Back to Journals » Cancer Management and Research » Volume 13
TNRC6C-AS1 Promotes Thyroid Cancer Progression by Upregulating LPAR5 via miR-513c-5p
Authors Tong C , Wang C, Wang Y, Xiao X
Received 25 March 2021
Accepted for publication 19 June 2021
Published 6 August 2021 Volume 2021:13 Pages 6141—6155
DOI https://doi.org/10.2147/CMAR.S312621
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Chuanming Tong,1 Chuan Wang,1 Yajie Wang,1 Xiongsheng Xiao2
1Department of General Surgery, People’s Hospital of Dongxihu District, Wuhan, Hubei, 430040, People’s Republic of China; 2Department of Thyroid and Breast Surgery, Affiliated Hospital of Guangdong Medical University, Zhanjiang, Guangdong, 524000, People’s Republic of China
Correspondence: Chuanming Tong
Department of General Surgery, People’s Hospital of Dongxihu District, Intersection of Linkonggang Avenue and Sandian Avenue, Dongxihu District, Wuhan, 430040, Hubei, People’s Republic of China
Tel +86 15327362565
Email [email protected]
Background: Considering the combined role of long non-coding RNA (lncRNAs)-microRNA (miRNA)-mRNA in tumorigenesis, the purpose of this study was to investigate how TNRC6C-AS1 regulates the expression of lysophosphatidic acid receptor 5 (LPAR5) by modulating miR-513c-5p, thus influencing the progression of thyroid cancer (THCA).
Methods: qRT-PCR and Western blotting were performed to detect the expression levels of TNRC6C-AS1, miR-513c-5p, and LPAR5 in THCA tissues and cell lines. The viability, proliferation, migration, and invasion were assessed using CCK-8, BrdU, wound healing, and transwell migration assays, respectively. Dual-luciferase reporter assay, RIP assay, and RNA pull-down assay were used to evaluate the relationship between TNRC6C-AS1, miR-513c-5p, and LPAR5.
Results: TNRC6C-AS1 was highly expressed in THCA tissues, and knockout of TNRC6C-AS1 reduced the viability, proliferation, migration, and invasion of THCA cells. TNRC6C-AS1 competitively adsorbed miR-513c-5p. In addition, the biological function of TNRC6C-AS1 was blocked by knocking down the thyroid cell line TNRC6C-AS1 with miR-513c-5p inhibitor transfection. LPAR5 is the target gene for miR-513c-5p, which has the ability to eliminate the influence of miR-513c-5p on THCA cells.
Conclusion: The TNRC6C-AS1/miR-513c-5p/LPAR5 axis is a novel signaling pathway that modulates THCA progression and may be a potential target for cancer therapy.
Keywords: TNRC6C-AS1, THCA, proliferation, migration, viability
Introduction
Thyroid cancer (THCA) is a common endocrine cancer, with increased morbidity and mortality.1,2 Compared to those with other cancers, the prognosis is unsatisfactory among patients with aggressive metastatic THCA.3 Therefore, there is an urgent need to identify biomarkers for THCA diagnosis. In addition, the exploitation of new therapeutic targets for THCA may assist in the treatment of specific THCA.4
Over the last few decades, it is well known that long non-coding RNAs (lncRNAs) are key regulators of many cancers and act as tumor suppressors or carcinogens under different conditions.5 Abnormal expression of lncRNAs is usually associated with maladjustment of proliferation, apoptosis, drug resistance, metastasis, and invasion of cancer cells.6 It is worth noting that the development of THCA has been associated with the expression of a variety of lncRNAs. For example, TNRC6C-AS1 is highly expressed in THCA tissues and cells, and its low expression reduces the proliferation and metastasis of THCA cells and inhibits tumorigenesis in vivo.7 In addition, suppression of TNRC6C-AS1 leads to inhibition of proliferation, while promoting apoptosis and autophagy through STK4 methylation via the Hippo signaling pathway in THCA.8,9 These preliminary studies suggest that TNRC6C-AS1 is involved in the etiology of THCA.
Interestingly, lncRNAs interact with microRNAs (miRNAs) to exert their biological effects on cancer.10 For example, inhibition of lncRNA NEAT1 inhibits the occurrence of papillary thyroid cancer by upregulating miR-129-5p.11 TNRC6C-AS1 has been shown to mediate biological processes in THCA cells by sponging miR-129-5p.12 DLX6-AS1 promotes malignant progression of neuroblastoma cells by sponging miR-513c-5p.13 However, it is still unclear whether TNRC6C-AS1 plays a valuable role in THCA by sponging miR-513c-5p.
LncRNAs bind to the response elements of target miRNAs, limiting their activity and leading to changes in mRNA levels.14 For example, lysophosphatidic acid receptor 5 (LPAR5) is a key factor in various organs and has a variety of biological functions related to apoptosis protection, cell morphogenesis, proliferation, and differentiation.15,16 Studies have shown that LPAR5 is strongly overexpressed in THCA cells and affects the metastasis of THCA cells by modulating the expression of EMT-related proteins.17 However, few studies have clarified whether TNRC6C-AS1 can alter the pathogenesis of THCA through miR-513c-5p and LPAR5.
Therefore, this study aimed to clarify the potential mechanism of the TNRC6C-AS1/miR-513c-5p/ LPAR5 axis and the development of THCA, and to identify potential biomarkers for the diagnosis and treatment of THCA.
Method
Patients
Clinical samples were collected from patients with THCA undergoing surgical resection, with the approval of the Ethics Committee of our hospital. Participants who had not received any radiation or chemotherapy prior to tissue collection were excluded, and a written consent form was signed prior to the study. The baseline characteristics of the patients are shown in Table 1.
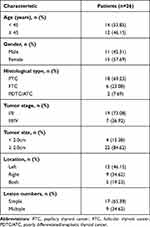 |
Table 1 Baseline Characteristics of THCA Patients |
Cell Culture
Human thyroid cell line Nthy-ori 3–1 and THCA cell lines SW579, BCPAP, and TPC-1, purchased from ATCC (USA), were cultured in DMEM medium containing 10% FBS and incubated in an incubator containing 5% CO2 at 37 °C. Passage was performed when the confluence rate reached 90%.
Cell Transfection
TNRC6C-AS1 short interfering RNA (siRNA) (si-TNRC6C-AS1; 50nM), si-TNRC6C-AS1 negative control (si-lnc-NC; 50nM), miR-513c-5p inhibitor (50nM), inhibitor negative control (inhibitor-NC), LPAR5 siRNA (si-LPARS, 2 µg), and LPAR5 siRNA negative control (si-LPARS-NC, 2 µg) were designed and synthesized by RiboBio (Guangzhou, China). Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, MA, USA) was used for cell transfection. After transfection for 48 h, SW579 and TPC-1 cell lines were used for the follow-up assay.
qRT-PCR
RNAiso Plus (Takara, Tokyo, Japan) was used for total RNA extraction. M-MLV reverse transcriptase (Takara) was used for reverse transcription. PCR was performed using SYBR Green qPCR Mix (Biorad, CA, USA) on a Stratagene MX3000P system (Agilent Technologies, CA, USA). TNRC6C-AS1 and LPAR5 were standardized using GAPDH.
miRNAs were extracted using a mirVana miRNA isolation kit (Ambion, CA, USA). Stem-loop RT-PCR was used to detect the expression of miR-513c-5p. SYBR®Premix Ex Taq™ and PrimeScript®RT reagent kit were obtained from Takara (Otsu, Japan). The U6 primer and miR-513c-5p stem-loop primers were purchased from Beijing Microread Gene Technology (Beijing, China). The PCR cycling conditions were as follows: 37 °C for 15 min, 85 °C for 5 s, for reverse transcription, 95 °C for 10 s, 95 °C for 5 s, and 61 °C for 20 s, for 40 cycles. The 2−ΔΔCt method and SNORD61 were used as normalization programs to calculate the change in miR-513c-5p expression folding. The PCR sequences of the primers used in this study are shown in Table 2.
 |
Table 2 PCR Sequences of the Primers in This Study |
Nuclear-Cytoplasmic Fractionation
The PARISTM kit (Invitrogen) was used to extract the cytoplasmic and nuclear RNA. Briefly, the cells were digested with trypsin, lysed with 500 μL ice-cold cell fractionation buffer, and incubated on ice for 10 min. The lysate was centrifuged for 3 min at 500 g to separate the nucleus and cytoplasm, and the supernatant was collected. The remaining lysate was washed with ice-cold cell fractionation buffer, and 450 μL ice-cold cell disruption buffer was added after 1 min of centrifugation at 500 g until the lysate was homogenized. The lysate and supernatant were mixed with 2 × lysate/binder and filtered by adding an equal volume of 100% ethanol. RNA from the cytoplasm and nucleus was eluted with a 95 °C elution solution. All steps were carried out at 4 °C.
CCK-8 Assay
The CCK-8 kit (Tongren, Shanghai, China) was used to evaluate the proliferation of SW579 and TPC-1 cells at 80% confluence, that were seeded in 96-well plates at 100μL/well for 24, 48, 72, and 96 h. Then, 10μL of CCK-8 solution was added to each well and incubated for 1 h. Finally, the optical density (OD) of each well was evaluated using a microplate reader (BioTek Instruments, VT, USA) at 450 nm.
BrdU Assay
SW579 and TPC-1 cells after transfection for 48 h were seeded into 96-well plates at 2×103 cells/well and incubated for 2 h with 10 μmol/L BrdU solution. The cells were then fixed at room temperature for 15 min with 4% paraformaldehyde before treatment with DNase for 15 min. The cells were then washed with PBS and incubated for 60 min at room temperature with a peroxidase-conjugated anti-BrdU antibody (Sigma-Aldrich, MO, USA). After washing with PBS, the cells were incubated for 30 min with tetramethylbenzidine, and absorbance was measured at 450 nm.
Wound Healing Assay
TPC-1 and SW579 cells were cultured to 90% confluence in a 6-well culture dish. A wound was created using the tip of a sterile 10 μL pipette and cleaned with PBS. Next, the medium containing 2% serum was added to each well, and the migration of the cells was monitored at 0 and 24 h with a microscope (Nikon, NY, USA).
Transwell Invasion Assay
A transwell chamber coated with 100 μg Matrigel (Becton-Dickinson, CA, USA) was used to evaluate cell invasion ability. TPC-1 and SW579 cells (1 × 105) were resuspended in 500 μL of serum-free medium and placed in the upper chamber of the transwell. Five hundred microliters of DMEM containing 10% FBS was added to the lower chamber. After 48 h incubation, cells in the lower chamber were immobilized with 4% DEPC for 10 min and stained with 1% crystal violet at room temperature for 30 min. An optical microscope was used to count the number of invasive cells.
Dual-Luciferase Reporter Assay
Using the human genome DNA as a template, a full-length TNRC6C-AS/LPAR5 3′ UTR was amplified by PCR and linked to the pMIR-REPORT vector (Ambion). After restriction endonuclease digestion, PCR products were introduced into the pMIR-REPORT vector at the SpeI and HindIII sites. To create the mutant 3′ UTR, we introduced point mutations on all miRNA-oligonucleotides in the selected putative vaccination sequence region. Next, 0.2 μg TNRC6C-AS1 3′UTR/LPAR5 3′UTR luciferase, 0.04 μg plasmid pRLSV40, 0.6 μg of miR-513c-5p or pGreenPuro control vector were transfected into SW579 and TPC-1 cells and incubated for 58 h. Dual‐luciferase reporter assay Kit (Promega WI, USA) was used to measure the activities of firefly and renilla luciferases. The activity of firefly luciferase was normalized to renilla activity and expressed as a relative value.
RIP Assay
This assay was conducted using the EZ Magna RIP Kit (Millipore, MA, USA). The miR-513c-5p mimic-transfected TPC-1 and SW579 cells were lysed in RIP lysis buffer. Next, the extract containing magnetic beads bound to proteinA/G (Thermo Fisher Scientific) was incubated with IgG or Ago2 antibody (Millipore) at 4 °C for 6 h. The beads were then incubated with protease K. Finally, qRT-PCR was used to analyze the purified RNA with TNRC6C-AS1 specific primer.
RNA Pull-Down
Biotin-labeled miR-513c-5p mimic (Bio-mimic) and negative control (Bio-NC) were obtained from Genechem (Shanghai, China). The above biotinylated sequence was transfected into SW579 and TPC-1 cells for 48 h. The cell lysate was treated with streptavidin agarose beads (Invitrogen) and washed 5 times. The PCR method was used to detect the co-deposited RNA.
Western Blotting
Protein extraction from cells was performed using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) containing protease inhibitors (Roche Diagnostics, Indianapolis, IN, USA), and its concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific). Protein extracts (30 µg) were separated by 10% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membrane. The membrane was sealed with 5% skimmed milk in Tris-buffered saline at room temperature for 1 h and then incubated overnight with antibodies against LPAR5 (1:200; Cat #PA5-77480; Thermo Fisher Scientific) and GAPDH (1:1000; Cat #PA1-987; Thermo Fisher Scientific) at 4 °C. The corresponding secondary antibodies were added at room temperature and incubated for 1 h. An ECL detection reagent (EMD Millipore, MA, USA) was used for imaging, and band images were analyzed using the Gene Genius gel imaging system (Syngene Europe, Cambridge, UK).
Statistical Analyses
The results are presented as mean ± standard deviation and analyzed by Prism statistical software (GraphPad Software, CA, USA). The paired t-test was used to compare two groups of data, and one-way ANOVA was used to compare multiple groups of data. Pearson’s correlation coefficient was used to analyze the association between TNRC6C-AS1, miR-513c-5p, and LPAR5 in cancer tissues. All experiments were performed in triplicate. Statistical significance was set at p < 0.05.
Result
LPAR5 and miR-513c-5p Were Found to Be Potential Participants in THCA
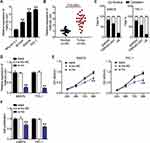
By analyzing the GSE6004 and GSE3678 data series, 77 and 325 DEGs were identified, respectively. A total of 31 common DEGs were screened out (Figure 1A) and input to the STRING database for PPI analysis. The PPI analysis result (at medium confidence of 0.4) showed that 9 of the 31 genes interacted closely with one another (Figure 1B). Among them, it was noticed that LPAR5 was identified as a potential upregulated gene in THCA during bioinformatics analyses for multiple times18–21 and was once reported to be a proto-oncogene in THCA.17 Multiple bioinformatics analyses suggest that LPAR5 could be a robust regulator in THCA progression; thus, we chose LPAR5 to be studied herein. In addition, LPAR5 was shown to be significantly overexpressed in THCA (Figure 1C), and the upregulated LPAR5 was closely correlated with worse overall survival of THCA patients (Figure 1D). Then, we identified one potential miRNA, miR-513c-5p, that linked TNRC6C-AS1 and LPAR5, by interrogating the starbase database (http://starbase.sysu.edu.cn/, Figure 1E). miR-513c-5p has been reported to be a significant cancer suppressor in various cancers13,22–26 other than THCA, suggesting its potential suppressive role in THCA.
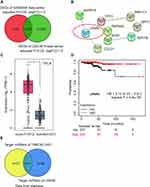 |
Figure 1 LPAR5 and miR-513c-5p were found to be potential participants in THCA. (A) The intersection of DEGs from GSE6004 and GSE3678 data series with adjusted P<0.05 and |logFC|>1.5. (B) The PPI analysis of the intersected DEGs by STRING database. (C) The expression of LPAR5 in THCA from GEPIA database (http://gepia2.cancer-pku.cn/#analysis). * P < 0.01. (D) The relationship between LPAR5 expression level and the overall survival outcome (http://kmplot.com/analysis/). (E) The identification of the miRNA that links TNRC6C-AS1 and LPAR5 using starbase database. |
Silencing TNRC6C-AS1 Inhibits Proliferation, Invasion, and Tumorigenesis of THCA Cells
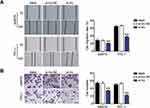
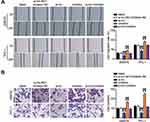
In order to investigate the expression of TNRC6C-AS1 in THCA, TNRC6C-AS1 expression levels were determined and it was found that the expression of TNRC6C-AS1 in the THCA cell line (SW579, BCPAP, and TPC-1) was significantly higher than that in the immortalized thyroid epithelial cell line Nthy-ori 3–1, and the expression of TNRC6C-AS1 was higher in SW579 and TPC-1 cells (Figure 2A). The expression of TNRC6C-AS1 in THCA and surrounding normal tissues was examined. The expression of TNRC6C-AS1 was approximately 2.5-fold higher in THCA tissues than in the corresponding non-cancerous tissues (Figure 2B). These results suggest that NRC6C-AS1 is highly expressed in THCA tissues and cell lines. Next, we evaluated the subcellular localization of TNRC6C-AS1, and the results showed that TNRC6C-AS1 was expressed in both the nucleus and cytoplasm (Figure 2C). This indicates that TNRC6C-AS1 plays a role in the nucleus and cytoplasm. To reveal the biological function of TNRC6C-AS1 in THCA, TNRC6C-AS1 was expressed at low levels in SW579 and TPC-1 cells. qRT-PCR assay confirmed that the expression of TNRC6C-AS1 in si-lnc-transfected cells was approximately 20% lower than that in control vector-transfected cells (Figure 2D). Notably, TNRC6C-AS1 knockdown significantly inhibited the activity of SW579 and TPC-1 cells by approximately 40% (Figure 2E). BrdU assay was performed to evaluate the proliferation, and it was found that the proliferation of SW579 and TPC-1 cells transfected with the si-lnc group was 70% and 60%, respectively, as compared with the si-lnc-NC group (Figure 2F). In the wound-healing experiment, low expression of TNRC6C-AS1 was found to significantly reduce the migration rate of SW579 and TPC-1 cells (Figure 3A). Transwell invasion assay showed that the invasion rate of TNRC6C-AS1 knockdown SW579 and TPC-1 cells was significantly lower than that of control vector-transfected cells (Figure 3B). In conclusion, TNRC6C-AS1 knockdown inhibited the growth and invasion of THCA cells.
TNRC6C-AS1 Interacts with and Downregulates miR-513c-5p
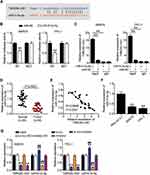
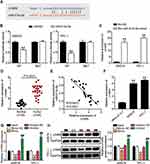
To elucidate the mechanism by which TNRC6C-AS1 inhibits the invasiveness of THCA, we selected a potential target miRNA of TNRC6C-AS1, namely miR-513c-5p. The binding site of TNRC6C-AS1 to miR-513c-5p is shown in Figure 4A. To identify the interaction between miR-513c-5p and TNRC6C-AS1 in THCA cells, a dual-luciferase reporter assay was performed. The activity of the NRC 6C-AS1 reporter gene was significantly inhibited by miR-513c-5p overexpression, while the miR-513c-5p binding site mutation prevented the activity of the NRC 6C-AS1 reporter gene, which was mediated by miR-513c-5p (Figure 4B). Moreover, an anti-argonaute 2 (AGO2) RNA RIP assay was performed. The results showed that both miR-513c-5p and TNRC6C-AS1 were enriched in AGO2 RIP samples from SW579 and TPC-1 cells (Figure 4C). Further investigation of miR-513c-5p expression in THCA revealed that miR-513c-5p expression in cancer tissue was 45% lower than that in normal tissues (Figure 4D). Pearson correlation analysis showed a negative correlation between miR-513c-5p and TNRC6C-AS1 in THCA tissues (Figure 4E). These results suggest that miR-513c-5p is an miRNA that mediates the role of TNRC6C-AS1 in THCA. The expression of miR-513c-5p in the cell lines was further detected by qRT-PCR, which showed that the expression level of miR-513c-5p in SW579 and TPC-1 cells was nearly 70% lower than that in Nthy-ori 3–1 cells (Figure 4F). Furthermore, to clarify the degree of regulation of TNRC6C-AS1 on miR-513c-5p, we knocked down TNRC6C-AS1 and miR-513c-5p in both SW579 and TPC-1 cells. The results showed that the expression level of miR-513c-5p increased by approximately 1.5 times after the knockdown of TNRC6C-AS1, and that of miR-513c-5p decreased by approximately 80% after knock-out of miR-513c-5p and reversed the effect of TNRC6C-AS1 knockdown (Figure 4G). Taken together, these data suggest a functional relationship between miR-513c-5 and TNRC6C-AS1 in THCA.
MiR-513c-5p Acts as a Tumor Suppressor Gene in THCA and Reverses the Effect of TNRC6C-AS1
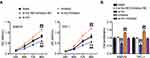
Next, we studied the role of miR-513c-5p in THCA. The results of CCK-8 and BrdU assays indicated that low expression of miR-513c-5p significantly increased the viability and proliferation of SW579 and TPC-1 cells in vitro, and reversed the effect of TNRC6C-AS1 knockdown on the viability and proliferation of THCA cells (Figure 5A, B). Further analysis of cell migration rate by wound healing assay showed that the migration rate of SW579 and TPC-1 cells was increased by more than 1.3 times after transfection with miR-513c-5p inhibitor, which partially eliminated the effect of transfection of si-lnc (Figure 6A). In addition, the transwell assay was used to evaluate the effect of miR-513C-5p on the invasion of THCA cells. Consistent with this hypothesis, the results revealed that the low expression of miR-513c-5p enhanced the invasion levels of SW579 and TPC-1 cells, and partially eliminated the effect of TNRC6C-AS1 knockdown (Figure 6B). These results indicated that miR-513c-5p knockdown inhibited the cell activity, proliferation, migration, and invasion of THCA cells, and reversed the inhibitory effect of TNRC6C-AS1 knockdown on tumor development.
LPAR5 is the Target Gene for miR-513c-5p
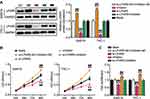
Next, we explored the downstream factors of miR-513c-5p in THCA. First, we searched for the binding site of miR-513c-5p to a specific mRNA, LPAR5. The query results are shown in Figure 7A. In addition, the binding site of miR-513c-5p on LPAR5 was mutated to produce the LPAR5-Mut reporter gene. Overexpression of miR-513c-5p reduced the luciferase activity of LPAR5-Wt but did not affect the activity of LPAR5-Mut (Figure 7B). The results showed that miR-513c-5p interacted with LPAR5 at the predicted site. Moreover, the RNA pull-down assay showed that miR-513c-5p was enriched in LPAR5 (Figure 7C). Later, we confirmed that LPAR5 was overexpressed in THCA tissues compared to that in normal tissues (Figure 7D). Moreover, Pearson analysis showed a negative correlation between miR-513c-5p and LPAR5 in cancer tissues (Figure 7E). Furthermore, the expression of LPAR5 in SW579 and TPC-1 cells was approximately four times higher than that in NTHY-ORI 3–1 cells (Figure 7F). In addition, upregulation of miR-513c-5p decreased LPAR5 mRNA and protein expression levels, while downregulation of miR-513c-5p increased LPAR5 mRNA and protein levels. (Figure 7G and H). Thus, LPAR5 is a target of miR-513c-5p in THCA cells.
MiR-513c-5p Inhibits THCA Through LPAR5
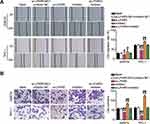
Next, we wanted to determine whether LPAR5 is involved in the inhibition of THCA mediated by miR-513c-5p. First, low expression of LPAR5 in SW579 and TPC-1 cells was found to decrease the expression of LPAR5 protein, and low expression of miR-513c-5p reversed the increase in LPAR5 expression (Figure 8A). Functionally, we found that the cell activity and proliferation levels were inhibited in the LPAR5 knockdown group, and that LPAR5 knockdown reversed the effect of the miR-513c-5p inhibitor on the proliferation of SW579 and TPC-1 cells (Figure 8B and C). It is suggested that LPAR5 interferes with the proliferation inhibition effect of miR-513c-5p on THCA cells. Next, we tested whether the anti-invasion mediated by miR-513c-5p was dependent on LPAR5. The results showed that invasion was inhibited in LPAR5-deficient THCA cells, and the invasion promotion by miR-513c-5p inhibitor was significantly reduced (Figure 9A and B). In conclusion, miR-513c-5p inhibitor-mediated inhibition of THCA cell biological function is attributed to the inhibition of LPAR5 knockdown.
Discussion
THCA, one of the most malignant tumors, is usually associated with specific genetic abnormalities and environmental factors.27 Therefore, early diagnosis and treatment is urgently required. With the development of high-throughput sequencing or new generation sequencing technology, an increasing number of new lncRNAs have been identified in many diseases, including tumor progression.28,29 Therefore, we investigated the role of TNRC6C-AS1/miR-513c-5p/LPAR5 in regulating THCA cell viability, proliferation, migration, and invasion.
TNRC6C-AS1 has long been identified as a key gene in THCA development. The expression of TNRC6C-AS1 in THCA tissues has been found to be more than 2 times higher than that in the adjacent normal tissues.12 Similar results were obtained in this study, which also demonstrated that interfering TNRC6C-AS1 inhibits THCA cell viability, proliferation, and metastasis in vitro. Notably, previous studies have found that TNRC6C-AS1 regulates the biological function of THCA cells by inhibiting miR-129-5p expression.12 In this study, we found that TNRC6C-AS1 inhibited the expression of miR-513c-5p, resulting in an increase in tumor cell proliferation and metastasis. These results suggest that TNRC6C-AS1 interacts with multiple miRNAs and affects the activity of THCA cells, and other downstream molecules need to be further studied.
Interestingly, a growing number of studies have suggested that the lncRNA-miRNA-mRNA interaction plays a crucial role in various tumorigenesis processes.30–32 Consistent with this view, TNRC6C-AS1 was found to restrict the expression of miR-513c-5p by competitive adsorption. In addition, miR-513c-5p was identified as a tumor suppressor gene of THCA that regulates cell proliferation, migration, and invasion. This study concluded that TNRC6C-AS1 sponges miR-513c-5p to inhibit the proliferation, migration, and invasion of THCA cells.
In addition, this study also showed that miR-513c-5p targets the LPAR5 3′ UTR and inhibits its expression. LPAR5 plays a key role in carcinogenesis and progression in the tumor microenvironment. More specifically, high LPAR5 levels predict higher lymphatic and distant metastasis rates, as well as lower overall survival and progression-free survival in breast cancer.33 LPAR5 knockdown impairs non-small-cell lung carcinoma cell proliferation and migration in vitro.34 Similarly, this study showed that knocking out LPAR5 promoted the proliferation of THCA cells. Furthermore, the effect of LPAR5 is regulated by TNRC6C-AS1/miR-513c-5p.
However, this study has some limitations. Our study was primarily cell-based, and the function of the TNRC6C-AS1/miR-513c-5p/LPAR5 axis needs to be validated in vivo. In addition, whether LPAR5 plays a role in THCA through the phosphatidylinositol 3-Kinase/Akt/Mammalian target of rapamycin (mTOR) pathway remains to be verified.17 Moreover, we will evaluate the correlation between TNRC6C-AS1, LPAR5, or miR-513c-5p and the clinicopathological features of tumor tissue and the prognosis of patients in the future.
Conclusions
In summary, our study suggests that the TNRC6C-AS1/miR-513c-5p/LPAR5 axis is a novel signaling pathway that modulates THCA progression and may be a potential therapeutic target for cancer therapy. TNRC6C-AS1 competes with LPAR5T for miR-513c-5p, ultimately regulating THCA cell proliferation and tumor growth. These findings may provide a solid experimental basis for further understanding of THCA pathology.
Data Sharing Statement
The intersection of DEGs from GSE6004 and GSE3678 data series with adjusted P<0.05 and |logFC|>1.5. The PPI analysis of the intersected DEGs by STRING database. The expression of LPAR5 in THCA from GEPIA database (http://gepia2.cancer-pku.cn/#analysis). The relationship between LPAR5 expression level and the overall survival outcome (http://kmplot.com/analysis/). The identification of the miRNA that links TNRC6C-AS1 and LPAR5 using starbase database.
Ethics Approval and Informed Consent
The present study was approved by the Ethics Committee of the People’s Hospital of Dongxihu District (Wuhan, China). The processing of clinical tissue samples is in strict compliance with the ethical standards of the Declaration of Helsinki. All patients signed written informed consent.
Consent for Publication
Consent for publication was obtained from the participants.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
Funding information is not applicable.
Disclosure
The authors declare that they have no conflict of interests.
References
1. Zaballos MA, Santisteban P. Key signaling pathways in thyroid cancer. J Endocrinol. 2017;235(2):R43–r61. doi:10.1530/joe-17-0266
2. Wang X, Zhang Q, Cai Z, Dai Y, Mou L. Identification of novel diagnostic biomarkers for thyroid carcinoma. Oncotarget. 2017;8(67):111551–111566. doi:10.18632/oncotarget.22873
3. Lundgren CI, Hall P, Dickman PW, Zedenius J. Clinically significant prognostic factors for differentiated thyroid carcinoma: a population-based, nested case-control study. Cancer. 2006;106(3):524–531. doi:10.1002/cncr.21653
4. Xing M, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet (London, England). 2013;381(9871):1058–1069. doi:10.1016/s0140-6736(13)60109-9
5. Sedaghati M, Kebebew E. Long noncoding RNAs in thyroid cancer. Curr Opin Endocrinol Diabetes Obes. 2019;26(5):275–281. doi:10.1097/med.0000000000000497
6. Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222. doi:10.1007/978-981-10-5203-3_7
7. Muhanhali D, Zhai T, Jiang J, Ai Z, Zhu W, Ling Y. Long non-coding antisense RNA TNRC6C-AS1 Is activated in papillary thyroid cancer and promotes cancer progression by suppressing TNRC6C expression. Front Endocrinol (Lausanne). 2018;9:360. doi:10.3389/fendo.2018.00360
8. Yang LX, Wu J, Guo ML, Zhang Y, Ma SG. Suppression of long non-coding RNA TNRC6C-AS1 protects against thyroid carcinoma through DNA demethylation of STK4 via the Hippo signalling pathway. Cell Prolif. 2019;52(3):e12564. doi:10.1111/cpr.12564
9. Peng X, Ji C, Tan L, et al. Long non-coding RNA TNRC6C-AS1 promotes methylation of STK4 to inhibit thyroid carcinoma cell apoptosis and autophagy via Hippo signalling pathway. J Cell Mol Med. 2020;24(1):304–316. doi:10.1111/jcmm.14728
10. Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8(2):e53823. doi:10.1371/journal.pone.0053823
11. Zhang H, Cai Y, Zheng L, Zhang Z, Lin X, Jiang N. Long noncoding RNA NEAT1 regulate papillary thyroid cancer progression by modulating miR-129-5p/KLK7 expression. J Cell Physiol. 2018;233(10):6638–6648. doi:10.1002/jcp.26425
12. Hou S, Lin Q, Guan F, Lin C. LncRNA TNRC6C-AS1 regulates UNC5B in thyroid cancer to influence cell proliferation, migration, and invasion as a competing endogenous RNA of miR-129-5p. J Cell Biochem. 2018;119(10):8304–8316. doi:10.1002/jcb.26868
13. Jia P, Wei E, Liu H, Wu T, Wang H. Silencing of long non-coding RNA DLX6-AS1 weakens neuroblastoma progression by the miR-513c-5p/PLK4 axis. IUBMB Life. 2020;72(12):2627–2636. doi:10.1002/iub.2392
14. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19. doi:10.3390/ijms19051310.
15. Zhu X, Jiao J, Zhou C, et al. LPAR5, GNAT3 and partial amino acid transporters messenger RNA expression patterns in digestive tracts, metabolic organs and muscle tissues of growing goats. Animal. 2019;13(7):1394–1402. doi:10.1017/s1751731118002823
16. Araki M, Kitayoshi M, Dong Y, et al. Inhibitory effects of lysophosphatidic acid receptor-5 on cellular functions of sarcoma cells. Growth Factors (Chur, Switzerland). 2014;32(3–4):117–122. doi:10.3109/08977194.2014.911294
17. Wu CY, Zheng C, Xia EJ, et al. Lysophosphatidic acid Receptor 5 (LPAR5) plays a significance role in papillary thyroid cancer via phosphatidylinositol 3-Kinase/Akt/Mammalian Target of Rapamycin (mTOR) pathway. Med Sci Monitor. 2020;26:e919820. doi:10.12659/msm.919820
18. Zhang S, Wang Q, Han Q, Han H, Lu P. Identification and analysis of genes associated with papillary thyroid carcinoma by bioinformatics methods. Biosci Rep. 2019;39(4):Apr. doi:10.1042/bsr20190083
19. Liu L, He C, Zhou Q, Wang G, Lv Z, Liu J. Identification of key genes and pathways of thyroid cancer by integrated bioinformatics analysis. J Cell Physiol. 2019;234(12):23647–23657. doi:10.1002/jcp.28932
20. Tang J, Kong D, Cui Q, et al. Bioinformatic analysis and identification of potential prognostic microRNAs and mRNAs in thyroid cancer. Peer J. 2018;6:e4674. doi:10.7717/peerj.4674
21. Xue G, Lin X, Wu JF, et al. Identification of key genes of papillary thyroid carcinoma by integrated bioinformatics analysis. Biosci Rep. 2020;40:8. doi:10.1042/bsr20201555
22. Xia HL, Lv Y, Xu CW, et al. MiR-513c suppresses neuroblastoma cell migration, invasion, and proliferation through direct targeting glutaminase (GLS). Cancer Biomarkers. 2017;20(4):589–596. doi:10.3233/cbm-170577
23. Wang O, Huang Y, Wu H, Zheng B, Lin J, Jin P. LncRNA LOC728196/miR-513c axis facilitates glioma carcinogenesis by targeting TCF7. Gene. 2018;679:119–125. doi:10.1016/j.gene.2018.08.081
24. Xu J, Sun T, Hu X. microRNA-513c suppresses the proliferation of human glioblastoma cells by repressing low-density lipoprotein receptor-related protein 6. Mol Med Rep. 2015;12(3):4403–4409. doi:10.3892/mmr.2015.3913
25. Zhang K, Zhao Z, Yu J, Chen W, Xu Q, Chen L. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma. J Cell Biochem. 2018;119(7):6045–6056. doi:10.1002/jcb.26802
26. Liu X, Peng D, Cao Y, et al. Upregulated lncRNA DLX6-AS1 underpins hepatocellular carcinoma progression via the miR-513c/Cul4A/ANXA10 axis. Cancer Gene Ther. 2020. doi:10.1038/s41417-020-00233-0
27. Li X, Abdel-Mageed AB, Mondal D, Kandil E. The nuclear factor kappa-B signaling pathway as a therapeutic target against thyroid cancers. Thyroid. 2013;23(2):209–218. doi:10.1089/thy.2012.0237
28. Wen C, Yang S, Zheng S, Feng X, Chen J, Yang F. Analysis of long non-coding RNA profiled following MC-LR-induced hepatotoxicity using high-throughput sequencing. J Toxicol Environ Health A. 2018;81(22):1165–1172. doi:10.1080/15287394.2018.1532717
29. Chuang TD, Khorram O. Expression profiling of lncRNAs, miRNAs, and mRNAs and their differential expression in leiomyoma using next-generation RNA sequencing. Reprod Sci. 2018;25(2):246–255. doi:10.1177/1933719117711265
30. He JH, Han ZP, Zou MX, et al. Analyzing the LncRNA, miRNA, and mRNA regulatory network in prostate cancer with bioinformatics software. J Computational Biol. 2018;25(2):146–157. doi:10.1089/cmb.2016.0093
31. Li DY, Chen WJ, Luo L, et al. Prospective lncRNA-miRNA-mRNA regulatory network of long non-coding RNA LINC00968 in non-small cell lung cancer A549 cells: a miRNA microarray and bioinformatics investigation. Int J Mol Med. 2017;40(6):1895–1906. doi:10.3892/ijmm.2017.3187
32. Mao Y, Liu R, Zhou H, et al. Transcriptome analysis of miRNA-lncRNA-mRNA interactions in the malignant transformation process of gastric cancer initiation. Cancer Gene Ther. 2017;24(6):267–275. doi:10.1038/cgt.2017.14
33. Zheng YQ, Miao X, Li J, et al. Trichostatin A alleviates the process of breast carcinoma by downregulating LPAR5. Eur Rev Med Pharmacol Sci. 2020;24(11):6417–6425. doi:10.26355/eurrev_202006_21540
34. Zhang HP, Chen QK, Xu JF. LPAR5 stimulates the malignant progression of non-small-cell lung carcinoma by upregulating MLLT11. Eur Rev Med Pharmacol Sci. 2020;24(17):8902–8910. doi:10.26355/eurrev_202009_22831
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.