500 xg for 30 min (no brake). Using a transfer pipette, the cell interface (~2 mL) was removed and placed in a clean and sterile 15 mL centrifuge tube. The cell interface tube was filled with R10 media to a maximum of 14 mL and centrifuged at 350 xg for 10 min with full brake. The supernatants were discarded and the cell pellet was resuspended into 2 mL R10 media. The cells were counted, aliquoted (3x106 cells/mL), and preserved by using freezing media (90% FBS and 10% DMSO).
The cryopreserved cynomolgus monkeys tissue cells, human and monkey PBMCs, and human cell lines were thawed on ice and centrifuged at 2000 xg for 6 min at 4° C. The supernatants were discarded and 1 mL RPMI 1640 medium (Sigma-Aldrich, Oakville, Canada) without FBS (fetal bovine serum) was added. The cells were gently mixed and centrifuged at 2000×g for 6 min at 4° C. The washing of cells with RPMI 1640 medium was repeated once and then washed once with PBS pH 7.2. After final washing, RIPA lysis buffer (50 µL/106 cells; Thermo Fisher Scientific, Toronto, Canada) containing protease inhibitor (10 µL per mL of buffer) was added and vigorously vortexed. The lysates were then transferred to QIA Shredder (Qiagen, Toronto, Canada) and centrifuged at 17500×g for 2 min at 4° C. The flow-through was collected and the concentration of protein in lysates was determined using BCA (bicinchoninic acid) protein assay (ThermoFisher Scientific, Toronto, Canada).
Determination of the Protein Concentration in the Lysates of Tissues, PBMCs, and Cell Lines
The concentration of protein in tissues, PBMCs, and cell line lysates was determined by Pierce BCA protein assay as described elsewhere.7 In brief, a 10 µL of diluted bovine serum albumin (BSA) standard (0.25 mg/mL [standard 1] to 2.0 mg/mL [standard 8]) and unknown lysates were separately dispensed into each well of the microtitre plate (flat-bottom) followed by the addition of 200 µL of BCA working solution. The plate was incubated at 37° C for 30 min and the protein concentration was measured by using a plate reader (SpectraMax M2e; Molecular Devices, California, USA).
Western Blot Analysis
Western blot analysis of tissues, PBMCs, and cell line lysates was performed as described previously.7,36 In brief, NuPAGE precast gel (4–12% Bis-Tris 1.0 mm x 15 well) (Thermo Fisher Scientific, Toronto, Canada) was used to run each lysate separately. The loading concentration of each lysate was optimized and each lane of the gel was loaded with 2.5 µg (human tissue lysates), 5.0 µg (monkey tissue cell lysates), 14.0 µg (human/monkey PBMCs lysates), and 7.0 µg (human cell line lysates) protein, respectively. The gel was run for 45 min with 200 V. TILRR protein was probed with in-house developed mouse anti-TILRR mAb (F218G4; 5 µg/mL) and rabbit anti-β-actin mAb (1:1000; LI-COR, Nebraska, USA). In parallel, the membrane was also probed with mouse IgG1 isotype control antibody (1µg/mL; Abcam, Ontario, Canada) and rabbit anti-β-actin mAb (1:1000; LI-COR, Nebraska, USA). Mouse anti-TILRR F218G4 mAb was used to detect TILRR protein, whereas rabbit anti-β-actin mAb was used as a housekeeping control protein (loading control). IRDye(R) 800CW goat anti-mouse IgG (H+L) and IRDye(R) 680RD goat anti-rabbit IgG (H+L) secondary antibodies were used (1:10,000; LI-COR, Nebraska, USA) to detect the TILRR protein and β-actin, respectively. The image was taken by using an Odyssey CLx imager (LI-COR, Nebraska, USA). The signal intensity of TILRR protein was measured by Image Studio Lite version 5.2.
Coomassie Blue Staining
Coomassie blue gel staining was performed to quickly visualize the expected ~70 kDa TILRR protein band after iBlot transfer by BioSafeTM Coomassie stain (Bio-Rad, Mississauga, Canada) as described elsewhere.7 Briefly, after iBlot transfer, the gel was washed with 200 mL ddH2O (double-distilled water) followed by a further 3X with ddH2O for 5 min. After the complete removal of ddH2O, the gel was incubated with 20 mL BioSafeTM Coomassie stain for 1 h, rinsed for 30 min in 200 mL ddH2O, and examined in Odyssey CLx imager.
Statistical Analysis
All data were analyzed by GraphPad Prism version 9.0.0 (GraphPad Software Inc., California, USA). Image Studio Lite version 5.2 (https://www.licor.com/bio/image-studio-lite/) was used to process all blot and gel images. TILRR protein intensity is shown as relative fluorescence units (rfu). In some figures, the “rfu” is shown as median with an interquartile range (IQR). A t-test with 95% CI (confidence intervals) was used for statistical comparisons, and p<0.05 was reported as statistically significant.
Results
TILRR Protein is Highly Expressed in a Variety of Human and Monkey Tissues
To explore whether TILRR protein is expressed in normal adult human and animal tissues, we examined its expression in human tissue lysates and cynomolgus monkey tissue cell lysates. The human tissue lysates include the tissues from the respiratory tract (trachea and lungs), digestive tract (stomach, small intestine [ileum], cecum, colon, and rectum), and reproductive tract (female: vagina, cervix, and uterus; male: penis). Western blot analysis demonstrated that TILRR protein is expressed in all human tissue lysates examined (Figure 1). The quantity of TILRR protein (relative fluorescence intensity [rfu]) was higher in vaginal tissues (1648.07 rfu), followed by rectal (1629.17 rfu), cervical (1549.97 rfu), and uterine tissues (1025.03 rfu) (Figure 1, green bars; and Figure S1A). The quantity of TILRR protein was lower in small intestinal tissue (200.25 rfu), whereas the TILRR protein expressions in the stomach (832.99 rfu), large intestines (cecum [810.49 rfu] and colon [874.49 rfu]), trachea (816.81 rfu), lungs (856.71 rfu), and penile tissues (820.12 rfu) were higher than that of the small intestine (ileum), but lower than rectal and cervicovaginal tissues (Figure 1, green bars; and Figure S1A). Coomassie blue staining of the gel after iBlot transfer showed that a portion of proteins from all tissue lysates remained in the gel, which prevents more accurate quantification of TILRR protein in the tissue lysates following Western blot analysis. However, the highest level of un-transferred protein in cervical tissue lysate (Figure 1, red bars; and Figure S1B) is consistent with higher expression of TILRR protein in cervicovaginal tissues.
 |
Figure 1 TILRR protein expression in normal human tissues. Tissue lysates (2.5 µg/lane protein) were examined by Western blot (green color, left) and Coomassie blue staining (red color, right). TILRR protein was probed by primary mouse anti-TILRR F218G4 mAb followed by secondary goat anti-mouse IRDye 800CW antibody (1:10,000; LI-COR). TILRR protein bands (70 kDa, green color, cropped) are shown on the top of the bar graph (see Figure S1A for un-cropped blot image). Untransferred proteins in the gel after iBlot transfer were stained with Coomassie blue staining (G-250, Bio-Rad) and the protein bands (~70 kDa, red color, cropped) are shown on the top of the bar graph (see Figure S1B for un-cropped blot image). The bar graph shows the TILRR protein intensity (R.F.U) for each tissue lysate (n=1) where X-axis demonstrates the normal human tissue lysates and Y-axis indicates the signal intensity (R.F.U). kDa, kiloDalton; RFU, relative fluorescence units; S. intestine, small intestine (ileum). |
We also examined TILRR protein expression in normal lungs, colon, and rectal tissue cell lysates of adult female cynomolgus monkeys (Macaca fascicularis). Similar to the human tissue lysates, Western blot analysis detected TILRR protein in all the tissue cell lysates of the cynomolgus monkeys (Figure 2A). The highest TILRR protein level was found in colon tissues (median with interquartile range [IQR]: 5815.46 [5728.56–5902.36] rfu) followed by lungs (4031.57 [3840.04–4223.09] rfu) and rectal tissues (2100.38 [1690.41–2510.35] rfu) (Figure 2B, green bars; and Figure S1C). Coomassie blue staining also showed that a portion of protein remained in the gel after iBlot transfer (Figure 2B, red bars; and Figure S1D). Thus, TILRR protein is expressed in all normal human and cynomolgus monkey tissues examined, and its quantities are variable.
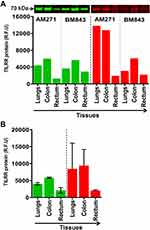 |
Figure 2 TILRR protein expression in normal Cynomolgus monkey tissues. (A) Tissue lysates (5.0 µg/lane proteins) were examined by Western blot (green color, left) and Coomassie blue staining (red color, right). TILRR protein was probed by primary mouse anti-TILRR F218G4 mAb followed by secondary goat anti-mouse IRDye 800CW antibody (1:10,000; LI-COR). TILRR protein bands (70 kDa, green color, cropped) are shown on the top of the bar graph (Figure S1C shows the un-cropped blot image). Untransferred proteins in the gel after iBlot transfer were stained with Coomassie blue staining (G-250, Bio-Rad) and the protein bands (~70 kDa, red color, cropped) are shown on the top of the bar graph (Figure S1D shows the un-cropped gel). The bar graph indicates the TILRR protein intensity (R.F.U) for each tissue lysate (n=1). (B) TILRR protein intensity (R.F.U) (median with interquartile range [IQR]) of each tissue cell lysate observed in two cynomolgus monkeys (n=2). The X-axis represents the normal Cynomolgus monkey tissue cell lysates and Y-axis shows the signal intensity (R.F.U). AM271 and BM843 are the monkeys’ identification numbers. kDa, kiloDalton; RFU, relative fluorescence units; n= number of subjects. |
TILRR Protein is Expressed in Human and Monkey PBMCs
Next, we examined the PBMC lysates of healthy adult humans and adult male and female monkeys (rhesus and cynomolgus). In the case of humans, the archived PBMCs from four women of PSWC were used. Western blot analysis showed that human PBMCs expressed TILRR protein (Figure 3A). The TILRR protein intensity (median [IQR]) was 2697.16 [2391.90–3187.02] rfu (Figure 3B, green bars; and Figure S2A). Similarly, a large proportion of protein from the human PBMC lysates remained in the gel following iBlot transfer (Figure 3B, red colors; and Figure S2B) that prevented the precise quantification of TILRR protein in PBMC lysates using Western blot analysis.
 |
Figure 3 TILRR protein expression in normal human and macaque PBMCs. (A) PBMC lysates (14.0 µg/lane proteins) of humans (n=4), rhesus monkeys (n=4), and cynomolgus monkeys (n=4) were analyzed by Western blot (green color, left) and Coomassie blue staining (red color, right). TILRR protein was probed by primary mouse anti-TILRR F218G4 mAb followed by secondary goat anti-mouse IRDye 800CW antibody (1:10,000; LI-COR). TILRR protein bands (70 kDa, green color, cropped) are shown on the top of the bar graph (see Figure S2A for un-cropped blot image). Untransferred proteins in the gel after iBlot transfer were stained with Coomassie blue staining (G-250, Bio-Rad), and the protein bands (~70 kDa, red color, cropped) are shown on the top of the bar graph (see Figure S2B for un-cropped gel). The bar graph shows the TILRR protein intensity (R.F.U). (B) TILRR protein intensity (R.F.U) (median with IQR) of PBMC lysates observed in four study subjects (n=4) including humans, rhesus monkeys, and cynomolgus monkeys. (C) TILRR protein expression between humans and monkeys. Data are shown as median with IQR in figures B and C. A t-test with 95% CI was used for statistical comparisons and all p<0.05 were reported as statistically significant and presented with asterisks. The X-axis of figure A indicates the patients’ and monkeys’ identification numbers and Y-axis shows the signal intensity (R.F.U). kDa, kiloDalton; RFU, relative fluorescence units; n= number of subjects; ID#, identification number, Rhesus M, rhesus monkey; Cyno M, cynomolgus monkey. |
Western blot analysis also detected TILRR protein in PBMCs of both adult male and female rhesus monkeys (Macaca mulatta) and adult female cynomolgus monkeys (Macaca fascicularis) (Figure 3A). The TILRR protein intensity (median with IQR) was 2038.44 (1726.37–2402.54) rfu and 2192.83 (1906.14–2760.48) rfu in rhesus and cynomolgus monkeys, respectively (Figure 3B, green bars; and Figure S2A). There was no significant difference in TILRR protein expression between PBMCs of humans and monkeys (p= 0.2858) (Figure 3C).
In addition to the TILRR protein, we found that the variants of FREM1 (~250 kDa and ~150 kDa) and variants of β-actin (<42 kDa) expressed in monkey PBMCs are different from the ones expressed in human PBMCs based on the size and the pattern of the Western blot (Figure S2A).
Few Examined Human Cell Lines Expressed TILRR Protein
We further examined the TILRR protein expression in different human cell lines, including HeLa, VK2, Ect1, End1, THP-1, MOLT-4, SupT1, and Jurkat. These cell lines could be used as in vitro model system to study the effect of TILRR protein. Western blot analysis demonstrated that TILRR protein was not detected in all cell lines examined, except for the Jurkat cell line (Figure 4). The intensity of TILRR protein in the Jurkat cell line was 646.00 (rfu) (Figure 4, green bars; and Figure S3A). Similarly, the iBlot transfer system cannot transfer all proteins in the gel (Figure 4, red bars; and Figure S3B). Thus, most of the human cell lines examined expressed little TILRR protein, and these cell lines can be used as in vitro model system to overexpress TILRR protein to examine the effect of TILRR protein expression.
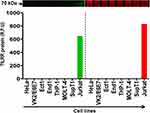 |
Figure 4 TILRR protein expression in human cell lines. Cell line lysates (7 µg/lane protein) were examined by Western blot (green color, left) and Coomassie blue (red color, right). TILRR protein was probed by primary mouse anti-TILRR F218G4 mAb followed by secondary goat anti-mouse IRDye 800CW antibody (1:10,000; LI-COR). TILRR protein bands (70 kDa, green color, cropped) are shown on the top of the bar graph (Figure S3A shows the un-cropped blot image). Untransferred proteins in the gel after iBlot transfer were stained with Coomassie blue staining (G-250, Bio-Rad) and the protein bands (~70 kDa, red color, cropped) are shown on the top of the bar graph (Figure S3B shows the un-cropped gel). The bar graph shows the TILRR protein intensity (R.F.U) for each tissue lysate (n=1) where X-axis demonstrates human cell lines and Y-axis represents signal intensity (R.F.U). kDa, kiloDalton; RFU, relative fluorescence units. |
Anti-TILRR Monoclonal Antibodies (mAb) are Specific for TILRR Protein
We previously showed that the in-house developed anti-TILRR monoclonal antibodies are specific for the TILRR protein.7,37 The current study with Western blot analysis of human tissue lysates further confirmed the specificity of the anti-TILRR mAbs. Anti-TILRR mAb (F218G4) targets major epitopes on the Calx-β domain of TILRR protein.37 A mouse IgG1 mAb isotype control (Abcam, Canada) was used as a control for Western blot analysis of human vaginal and cervical tissue lysates. The Western blot analysis demonstrated that the anti-TILRR mAb detected TILRR protein (70 kDa) in the vagina (869.79 rfu) and cervix (834.13 rfu) tissue lysates, whereas no signal of the 70 kDa protein was observed with mouse IgG1 isotype control (Figure 5, green bars; Figure S4A). Coomassie blue staining showed un-transferred proteins in the gel following iBlot transfer to the nitrocellulose membrane (Figure 5, red bars; Figure S4B).
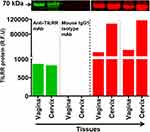 |
Figure 5 Anti-TILRR mAb is specific for TILRR protein. Tissues lysates (2.5 µg/lane protein) (n=2) were examined by Western blot (green color, left) and Coomassie blue staining (red color, right). TILRR protein was probed by either primary mouse anti-TILRR F218G4 mAb or primary mouse IgG1 mAb (isotype control) (Abcam, Canada) followed by secondary goat anti-mouse IRDye 800CW antibody (1:10,000; LI-COR). TILRR protein bands (70 kDa, green color, cropped) are shown on the top of the bar graph (see Figure S4A for un-cropped blot image). Untransferred proteins in the gel after iBlot transfer were stained with Coomassie blue staining (G-250, Bio-Rad), and the protein bands (~70 kDa, red color, cropped) are shown on the top of the bar graph (see Figure S4B for un-cropped gel). The bar graph shows the TILRR protein intensity (R.F.U) for each tissue lysate where X-axis represents human tissue lysates and Y-axis indicates the signal intensity (R.F.U). kDa, kiloDalton; RFU, relative fluorescence units. |
Discussion
TILRR regulates the MYD88-dependent amplification of NF-κB signaling and aberrant inflammatory responses.1,4 It is associated with inflammatory vascular disease development4 and found to be a biomarker in breast cancer.38 We previously showed that TILRR modulated many inflammation-responsive genes,5 promoted the secretion of inflammatory mediators,5 and the migration of immune cells.6 Recently, we showed that TILRR protein also circulates in the blood of healthy women, and the level of plasma TILRR protein is variable among different women.7 Also, the level of plasma TILRR protein is positively correlated with different plasma pro-inflammatory cytokines/chemokines8 and high plasma TILRR protein is a risk factor in HIV seroconversion.8 The level of TILRR protein expression may influence the inflammation status of tissues and cells. Studies have shown that TILRR mRNA is variably expressed in human and animal immune and epithelial cell lines.1,2,4 Although much is known about TILRR mRNA expression in cell lines and PBMCs, there is no information on protein level expression of TILRR in human and animal tissues and PBMCs. The knowledge of TILRR protein expression in tissues, PBMCs, and model cell lines is important to further study its role in some infectious and inflammatory diseases. The data reported in this study provided the much-needed information on TILRR protein expression in a number of human tissues and PBMCs that are relevant for some infectious and inflammatory diseases, such as SARS-CoV-2,9,10 HIV,15,16 and IBD.18 The cynomolgus and rhesus monkeys are often used as models for human diseases, including SARS-CoV-2, influenza, and HIV/SIV,39–42 and evaluation of vaccine and therapeutic studies.43–46 The expression of TILRR protein in some tissues and PBMCs of these two monkeys’ species could help future studies on the role of TILRR protein in these animal models.
Our study showed that TILRR protein is expressed in different healthy human and monkey tissues. The TILRR protein expression is high in human rectal and cervicovaginal tissues, and relatively high in the trachea, lungs, stomach, small intestine (ileum), cecum, colon, uterus, and penile tissues (Figure 1). It may suggest the importance of studying the TILRR protein in modulating inflammation and cytokine/chemokine production and pathogenesis of these tissues. Tissues with a low-level TILRR protein under normal physiological conditions may be less likely inflammation responsive. While tissues expressing a high-level TILRR protein may be more inflammation responsive. However, these need to be further studied. Studies showed that abnormal cytokine/chemokine production, rapid influx of immune cells, and inflammation in tissues and epithelial cells increase the rate of inflammation-induced sexually transmitted infections (STIs including HIV),14–16 IBD,18,19 respiratory viral infections, and common diseases (SARS-CoV-2, MERS-CoV, H5N1 influenza A virus, RSV, pneumonia, and COPD),9–13,17 and progression of cancer.20 Since TILRR promotes secretion of inflammatory cytokines/chemokines,5 immune cell migration,6 and inflammation,1,4 it could be involved in inflammation-related disease pathogenesis and cancer development. These possibilities need to be examined in future studies.
Like humans, the expression of TILRR protein was observed in the cynomolgus monkeys’ lungs, colon, and rectal tissues (Figure 2B). As cynomolgus monkeys (Macaca fascicularis) are a frequently used animal model for studying human diseases,39–42,47 the expression of TILRR protein in tissues of cynomolgus monkeys would help to conduct future studies on TILRR protein using cynomolgus monkeys as a model.
Our study also showed that TILRR protein is expressed in human and monkey PBMCs. PBMCs of humans, rhesus monkeys (Macaca mulatta), and cynomolgus monkeys (Macaca fascicularis) showed a similar level of expression of TILRR protein. The expression of TILRR protein in PBMCs is consistent with an earlier study showing TILRR mRNA expression in human PBMCs.1 The major immune cell subsets of PBMCs include monocytes, T cells [CD4+T and CD8+T-cells], B cells, and NK cells.48 Studies showed that infection or injury to the tissues or epithelial/endothelial cells attracts peripheral blood immune cells to the site of injury through secretion of inflammatory chemotactic mediators from damaged cells,49–52 resulting in a cycle of inflammation and pathogenesis. It would be interesting to know whether all immune cell subsets in PBMCs express the TILRR protein and this needs to be investigated in future studies.
Our study showed that most of the model cell lines (HeLa, VK2, Ect1, End1, THP-1, MOLT-4, SupT1) we examined do not express the TILRR protein, with the exception of Jurkat cells. The result is consistent with our previous study showing no TILRR variant RNA was detected by sequencing HeLa cell RNA using NGS (next-generation sequencing) technology.5 Thus, these cell lines can be used as in vitro model system to overexpress TILRR protein to examine the effect of TILRR protein expression.
Conclusions
Our study for the first time showed that TILRR protein is expressed in many human tissues and some monkey tissues, and PBMCs. The level of expression of TILRR protein in the tissues and cells may reflect the role of TILRR protein in the inflammation response of these tissues and cells. The information will help the further study of the potential role of TILRR in infectious and inflammatory diseases.
Abbreviations
ACK, ammonium-chloride-potassium; BCA, bicinchoninic acid; BSA, bovine serum albumin; CV, coefficient of variation; COPD, chronic obstructive pulmonary disease; EDTA, ethylenediaminetetraacetic acid; FBS, fetal bovine serum; HIV, human immunodeficiency virus; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; IBD, inflammatory bowel disease; IgG, immunoglobulin G; IL-1R1, interleukin-1 receptor type 1; IQR, interquartile range; KCl, potassium chloride; kDa, kilodalton; mAb, monoclonal antibody; MERS-CoV, middle eastern respiratory syndrome coronavirus; MgCl2, magnesium chloride; NF-κB, nuclear factor-kappa-light-chain-enhancer of activated B cells; NP-40, nonidet P-40; PBMCs, peripheral blood mononuclear cells; PBS, phosphatebuffered saline; PSWC, Pumwani sex worker cohort; RFU, relative fluorescence units; RIPA, radioimmunoprecipitation assay; RSV, respiratory syncytial virus; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; SIV, simian immunodeficiency virus; STIs, sexually transmitted infections; TILRR, toll-like interleukin 1 receptor regulator; WB, Western blot.
Ethics Approval and Informed Consent
The Helsinki Declaration on ethical principles for medical research involving human participants being served as a guide for this study. The University of Manitoba and the University of Nairobi/Kenyatta National Hospital Ethics Committee have approved the ethics to perform the study using human PBMCs. Written informed consent was given to all enrolled subjects.
The cynomolgus monkey (Macaca fascicularis) use document was approved by the Canadian Sciences Centre for Human and Animal Health Animal Care Committee (protocol number: H-12-014R2), and the rhesus monkey (Macaca mulatta) use protocol (AUP562) was approved by IACUC (institutional animal care and use committee) of ABL.
Acknowledgments
We thank Dr. Guillaume Poliquin, The Vice-President of NML (National Microbiology Laboratory), Winnipeg, Manitoba, Canada, for his continuous support to perform this study. We gratefully thank and acknowledge NML, Canada for its financial support.
Author Contributions
All authors made a significant contribution to the work reported, particularly in study design, acquisition of data, analysis, and interpretation; took part in critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The study was funded by an operating grant from the Canadian Institutes of Health Research (CIHR), operating grant- PA: CHVI Vaccine Discovery and Social Research (http://www.cihr-irsc.gc.ca/e/193.html), as well as the National Microbiology Laboratory of Canada.
Disclosure
The authors declare no competing interest.
References
1. Zhang X, Shephard F, Kim HB, et al. TILRR, a Novel IL-1RI Co-receptor, Potentiates MyD88 Recruitment to Control Ras-dependent Amplification of NF-κB. J Biol Chem. 2010;285(10):7222–7232. doi:10.1074/jbc.M109.073429
2. Kashem MA, Li H, Liu LR, et al. The Potential Role of FREM1 and Its Isoform TILRR in HIV-1 Acquisition through Mediating Inflammation. Int J Mol Sci. 2021;22(15):54.
3. Mac Gabhann F. TILRR Steers Interleukin-1 Signaling: co-Receptor Provides Context and a Therapeutic Target. JACC Basic Transl Sci. 2017;2(4):415–417.
4. Smith SA, Samokhin AO, Alfadi M, et al. The IL-1RI Co-Receptor TILRR (FREM1 Isoform 2) Controls Aberrant Inflammatory Responses and Development of Vascular Disease. JACC Basic Transl Sci. 2017;2(4):398–414.
5. Kashem MA, Li H, Toledo NP, et al. Toll-like Interleukin 1 Receptor Regulator Is an Important Modulator of Inflammation Responsive Genes. Front Immunol. 2019;10(272):1–16.
6. Kashem MA, Ren X, Li H, et al. TILRR Promotes Migration of Immune Cells Through Induction of Soluble Inflammatory Mediators. Front Cell Dev Biol. 2020;8(563):1–13.
7. Kashem MA, Yuan X-Y, Kimani J, Plummer F, Luo M. TILRR (Toll-like Interleukin-1 Receptor Regulator), an Important Modulator of Inflammatory Responsive Genes, is Circulating in the Blood. J Inflamm Res. 2021;2021(14):4927–4943.
8. Kashem MA, Lischynski J, Stojak B, et al. High level of plasma TILRR protein is associated with faster HIV seroconversion. EBioMedicine. 2022;78:103955.
9. Del Valle DM, Kim-Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26(10):1636–1643.
10. Garcia LF. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front Immunol. 2020;11:1441.
11. Min CK, Cheon S, Ha NY, et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep. 2016;6:25359.
12. de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12(10):1203–1207.
13. Tan KS, Lim RL, Liu J, et al. Respiratory Viral Infections in Exacerbation of Chronic Airway Inflammatory Diseases: novel Mechanisms and Insights From the Upper Airway Epithelium. Front Cell Dev Biol. 2020;8:99.
14. Arnold KB, Burgener A, Birse K, et al. Increased levels of inflammatory cytokines in the female reproductive tract are associated with altered expression of proteases, mucosal barrier proteins, and an influx of HIV-susceptible target cells. Mucosal Immunol. 2016;9(1):194–205.
15. Passmore JA, Jaspan HB, Masson L. Genital inflammation, immune activation and risk of sexual HIV acquisition. Curr Opin HIV AIDS. 2016;11(2):156–162.
16. Masson L, Passmore JA, Liebenberg LJ, et al. Genital inflammation and the risk of HIV acquisition in women. Clin Infect Dis. 2015;61(2):260–269.
17. Moldoveanu B, Otmishi P, Jani P, et al. Inflammatory mechanisms in the lung. J Inflamm Res. 2009;2:1–11.
18. Matricon J, Barnich N, Ardid D. Immunopathogenesis of inflammatory bowel disease. Self/Nonself. 2010;1(4):299–309.
19. Ribaldone DG, Pellicano R, Actis GC. Inflammation in gastrointestinal disorders: prevalent socioeconomic factors. Clin Exp Gastroenterol. 2019;12:321–329.
20. Di Caro G, Carvello M, Pesce S, et al. Circulating Inflammatory Mediators as Potential Prognostic Markers of Human Colorectal Cancer. PLoS One. 2016;11(2):e0148186.
21. Adler KB, Fischer BM, Wright DT, Cohn LA, Becker S. Interactions between respiratory epithelial cells and cytokines: relationships to lung inflammation. Ann N Y Acad Sci. 1994;725:128–145.
22. Onyiah JC, Colgan SP. Cytokine responses and epithelial function in the intestinal mucosa. Cell Mol Life Sci. 2016;73(22):4203–4212.
23. Roan F, Obata-Ninomiya K, Ziegler SF. Epithelial cell-derived cytokines: more than just signaling the alarm. J Clin Invest. 2019;129(4):1441–1451.
24. Stadnyk AW. Cytokine production by epithelial cells. FASEB J. 1994;8(13):1041–1047.
25. Fahey JV, Schaefer TM, Channon JY, Wira CR. Secretion of cytokines and chemokines by polarized human epithelial cells from the female reproductive tract. Hum Reprod. 2005;20(6):1439–1446.
26. Hernandez-Rodriguez J, Segarra M, Vilardell C, et al. Tissue production of pro-inflammatory cytokines (IL-1beta, TNFalpha and IL-6) correlates with the intensity of the systemic inflammatory response and with corticosteroid requirements in giant-cell arteritis. Rheumatology. 2004;43(3):294–301.
27. Mahapatro M, Erkert L, Becker C. Cytokine-Mediated Crosstalk between Immune Cells and Epithelial Cells in the Gut. Cells. 2021;10(1):w356.
28. Plummer FA, Simonsen JN, Cameron DW, et al. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J Infect Dis. 1991;163(2):233–239.
29. Li H, Omange RW, Czarnecki C, et al. Mauritian cynomolgus macaques with M3M4 MHC genotype control SIVmac251 infection. J Med Primatol. 2017;46(4):137–143.
30. Li H, Nykoluk M, Li L, et al. Natural and cross-inducible anti-SIV antibodies in Mauritian cynomolgus macaques. PLoS One. 2017;12(10):e0186079.
31. Li H, Li L, Liu LR, et al. Hypothetical endogenous SIV-like antigens in Mauritian cynomolgus macaques. Bioinformation. 2018;14(2):48–52.
32. Ravindran A, Ronnberg E, Dahlin JS, et al. An Optimized Protocol for the Isolation and Functional Analysis of Human Lung Mast Cells. Front Immunol. 2018;9:2193.
33. Fuss IJ, Kanof ME, Smith PD, Zola H. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr Protocols Immunol. 2009;1:523.
34. Riedhammer C, Halbritter D, Weissert R. Peripheral Blood Mononuclear Cells: isolation, Freezing, Thawing, and Culture. Methods Mol Biol. 2016;1304:53–61.
35. Budzko D, Madden DL, London WT, Sever JL. Improved separation of rhesus monkey lymphocytes with Percoll. Lab Invest. 1985;53(5):586–588.
36. Li H, Hai Y, Lim SY, et al. Mucosal antibody responses to vaccines targeting SIV protease cleavage sites or full-length Gag and Env proteins in Mauritian cynomolgus macaques. PLoS One. 2018;13(8):e0202997.
37. Yuan XY, Liu LR, Krawchenko A, et al. Development of monoclonal antibodies to interrogate functional domains and isoforms of FREM1 protein. Monoclon Antib Immunodiagn Immunother. 2014;33(2):129–140.
38. Xu XY, Guo WJ, Pan SH, et al. TILRR (FREM1 isoform 2) is a prognostic biomarker correlated with immune infiltration in breast cancer. Aging. 2020;12(19):19335–19351.
39. Kuiken T, Fouchier RA, Schutten M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270.
40. Gardner MB, Luciw PA. Macaque models of human infectious disease. ILAR j. 2008;49(2):220–255.
41. Putkonen P, Thorstensson R, Albert J, et al. Infection of cynomolgus monkeys with HIV-2 protects against pathogenic consequences of a subsequent simian immunodeficiency virus infection. AIDS. 1990;4(8):783–789.
42. Urano E, Okamura T, Ono C, et al. COVID-19 cynomolgus macaque model reflecting human COVID-19 pathological conditions. Proc Natl Acad Sci U S A. 2021;118(43):865.
43. Antony JM, MacDonald KS. A critical analysis of the cynomolgus macaque, Macaca fascicularis, as a model to test HIV-1/SIV vaccine efficacy. Vaccine. 2015;33(27):3073–3083.
44. Baroncelli S, Negri DR, Michelini Z, Cara A. Macaca mulatta, fascicularis and nemestrina in AIDS vaccine development. Expert Rev Vaccines. 2008;7(9):1419–1434.
45. Shedlock DJ, Silvestri G, Weiner DB. Monkeying around with HIV vaccines: using rhesus macaques to define ‘gatekeepers’ for clinical trials. Nat Rev Immunol. 2009;9(10):717–728.
46. Almond N, Berry N, Stebbings R, et al. Vaccination of Macaques with DNA Followed by Adenoviral Vectors Encoding Simian Immunodeficiency Virus (SIV) Gag Alone Delays Infection by Repeated Mucosal Challenge with SIV. J Virol. 2019;93(21):53662.
47. Saito A, Nomaguchi M, Iijima S, et al. Improved capacity of a monkey-tropic HIV-1 derivative to replicate in cynomolgus monkeys with minimal modifications. Microbes and Infection. 2011;13(1):58–64.
48. Corkum CP, Ings DP, Burgess C, Karwowska S, Kroll W, Michalak TI. Immune cell subsets and their gene expression profiles from human PBMC isolated by Vacutainer Cell Preparation Tube (CPT) and standard density gradient. BMC Immunol. 2015;16:48.
49. Beringer A, Molle J, Bartosch B, Miossec P. Two phase kinetics of the inflammatory response from hepatocyte-peripheral blood mononuclear cell interactions. Sci Rep. 2019;9(1):8378.
50. Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: present and future therapeutic targets. Nat Immunol. 2005;6(12):1182–1190.
51. Nowarski R, Gagliani N, Huber S, Flavell RA. Innate immune cells in inflammation and cancer. Cancer Immunol Res. 2013;1(2):77–84.
52. Chen L, Deng H, Cui H, et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2018;9(6):7204–7218.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
