Back to Journals » Drug Design, Development and Therapy » Volume 15
Thiazole-Based Thiosemicarbazones: Synthesis, Cytotoxicity Evaluation and Molecular Docking Study
Authors Gomha SM , Abdelhady HA, Hassain DZH, Abdelmonsef AH , El-Naggar M , Elaasser MM , Mahmoud HK
Received 20 November 2020
Accepted for publication 20 January 2021
Published 17 February 2021 Volume 2021:15 Pages 659—677
DOI https://doi.org/10.2147/DDDT.S291579
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Manfred Ogris
Sobhi M Gomha,1,2 Hyam A Abdelhady,2 Doaa ZH Hassain,2 Aboubakr H Abdelmonsef,3 Mohamed El-Naggar,4 Mahmoud M Elaasser,5 Huda K Mahmoud2
1Chemistry Department, Faculty of Science, Islamic University in Almadinah Almonawara, Almadinah Almonawara, 42351, Saudi Arabia; 2Chemistry Department, Faculty of Science, University of Cairo, Giza, Egypt; 3Chemistry Department, Faculty of Science, South Valley University, Qena, 83523, Egypt; 4Chemistry Department, Faculty of Sciences, University of Sharjah, Sharjah, 27272, United Arab Emirates; 5The Regional Center for Mycology and Biotechnology, Al-Azhar University, Cairo, 11371, Egypt
Correspondence: Sobhi M Gomha Email [email protected]
Aboubakr H Abdelmonsef Email [email protected]
Introduction: Hybrid drug design has developed as a prime method for the development of novel anticancer therapies that can theoretically solve much of the pharmacokinetic disadvantages of traditional anticancer drugs. Thus a number of studies have indicated that thiazole-thiophene hybrids and their bis derivatives have important anticancer activity. Mammalian Rab7b protein is a member of the Rab GTPase protein family that controls the trafficking from endosomes to the TGN. Alteration in the Rab7b expression is implicated in differentiation of malignant cells, causing cancer.
Methods: 1-(4-Methyl-2-(2-(1-(thiophen-2-yl) ethylidene) hydrazinyl) thiazol-5-yl) ethanone was used as building block for synthesis of novel series of 5-(1-(2-(thiazol-2-yl) hydrazono) ethyl) thiazole derivatives. The bioactivities of the synthesized compounds were evaluated with respect to their antitumor activities against MCF-7 tumor cells using MTT assay. Computer-aided docking protocol was performed to study the possible molecular interactions between the newly synthetic thiazole compounds and the active binding site of the target protein Rab7b. Moreover, the in silico prediction of adsorption, distribution, metabolism, excretion (ADME) and toxicity (T) properties of synthesized compounds were carried out using admetSAR tool.
Results: The results obtained showed that derivatives 9 and 11b have promising activity (IC50 = 14.6 ± 0.8 and 28.3 ± 1.5 μM, respectively) compared to Cisplatin (IC50 = 13.6 ± 0.9 μM). The molecular docking analysis reveals that the synthesized compounds are predicted to be fit into the binding site of the target Rab7b. In summary, the synthetic thiazole compounds 1– 17 could be used as potent inhibitors as anticancer drugs.
Conclusion: Promising anticancer activity of compounds 9 and 11 compared with cisplatin reference drug suggests that these ligands may contribute as lead compounds in search of new anticancer agents to combat chemo-resistance.
Keywords: thiazoles, hydrazones, hydrazonoyl halides, docking, Rab7b, MCF-7
Introduction
Cancer is a broad concept that encompasses a wide variety of diseases, essentially marked by spontaneous growth and cell proliferation, with failures in the division routes known as the cell cycle. It is a major global public health problem as it is the world’s second-largest cause of death, with approximate 9.6 million deaths in 2018.1 Investigation of novel compounds that may be of use in designing new less toxic, selective, and potent anti-cancer agents is still the main challenge for medical chemists. Cisplatin is one of the most effective anticancer agents widely used in the treatment of breast cancer. It prevents DNA replication in cancer cells by a ligand replacement reaction with DNA in which a bond is formed between platinum and a nitrogen atom on guanine.2 Several studies on various diseases have been performed with several sulfur heterocycles, including thiophene and thiazole. The literature reports that thiophene core compounds have drawn significant attention in the areas of drug discovery because of their versatile and wide range of biological activities, which include antimicrobial,3 anti-inflammatory,4 antidepressant,5 analgesic,6 anticonvulsant.7 In addition, thiophene, being one of the main scaffolds, is continuously being sought by several researchers to develop potential cancer agents. Thiophene derivatives have been identified as anti-cancer agents for several years and exhibit their influence via different cancer pathways.8–13 Thiazole-containing drugs, on the other hand, have demonstrated their involvement in a variety of commercially available anti-cancer medications, such as tiazofurin (inhibitor of IMP dehydrogenase),14 dasatinib (Bcr-Abl tyrosine kinase inhibitor),15 dabrafenib (inhibitor of enzyme B-RAF),16 ixabepilone (stabilization of microtubules),17 and epothilone (inhibition of microtubule function)18 (Figure 1). Thiazole-containing compounds depict anticancer activity profile through diverse mechanisms.19–27
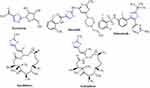 |
Figure 1 Examples of thiazole bearing anticancer drugs. |
Molecular hybridization is a beneficial approach to structural alteration involving the integration in a single species of two or more pharmacophores. Over the last several years, hybrid drug design has developed as a prime method for the development of novel anticancer therapies that can theoretically solve much of the pharmacokinetic disadvantages of traditional anticancer drugs.28,29 Thus, a number of studies have indicated that thiazole-thiophene hybrids and their bis derivatives have important anticancer activity.30–34 Based on the above-mentioned promising aspects, the strategy of this work includes gathering the two bioactive entities thiophene-thiazole in one compact structure for the purpose of synergism and examined the prepared compounds as anticancer agents. Activity against anticancer cell lines would expect to show remarkable activity. The in vitro cytotoxic potential of the newly synthesized compounds was examined against the human breast cancer cell line (MCF-7) using the MTT assay and the results showed compounds 9 and 11b have promising activity.
Rab proteins are crucial regulators of all aspects of membrane trafficking in all cell types.35,36 Rab7b belongs to Ras superfamily of small GTPases, which is vital for various cellular processes.37 Human Rab7b is involved in regulating membrane transport from early to late endosomes; in addition, Rab7b has important roles in lipid metabolism, growth factor signaling, autophagy, and phagolysosome biogenesis.38 Moreover, the biochemical pathway of Homo sapiens Rab7b is adapted from BioRender web-based tool, as represented in Figure S1, in the supplementary data section, declared the critical role of the target protein in regulating signal transduction processes leading to cytoskeletal – dependent responses.39 The cycle of Rab7b protein declares that the target is activated to GTP bound form by GEF (guanine nucleotide exchange factor) and deactivated to GDP bound form by GAP (GTPase-activating protein).40,41 The active form of Rab7b protein plays a critical role in endocytic trafficking and other degradative pathways like phagocytosis and autophagy.42 The overexpression of Rab7b is characterized in human cancer progression.43–46 Finally, human Rab7b protein is targeted for the identification of potent drug candidates against cancer. The molecular docking studies were carried out to understand the molecular interactions between the active site of the target Rab7b and the examined compounds.
Materials and Methods
Chemistry
All melting points were determined on an electrothermal apparatus and were left uncorrected. IR spectra were recorded (KBr discs) on a Shimadzu FT-IR 8201 PC spectrophotometer. 1H-NMR and 13C-NMR spectra were recorded in DMSO solutions on BRUKER 400 FT-NMR system spectrometer and chemical shifts were expressed in ppm units using TMS as an internal reference. Mass spectra were recorded on a GC-MS QP1000 EX Shimadzu. Elemental analyses were carried out at the Microanalytical Center of Cairo University.47 The spectral data are shown in Figure S2, in the supplementary data section.
Synthesis of Thiazole Derivative 3
A mixture of acetyl thiazole derivative 1 (0.279 g, 1 mmol) and thiosemicarbazide (2) (0.091 g, 1 mmol) in EtOH (20 mL) containing drops of HCl was refluxed for 2–4 h (monitored by TLC, using n-Hexane/ethyl acetate (2:1) as elution solvent). The formed solid was filtered and recrystallized from dioxane solvent to give 2-(1-(4-methyl-2-(2-(1-(thiophen-2-yl) ethylidene) hydrazinyl) thiazol- 5-yl) ethylidene) hydrazinecarbothio-amide (3). Yellow solid, 80% yield, m.p. 220–222°C (DMF); 1H-NMR (DMSO-d6): δ 2.39 (s, 3H, CH3), 2.35 (s, 3H, CH3), 2.37 (s, 3H, CH3), 7.08–7.56 (m, 4H, Ar-H, NH), 8.73 (br s, 2H, NH2), 10.41 (br s, 1H, NH) ppm; 13C-NMR (DMSO-d6): δ 16.1, 17.9, 22.1 (CH3), 114.8, 114.9, 115.0, 130.0, 130.1, 134.3, 146.1, 149.4, 160.4 (Ar-C and C=N), 185.1 (C=S) ppm; IR (KBr): v 3409 (br. 2NH) cm−1; MS m/z (%): 354 (M+ +1, 2), 353 (M+, 1), 313 (5), 281 (7), 239 (7), 199 (8), 182 (12), 155 (6), 140 (7), 129 (17), 124 (22), 111 (29), 101 (32), 97 (39), 83 (45), 69 (56), 57 (100). Anal. Calcd for C13H16N6S3 (352.50): C, 44.29; H, 4.58; N, 23.84. Found: C, 44.33; H, 4.45; N, 23.65%.
Synthesis of Thiocarbohydrazone Derivative 5
A mixture of acetylthiazole 1 (2.79 g, 10 mmol) and thiocarbohydrazide (4) (1.06 g, 10 mmol) in 20 mL EtOH and HCl (2 drops) for 2–4 h (monitored by TLC). The formed solid product was filtered and crystallized from DMF to afford thiocarbohydrazone derivative 5 as yellow solid, 79% yield, m.p. 204–206 oC; 1H-NMR (DMSO-d6): δ 2.11 (s, 3H, CH3), 2.33 (s, 3H, CH3), 2.43 (s, 3H, CH3), 5.40 (br.s, 2H, NH2), 7.02–7.62 (m, 4H, Ar-H and NH), 8.68 (br.s, 1H, NH), 9.32 (br.s, 1H, NH) ppm; IR (KBr): v 3426–3239 (3NH + NH2), 1601 (C=N) cm−1; MS m/z (%): 368 (M+ +1, 2), 367 (M+, 2), 313 (3), 302 (2), 267 (2), 256 (4), 232 (7), 191 (4), 178 (8), 165 (8), 139 (10), 128 (35), 125 (28), 110 (47), 97 (46), 84 (48), 69 (70), 57 (100). Anal. Calcd for C13H17N7S3 (367.52): C, 42.48; H, 4.66; N, 26.68. Found: C, 42.36; H, 4.47; N, 26.59%.
Synthesis of Thiazole Derivatives 7, 9 and 11a, b
General procedure. A mixture of thiosemicarbazone derivative 3 (0.352 g, 1 mmol) and the appropriate α-halocarbonyl compound 6, 8 and 10a, b (1 mmol) in EtOH (20 mL) was refluxed for 2–4 h (monitored by TLC). The solvent was evaporated under vacuum pressure and the formed solid was crystallized from the appropriate solvent to give 7, 9 and 11a, b respectively.
Compound 7. Dark green solid, 71% yield, m.p. 210–212 oC; (Dioxane); 1H-NMR (DMSO-d6): δ 2.07 (s, 3H, CH3), 2.24 (s, 3H, CH3), 2.32 (s, 3H, CH3), 2.39 (s, 3H, CH3), 2.47 (s, 3H, CH3), 7.06–7.10 (t, 1H, Ar-H), 7.23 (d, 1H, Ar-H), 7.31 (br.s, 1H, NH), 7.37 (d, 1H, Ar-H), 7.88 (br.s, 1H, NH) ppm; IR (KBr): v 3414, 3229 (br. 2NH), 1697 (C=O), 1609 (C=N) cm−1; MS m/z (%): 433 (M++1, 2), 432 (M+, 7), 278 (26), 256 (9), 239 (38), 225 (58), 186 (20), 154 (20), 141 (47), 124 (79), 110 (82), 83 (52), 71 (100). Anal. Calcd for C18H20N6OS3 (432.59): C, 49.98; H, 4.66; N, 19.43. Found: C, 49.88; H, 4.51; N, 19.35%.
Compound 9. Dark green solid, 75% yield, m.p. 140–142 oC (Dioxane); 1H-NMR (DMSO-d6): δ 1.24 (t, 3H, CH3CH2), 2.08 (s, 3H, CH3), 2.28 (s, 3H, CH3), 2.32 (s, 3H, CH3), 2.35 (s, 3H, CH3), 4.18 (q, 2H, CH2CH3), 7.08–7.51 (m, 3H, Ar-H), 8.82 (br.s, 1H, NH), 9.54 (br.s, 1H, NH) ppm; 13C-NMR (DMSO-d6): δ 13.7, 16.3, 16.8, 18.3, 23.8 (CH3), 60.6 (CH2), 114.5, 116.6, 116.7, 127.9, 128.0, 128.2, 128.3, 128.6, 142.7, 142.9, 149.1, 150.2 (Ar-C and C=N), 168.9 (C=O) ppm; IR (KBr): v 3427, 3279 (br. 2NH), 1697 (C=O), 1604 (C=N) cm−1; MS m/z (%): 463 (M++1, 2), 462 (M+, 5), 420 (55), 309 (19), 289 (47), 278 (45), 195 (13), 141 (70), 124 (88), 110 (100), 71 (95). Anal. Calcd for C19H22N6O2S3 (462.61): C, 49.33; H, 4.79; N, 18.17. Found: C, 49.13; H, 4.99; N, 18.27%.
Compound 11a. Red solid, 71% yield, m.p. 260–262 oC (Dioxane); 1H-NMR (DMSO-d6): δ 2.27 (s, 3H, CH3), 2.35 (s, 3H, CH3), 2.42 (s, 3H, CH3), 7.07–8.15 (m, 8H, Ar-H and thiazole-H5), 8.41 (br.s, 1H, NH), 9.87 (br.s, 1H, NH) ppm; IR (KBr): v 3394, 3238 (br. 2NH), 1604 (C=N) cm−1; MS m/z (%): 488 (M++1, 1), 487 (M+, 11), 431 (31), 414 (53), 386 (16), 333 (58), 289 (36), 235 (57), 210 (36), 168 (29), 139 (66), 110 (100), 71 (41). Anal. Calcd for C21H19ClN6S3 (487.06): C, 51.78; H, 3.93; N, 17.25 Found: C, 51.65; H, 3.82; N, 17.15%.
Compound 11b. Dark brown solid, 75% yield, m.p. 180–182 oC (Dioxane); 1H-NMR (DMSO-d6): δ 2.11 (s, 3H, CH3), 2.28 (s, 3H, CH3), 2.35 (s, 3H, CH3), 7.06–8.36 (m, 8H, Ar-H and thiazole-H5), 8.81 (br.s, 1H, NH), 9.87 (br.s, 1H, NH) ppm; 13C-NMR (DMSO-d6): δ 16.2, 19.2, 21.9 (CH3), 116.5, 118.9, 119.2, 124.6, 125.4, 127.4, 128.3, 128.8, 129.8, 132.3, 144.0, 144.6, 150.6, 152.8, 159.6, 159.8 (Ar-C and C=N) ppm; IR (KBr): v 3406, 3236 (br. 2NH), 1598 (C=N) cm−1; MS m/z (%): 498 (M++1, 4), 497 (M+, 6), 383 (53), 344 (61), 329 (42), 289 (34), 247 (43), 234 (46), 221 (18), 150 (13), 141 (11), 124 (52), 97 (30), 71 (31), 58 (100). Anal. Calcd for C21H19N7O2S3 (497.62): C, 50.69; H, 3.85; N, 19.70. Found: C, 50.58; H, 3.78; N, 19.66%.
Synthesis of Thiazole-4-one Derivative 13
A mixture of 3 (0.352 g, 1 mmol) and ethylchloroacetate 12 (1 mmol) in AcOH (20 mL) containing anhydrous sodium acetate (1 mmol) was refluxed for 2–4 h. (monitored by TLC, using n-Hexane/ethyl acetate (2:1) as elution solvent). The solvent was evaporated under vacuum pressure and the formed solid was crystallized from AcOH to give 13 as Brown solid, 80% yield, m.p. 220–222 oC (Dioxane); 1H-NMR (DMSO-d6): δ 2.10 (s, 3H, CH3), 2.16 (s, 3H, CH3), 2.25 (s, 3H, CH3), 4.27 (s, 2H, thiazole-CH2), 6.99–7.08 (t, 1H, Ar-H), 7.51 (d, 2H, Ar-H), 10.53 (br.s, 1H, NH), 11.74 (br.s, 1H, NH) ppm; IR (KBr): v 3448, 3261 (br. 2NH), 1697 (C=O), 1612 (C=N) cm−1; MS m/z (%): 393 (M++1, 3), 392 (M+, 13), 380 (40), 309 (33), 293 (22), 248 (57), 183 (16), 166 (16), 156 (24), 124 (100), 110 (98), 97 (54), 71 (73). Anal. Calcd for C15H16N6OS3 (392.52): C, 45.90; H, 4.11; N, 21.41. Found: C, C, 45.88; H, 4.21; N, 21.33%.
Synthesis of Thiazole Derivatives 14, 15 and 16a, b
General procedure. A mixture of thiocarbohydrazone derivative 5 (0.367 g, 1 mmol) and the appropriate α-halocarbonyl compound 6, 8 and 10a, b (1 mmol) in EtOH (20 mL) was refluxed for 2–4 h. (monitored by TLC, using n-Hexane/ethyl acetate (2:1) as elution solvent). The solvent was evaporated under vacuum pressure and the formed solid was crystallized from appropriate solvent to give 14, 15 and 16a, b respectively.
Compound 14. Dark brown solid, 85% yield, m.p. 180–182 oC; (Dioxane); 1H-NMR (DMSO-d6): δ 2.12 (s, 3H, CH3), 2.26 (s, 3H, CH3), 2.31 (s, 3H, CH3), 2.38 (s, 3H, CH3), 2.42 (s, 3H, CH3), 3.36 (br.s, 2H, NH2), 7.07 (t, 1H, Ar-H), 7.36–7.53 (d, 2H, Ar-H), 11.73 (br.s, 1H, NH) ppm; IR (KBr): v 3464, 3387, 3209 (NH2 and NH), 1695 (C=O), 1603 (C=N) cm−1; MS m/z (%): 449 (M+ +2, 8), 447 (M+, 5), 432 (58), 416 (4), 401 (3), 390 (2), 370 (2), 358 (5), 330 (2), 320 (3), 313 (3), 309 (3), 302 (7), 294 (4), 287 (6), 278 (25), 267 (13), 196 (8), 166 (14), 141 (28), 124 (100), 110 (85), 97 (46), 67 (37). Anal. Calcd for C18H21N7OS3 (447.60): C, 48.30; H, 4.73; N, 21.90. Found: C, 48.19; H, 4.60; N, 21.74%.
Compound 15. Dark brown solid, 70% yield, m.p. 175–177 oC (Dioxane); 1H-NMR (DMSO-d6): δ 1.27 (t, 3H, CH3CH2), 2.10 (s, 3H, CH3), 2.25 (s, 3H, CH3), 2.32 (s, 3H, CH3), 2.39 (s, 3H, CH3), 3.51 (br.s, 2H, NH2), 4.23 (q, 2H, CH2CH3), 7.06 (t, 1H, Ar-H), 7.38–7.54 (d, 2H, Ar-H), 9.88 (br.s, 1H, NH) ppm; IR (KBr): v 3464, 3326, 3176 (NH2 and NH), 1697 (C=O), 1607 (C=N) cm−1; MS m/z (%): 477 (M+, 4), 462 (18), 444 (1), 426 (1), 388 (12), 342 (7), 322 (2), 289 (6), 278 (25), 267 (4), 237 (9), 182 (11), 166 (13), 141 (14), 124 (99), 110 (100), 71 (11). Anal. Calcd for C19H23N7O2S3 (477.63): C, 47.78; H, 4.85; N, 20.53. Found: C, 47.55; H, 4.70; N, 20.42%.
Compound 16a. Dark brown solid, 77% yield, m.p. 142–144 oC (Dioxane); 1H-NMR (DMSO-d6): δ 2.09 (s, 3H, CH3), 2.24 (s, 3H, CH3), 2.41 (s, 3H, CH3), 3.47 (br.s, 2H, NH2), 7.01–8.06 (m, 8H, Ar-H and thiazole-H5), 11.50 (br.s, 1H, NH) ppm; IR (KBr): v 3387, 3325, 3186 (NH2 and NH), 1601 (C=N) cm−1; MS m/z (%): 502 (M+, 5), 469 (22), 488 (13), 485 (8), 472 (2), 470 (4), 431 (2), 400 (7), 363 (4), 332 (7), 303 (14), 278 (15), 236 (11), 209 (11), 168 (11), 139 (62), 124 (100), 97 (43). Anal. Calcd for C21H20ClN7S3 (502.08): C, 50.24; H, 4.02; N, 19.53. Found: C, 50.37; H, 3.85; N, 19.41%.
Compound 16b. Dark brown solid, 80% yield, m.p. 180–182 oC (Dioxane); 1H-NMR (DMSO-d6): δ 2.07 (s, 3H, CH3), 2.27 (s, 3H, CH3), 2.38 (s, 3H, CH3), 3.57 (br.s, 2H, NH2), 7.08–8.43 (m, 8H, Ar-H and thiazole-H5), 11.52 (br.s, 1H, NH) ppm; IR (KBr v 3448, 3379, 3217 (NH2 and NH), 1597 (C=N) cm−1; MS m/z (%): 513 (M+ + 1, 1), 512 (M+, 3), 434 (2), 372 (2), 336 (2), 316 (2), 297 (3), 274 (3), 267 (2), 253 (4), 243 (3), 224 (3), 197 (4), 181 (5), 120 (31), 97 (43), 60 (68), 55 (100). Anal. Calcd for C21H20N8O2S3 (512.63): C, 49.20; H, 3.93; N, 21.86. Found: C, 49.05; H, 3.73; N, 21.71%.
Synthesis of Thiazole-4-one Derivative 17
A mixture of thiosemicarbazone derivative 5 (0.367 g, 1 mmol) and ethylchloroacetate 12 (1 mmol) in AcOH (20 mL) containing anhydrous sodium acetate (1 mmol) was refluxed for 2–4 h. (monitored by TLC, using n-Hexane/ethyl acetate (2:1) as elution solvent). The solvent was evaporated under vacuum pressure and the formed solid was crystallized from AcOH to give 17 as green solid, 77% yield, m.p. 190–192 oC (Dioxane); 1H-NMR (DMSO-d6): δ 2.16 (s, 3H, CH3), 2.33 (s, 3H, CH3), 2.69 (s, 3H, CH3), 3.39 (br.s, 2H, NH2), 4.12 (s, 2H, CH2), 7.09–7.54 (m, 3H, Ar-H), 11.89 (br.s, 1H, NH) ppm; IR (KBr): v 3436, 3318, 3212 (NH2 and NH), 1705 (C=O), 1607 (C=N) cm−1; MS m/z (%): 408 (M++ 1, 3), 407 (M+, 6), 392 (6), 382 (2), 367 (2), 350 (3), 341 (2), 325 (2), 313 (5), 303 (10), 293 (10), 278 (11), 263 (4), 253 (5), 239 (8), 226 (8), 220 (6), 185 (11), 166 (11), 149 (7), 141 (11), 124 (63), 110 (44), 84 (39), 57 (100). Anal. Calcd for C15H17N7OS3 (407.54): C, 44.21; H, 4.20; N, 24.06. Found: C, 44.30; H, 4.17; N, 24.01%.
Anticancer Activity
The synthesized compounds have been cytotoxically assessed against MCF-7 cells with 24 hours incubation MTT examination at the Regional Center for Mycology and Biotechnology at Al-Azhar University, Cairo, Egypt.
In vitro Cytotoxic Activity
The two cell cultures for human breast cancer (MCF-7) cell lines were bought from the American Type Culture Collection (Rockville, MD) and preserved in DMEM medium, supplemented by 10%. (Fetal bovine serum), 100U/mL penicillin and 100 U/mL streptomycin. Cells grew at 37°C in a humid atmosphere of 5% CO2.
MTT Cytotoxicity Assay
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay was utilized to evaluate the cytotoxicity of imidazothiazole derivatives versus MCF-7 human cancer cell lines. This method is depending on the dissent the salt of tetrazole by mitochondrial dehydrogenases in the cells. Distributed of the cells in a (5 x 104 cells/well) of 96 well sterile microplate, and at 37°C were incubated in DMSO with series of different concentrations of each tested imidazothiazole derivatives or Doxorubicin (positive control) for 48 h in a serum-free medium prior to the MTT assay. After incubation, the media was carefully removed and (2.5 mg/mL) of 40 μL of MTT was added to each well and incubated for an additional 4 hours. Purple formazan-dye crystals were dissolved by adding 200 μL of DMSO. At 590 nm, the absorbance was measured utilizing Spectra-Max Paradigm Multi-Mode plate reader. The relative viability of the cell was expressed as a percentage of viable cells compared to untreated control cells. All trials were conducted in three versions and repeated on three different days. All values were represented as ± SD. IC50s were determined by probit analysis by SPSP Inc. Analysis (USA, NY, IBM Corp., Armonk).48
In silico Studies
3D Structure Generation, Protein Preparation, Active Site Identification
The 3D structure of the target is essential for discovering novel inhibitors against cancer via computer-based docking approach.49 In fact, to date, the 3D model has not been generated yet, so the homology modeling approach is used to determine the 3D structure of the target Rab7b. The stereochemical quality of the generated model is checked by Ramachandran plot.50 The active site pockets of the target were evaluated using Computed Atlas of Surface Topography of proteins CASTp51 web server.
Ligand Preparation
The 3D structures of the ligand molecules are generated using ChemDraw Ultra 7.0 and saved as SDF files format by using Open Babel 2.4.1 tool.52 In-house library of thirteen compounds is generated for study. Energy of the compounds was minimized using Universal Force Field (UFF),53 to obtain stable confirmations. The energy minimized compounds are then read as input for PyRx virtual screening tool,54 in order to perform the docking simulation.
In silico Docking Protocol
To understand ligand–protein interaction, a molecular docking study is performed for of thirteen compounds 1–17 against Rab7b protein using PyRx tool. During the screening process, a maximum of nine conformers is considered for each compound, and then, the conformer with more negative binding energy is elected for further study.55,56 Two and three-dimensional representations of ligand–protein interactions are visualized using Accelrys discovery studio 3.5 (Accelrys Discovery Studio Visualizer Software 2010).
ADME Screening
The drug-likeness and physicochemical properties57,58 of the newly synthesized compounds are predicted using web-based softwares, admetSAR59 and Mol inspiration.
Results and Discussion
Chemistry
Thiosemicarbazone derivative 3 and thiocarbohydrazone derivative 5 were prepared from reaction of 2-(2-benzylidenehydrazinyl)-4-methylthiazole (1)60 with the respective thiosemicarbazide 2 and thiocarbohydrazide 4 in EtOH/HCl under reflux, respectively (Scheme 1). The chemical structure of the compounds 3 and 5 was elucidated by both spectral data and elemental analysis.
The chemical reactivity of thiosemicarbazone 3 towards α-halo-compounds was investigated with the aim of synthesizing a series of new thiazole systems. Thus, treatment of thiosemicarbazone derivative 3 with 3-chloropentane-2,4-dione (6) in refluxing EtOH yielded the acetylthiazole derivative 7 (Scheme 2). The spectra and elemental analysis of compound 7 were in accordance with suggested structure. The 1H-NMR spectra of 7 showed the expected signals at δ = 2.07, 2.24, 2.32, 2.39, 2.47 (5s, 15H, 5CH3), 7.06–7.10 (t, 1H, Ar-H), 7.23 (d, 1H, Ar-H), 7.31 (br.s, 1H, NH), 7.37 (d, 1H, Ar-H), 7.88 (br.s, 1H, NH). The mass of compound 7 was determined by mass spectrometry is equal to the calculated value.
In a similar way, thiosemicarbazone 3 reacted with ethyl 2-chloro-3-oxobutanoate 8 in refluxing EtOH to afford the respective thiazole ester 9 (Scheme 2). The 1H-NMR spectrum exhibited two broad singlet signals of two NH protons at δ 8.82 and 9.54 ppm, 2signals at δ = 1.24 (CH3, t) and 4.18 (CH2, q) ppm corresponding to ethyl group, and five singlet signals assignable for 5 CH3 groups at δ = 2.08, 2.25, 2.28, 2.32 and 2.35 ppm, in addition to the three aromatic protons.
Also by analogy, when compound 3 was reacted with p-substituted phenacyl bromide derivatives 10a, b, it afforded the products 11a, b as inferred from their spectral data and elemental analysis (Scheme 2). The 1H-NMR spectra of compound 11a showed the expected three singlet signals for the 3CH3 at δ 2.27, 2.35, 2.42 ppm, multiplet signal at δ 7.07–8.15 ppm (8H), and also two broad singlet signals at δ 8.41 and 9.87 ppm due to 2NH groups. Its IR spectra revealed two NH absorption bands at υ = 3394 and 3238 cm−1. Moreover, the mass of compounds 11a, b determined by mass spectrometry is equal to the calculated values.
In addition, the reaction of compound 3 with ethyl 2-chloroacetate 12 was also studied aiming to prepare new bioactive thiazolone derivative. Thus, refluxing compound 3 with 12 in AcOH\AcONa yielded thiazolone 13 based on spectral data (1H-NMR, IR and mass) (Scheme 2). The mass spectrum of compound 13 exhibited peak at m/z 392. Its 1H-NMR spectrum showed the existence of a singlet signal at δ = 4.27 ppm referred to the CH2 protons of thiazolone ring, in addition to the signals of 3 methyl groups, 3 thiophene protons and the two NH protons (see Experimental).
Next, our study was extended to prepare bioactive N-aminothiazole derivatives from reaction of thiocarbohydrazone 5 with α-halo-compounds, thus when thiocarbohydrazone derivative 5 was allowed to react with 3-chloropentane-2,4-dione 6 and ethyl 2-chloro-3-oxobutanoate 8 in refluxing EtOH afford the respective N-aminothiazole derivatives 14 and 15 as depicted in Scheme 3. The chemical structures of derivatives 14 and 15 were elucidated by IR, 1H-NMR and MS. For example, the IR spectrum of derivative 15 showed the stretching bands at υ = 3464, 3326, 3176 cm−1 which are attributed to the NH and NH2 groups, in addition to another band for the conjugated ester carbonyl group at υ = 1697 cm−1. The mass spectrum of derivatives 14 and 15 showed a molecular ion peaks at m/z = 447, 477 which are compatible with C19H23N7O2S3 and C18H21N7OS3, respectively. 1H-NMR spectrum of compound 15 showed two signals at δ = 3.51and 9.88 ppm attributed to the NH2 and NH protons, in addition to the expected signals of the aromatic protons, ester group and 4 methyl groups (see Experimental).
Analogously, the thiocarbohydrazone 5 was allowed to react with phenacyl bromides 10a, b to yield N-aminothiazole derivatives 16a, b as final product (Scheme 3). The chemical structures of the derivatives 16a, b were confirmed from the spectral data and elemental analyses. The 1H-NMR spectra of compound 16a showed the expected three singlet signals for the 3CH3 at δ 2.09, 2.24, 2.41 ppm, multiplet signal at δ 7.01–8.06 ppm (8H), and also two broad singlet signals at δ 3.47 and 11.50 ppm due to NH2 and NH groups, respectively. Moreover, the molecular weight determination of the products 16a, b was observed in the expected region.
Finally, compound 5 was reacted with ethyl 2-chloroacetate 12 in AcOH\AcONa under reflux to afford N-aminothiazolone derivative 17 as presented in Scheme 3. The 1H-NMR spectra of compound 17 showed seven signals at δ 2.16, 2.33, 2.69 (3s, 3CH3), 3.39 (br.s, NH2), 4.12 (s, CH2), 7.09–7.54 (m, 3Ar-H), 11.89 (br.s, NH) ppm.
Anticancer Activity
As a continuation of our studies on 1.3-thiazole and thiophene derivatives as anticancer agents61–71 and in a search for new anticancer drugs, we herein report the synthesis of a series of new thiazolyl-hydrazono-ethylthiazoles incorporating thiophene moiety and evaluation for their anticancer activities. Thiosemicarbazones (TSCs) have a wide clinical antitumor spectrum with efficacy in various tumor types such as leukemia, breast cancer, pancreatic cancer, non-small cell lung cancer, cervical cancer, prostate cancer and bladder cancer. Several possible mechanisms have been implemented for the anticancer activity of thiosemicarbazones.72
Recently, some studies have reported that Rab7b overexpression accelerated the proliferation and growth of human breast cancer (MCF-7) cells.46 Rab7b protein is selected as promising target to identify potentially inhibitors as breast cancer therapeutics. Thus, the pharmacological activities of the synthesized thiosemicarbazone 3, thiocarbohydrazone 5, and thiazoles 7, 9, 11a, b, 13, 14, 15, 16a, b, and 17 were investigated for their MCF-7 using colorimetric MTT assay and reference drug as Cisplatin. The data have been used to plot an exposure response curve in which the concentrations needed to kill half of the cell population (IC50) of the tested samples were calculated. Cytotoxic activities were expressed as the mean IC50 of three independent experiments (see Figure S3a and S3b in the supplementary data section). The results are represented in Figure 2 and Table 1.
 |
Table 1 In vitro Cytotoxic Activity of the Newly Synthesized Compounds 3, 5, 7, 9, 11a, b, 13, 14, 15, 16a, b, and 17 Against MCF-7 |
 |
Figure 2 The most active compounds towards the MCF-7 cell line. |
The results showed that all derivatives displayed concentration-dependent inhibitory effect against tumor cells. Compounds 9 and 11b have promising cytotoxic activity (IC50 < 30 µM which is close to Cisplatin reference drug), and compounds 3, 7, 11a and 16b exhibited medium activity (IC50 ~ 30–80 µM) while the rest compounds 5, 13, 14, 15, 16a and 17 are inactive (IC50 > 80 µM).
Among the thiazole derivatives, thiazole 9 and 11b (IC50 = 14.6 ± 0.8 and 28.3 ± 1.5 µM, respectively) showed promising cytotoxic activity close to Cisplatin (IC50 = 13.6 ± 0.9 µM) towards the MCF-7 cell line. The cytotoxic activity of thiosemicarbazone derivative 3 (IC50 = 76.4 ± 0.52 µM) ⋙ thiocarbohydrazone derivative 5 (IC50 = 292.1 ± 6.3µM) due to the presence of amino group (electron-donating group). Generally, 1.3-thiazole derivatives have more cytotoxic activity than the respective N-amino-1,3-thiazole derivatives (7, 9, 11a, 11b > 14, 15, 16a, 16b) due to the presence of amino group. Finally, compound 11b has more cytotoxic activity than 11a and also N-amino-1,3-thiazole derivative 16b has more cytotoxic activity than 16a may due to the presence of nitro group (electron-withdrawing group) at position 4 of the phenyl ring.
Molecular Docking Studies
In silico modeling, study73 is essential to recognize the mechanism of actions of the synthetic compound against Ra7b protein. The FASTA sequence of the target (accession no: Q96AH8) was obtained from NCBI. The reference structure used for protein structure prediction is the crystal structure of the REP-1 protein in complex with monoprenylated Rab7b protein (PDB: 1VG0), which was obtained by using BLASTp (Basic Local Alignment Search Tool) webserver. The homology model of the target protein Rab7b was generated using Modeller 9.11 software. Twenty-five protein models were generated, and the best model was selected for further validation. The 3D protein structure was prepared for docking process via removing of water molecules, addition and elimination of polar hydrogen atoms. The homologue model of the target (see Figure S4, in the supplementary data section), consists of 8 α-helices and 6 β-strands obtaining from PDBsum server74 as represented in Figure S5, in the supplementary data section.
The physicochemical properties of the target protein were calculated using ProtParam tool.75 The protein sequence has 199 amino acid residues with molecular weight 22.510 kDa. The most abundant amino acid residues are LEU, ILE, SER, VAL, LYS, ASP, GLU, and GLN, respectively, in high percentages in Rab7b, as declared in Figure. Leucine has the highest abundance (10.6%), and Tryptophan has the lowest abundance (1.5%), as represented in Figure S6, in the supplementary data section. The physicochemical parameters predicted a negatively charged protein as the result of the high number of negatively charged residues (aspartic acid 6.5% and glutamic acid 6%) in contrast with positively charged ones (Arginine 5% and lysine 7%). Further, the molecular formula of Rab7b is C1007H1607N267O298S9. The atomic composition of the target protein is 3188, with 1007 carbon (C), 1607 hydrogen (H), 267 nitrogen (N), and 9 sulfur (S). In addition, the protein is acidic, with an isoelectric point (pI) of 6.31. The estimated half-life time of Rab7b represented that it can remain intact without being degraded for 30 h in humans, less than 20 h in yeast, and less than 10 h in E.coli, and its extinction coefficient is 26,930 M−1 cm−1. Finally, the generated aliphatic index was 98.44, with grand average of hydrophobicity (GRAVY) of −0.129 and instability index was computed to be 36.48, which indicates that the protein is stable.
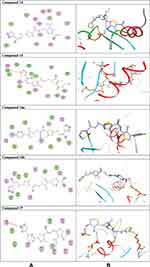
Once generated, the model was validated using Ramachandran plot, to check its stereochemical quality. RC plot (see Figure S7, in the supplementary data section) represents 89.4% (160 aa) of the total residues in most favored regions and 10.6% (19 aa) in additional allowed regions, indicating reasonable quality model. The active site prediction tools declared that the amino acid residues Gly18, Gly20, Lys21, Thr22, Ser23, Ala43, Ser44, Asp63, Glu68, and Lys125 are crucial for Rab7b protein. The grid box76 was then allocated over the predicted binding site region for specific docking, with dimension of 25 Aº X 25 Aº X 25 Aº, as shown in Figure S8, in the supplementary data section. Docking screening was performed for the compounds into active site of the target Rab7b, using PyRx virtual screening tool. Ligand–protein interactions are depicted in Figure 3. Table 2 shows the estimated binding energies which are in the range of – 6.7 to −5.0 kcal/mol. The molecule with the lowest binding energy (ie more negative) indicates highest binding affinity to the target protein.77,78 From the data gotten, the compounds exhibited respectable fitting to the binding pocket of the protein, through a network of non-covalent interactions like hydrogen bonds and π-cation. Compound 1 revealed binding energy of −5.4 kcal/mol and formed two hydrogen bonds with Thr22 and Ser23 at the distances of 2.78 and 2.98 Aº, respectively. Compound 3 displayed binding energy of −5.2 kcal/mol besides forming hydrogen bonding interactions Thr22 and Asp63 in distances 2.86 and 2.22 Aº, respectively. In addition, compound 5 interacted with the target through Asp63, Thr22 and Glu68 at distances of 1.97, 2.20 and 2.37 Aº, respectively.
 |
Table 2 Molecular Docking Results for the Screened Compounds and Rab7b |
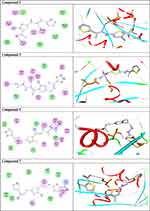 |
Figure 3a (Continued). |
 |
Figure 3b (Continued). |
Compound 7 was docked to the target through one H-bond with the residue Ser44. Moreover, compound 9 exhibited four H-bond interactions with the amino acid residues Gly18, Gly20, Lys21, and Thr22. The derivatives 11a (−5.8 kcal/mol) and 11b (−5.1 kcal/mol) exhibited H-bonds with Thr22 and Ser23, respectively. Introducing of electron-withdrawing groups such as -NO2 (strong) and -Cl (weak) on phenyl ring causing more cytotoxic activity of the compounds.79 The compound 13 represented two H-bond interactions with Thr22 at distances of 2.95 and 2.96 Aº, respectively. Compound 14 was successfully docked to the target protein with maximum binding energy - 6.7 kcal/mol, and exhibited two H-bond interactions with Gly20 and Glu68 at 2.66 and 2.31 Aº, respectively. Compound 15 interacted with the target at the residues Ser23, Ala43 and Lys125, forming three hydrogen bonds and one π-cation interactions at distance of 2.81, 2.98, 3.09 and 3.89 Aº, respectively. The derivative 16a (−6.3 kcal/mol) exhibited one H-bond with Thr22 at 2.57 Aº, while derivative 16b (−6.2 kcal/mol) showed two H-bonds with the amino acid residues Lys125 and Gly18. Finally, compound 17 showed hydrogen bonding and π-cation interactions with Ser23, Ala43, Asp63, and Lys125 at the distances of 2.95, 2.31, 2.96, 2.39 and 3.64 Aº, respectively. To further understand the nature of π-cation interactions; lysine Lys125 contains a positively charged amino on its side-chain (H3N+) that is involved in forming π-cation interactions with compounds 15 and 17. The synthetic thiazole derivatives 1–17 with heterocyclic moieties like thiophene and thiazole are noted to be common pharmacophore groups, which interact with the binding site pocket of the target Rab7b, through non-covalent interactions. The in silico molecular docking study results revealed that all the synthesized compounds having minimum binding energy and good affinity towards the active site pocket, thus, they may be considered as good drug-like small molecules for cancer treatment. On the other hand, the rule of 5 “RO5” methodology is an important way in defining drugability. The results tabulated in Tables 3 and 4 indicated that the synthesized compounds fit well with Lipinski rule of five (RO5).80 The results show that (a) all newly synthetic thiazole compounds have molecular weights within the limits of 180–500 g/mol, except 16a and 16b (b) the compounds have H-bond donating ability <6 (c) the compounds have H-bond accepting ability in the acceptable range (d) the topological polar surface area (TPSA) was found in the acceptable range ≤140 (e) the logp of the compounds indicates that they are not very lipophilic <5, except 11a, 11b and 16a. Also, ADMET properties declare that the newly synthetic analogues have better Human Intestinal Absorption (HIA) score, and good Blood-Brain Barrier (BBB) values, which means that they could be better absorbed by the human intestine.81 In addition, they showed negative toxicity and negative carcinogenicity test. As the newer compounds exhibited the fewest violations of Lipinski rule of five (RO5), our findings suggest that our synthesized compounds 1–17 could be pharmacologically efficient for preclinical use.
 |
Table 3 List of ADME Properties of Synthesized Molecules 1–17. The Pharmacokinetic Properties of Compounds are Predicted by Admet SAR Tool |
Conclusion
In this context, we herein present an efficient synthesis of a novel series of thiazole linked thiophene conjugates in good yields. The products were screened for their cytotoxic activity against MCF-7 cells and the results obtained showed that derivatives 9 and 11b have promising activity (IC50 = 14.6 ± 0.8 and 28.3 ± 1.5 µM, respectively) compared to Cisplatin (IC50 = 13.6 ± 0.9 µM). The molecular docking analysis reveals that the synthesized compounds are predicted to be fit into the binding site of the target Rab7b. In summary, the synthetic thiazole compounds 1–17 could be used as potent inhibitors as anticancer drugs.
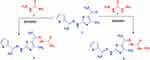 |
Scheme 1 Synthesis of thiosemicarbazone 3 and thiocarbohydrazone 5. |
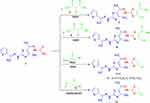 |
Scheme 2 Synthesis of thiazole derivatives 7, 9, 11a, b and 13. |
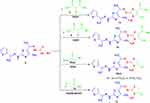 |
Scheme 3 Synthesis of thiazoles 14, 15, 16a, b and 17. |
Acknowledgments
The financial support by the Deanship of Scientific Research (Project Number 86), Islamic University, Saudi Arabia is gratefully acknowledged.
Author Contributions
All the authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or all in these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work and declare that there is no conflict of interests regarding the publication of this paper.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi:10.3322/caac.21254
2. Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;5:364–378. doi:10.1016/j.ejphar.2014.07.025
3. Harit T, Bellaouchi R, Asehraou A, Rahal M, Bouabdallah I, Malek F. Synthesis, characterization, antimicrobial activity and theoretical studies of new thiophene-based tripodal ligands. J Mol Struct. 2017;1133:74–79. doi:10.1016/j.molstruc.2016.11.051
4. Mishra P, Middha A, Saxena V, Saxena A. Synthesis and evaluation of anti-inflammatory activity of some cinnoline derivatives-4 (−2-amino-thiophene) cinnoline-3-carboxamide. J Pharm Biosci. 2016;4:64–68. doi:10.20510/ukjpb/4/i3/108388
5. Mathew B, Suresh J, Anbazhagan S. Synthesis, in silico preclinical evaluation, antidepressant potential of 5-substituted phenyl-3-(thiophen-2-yl)-4, 5-dihydro-1H-pyrazole- 1-carboxamides. Biomed Aging Pathol. 2014;4:327–333. doi:10.1016/j.biomag.2014.08.002
6. Ashour HM, Shaaban OG, Rizk OH, El-Ashmawy IM. Synthesis and biological evaluation of thieno[2′, 3′: 4, 5]pyrimido[1, 2-b][1, 2, 4]triazines and thieno [2,3-d][1, 2, 4] triazolo [1, 5-a]pyrimidines as anti-inflammatory and analgesic agents. Eur J Med Chem. 2013;62:341–351. doi:10.1016/j.ejmech.2012.12.003
7. Thirumurugan R, Sriram D, Saxena A, Stables J, Yogeeswari P. 2,4-Dimethoxyphenyl- semicarbazones with anticonvulsant activity against three animal models of seizures: synthesis and pharmacological evaluation. Bioorg Med Chem. 2006;14:3106–3112. doi:10.1016/j.bmc.2005.12.041
8. Archna P, Chawla S. Thiophene-based derivatives as anticancer agents: an overview on decade’swork. Bioorg Chem. 2020;101:1040262. doi:10.1016/j.bioorg.2020.104026
9. Gul HI, Yamali C, Sakagami H, et al. New anticancer drug candidates sulfonamides as selective hCA IX or hCA XII inhibitors. Bioorg Chem. 2018;77:411–419.
10. Kasibhatla S, Kuemmerle J, Kemnitzer O-MW, et al. Discovery and structure-activity relationship of 3-aryl-5-aryl-1, 2, 4-oxadiazoles as a new series of apoptosis inducers and potential anticancer agents. J Med Chem. 2005;48:5215–5223. doi:10.1021/jm050292k
11. Milik SN, Abdel-Aziz AK, Lasheen DS, Serya RA, Minucci S, Abouzid KA. Surmounting the resistance against EGFR inhibitors through the development of thieno [2, 3-d] pyrimidine-based dual EGFR/HER2 inhibitors. Eur J Med Chem. 2018;155:316–336. doi:10.1016/j.ejmech.2018.06.011
12. Hirsch FR, Witta S. Biomarkers for prediction of sensitivity to EGFR inhibitors in non-small cell lung cancer. Curr Opin Oncol. 2005;17:118–122. doi:10.1097/01.cco.0000155059.39733.9d
13. Gulipalli KC, Bodige S, Ravula P, et al. Design, synthesis, in silico and in vitro evaluation of thiophene derivatives: a potent tyrosine phosphatase 1B inhibitor and anticancer activity. Bioorg Med Chem Lett. 2017;27:3558–3564. doi:10.1016/j.bmcl.2017.05.047
14. Cappellacci L, Grifantini M, Barzi A, et al. Furanfurin and thiophenfurin: two novel tiazofurin analogues. Synthesis, structure, antitumour activity, and interactions with inosine monophosphate dehydrogenase. J Med Chem. 1995;38:3829–3837. doi:10.1021/jm00019a013
15. Li X, He Y, Ruiz CH, Koenig M, Cameron MD. Characterization of dasatinib and its structural analogs as CYP3A4 mechanism-based inactivators and the proposed bioactivation pathways. Drug Metab Dispos. 2009;37:1242–1250. doi:10.1124/dmd.108.025932
16. Hu-Lieskovan S, Mok S, Homet Moreno B, et al. Improved antitumour activity of immunotherapy with B-RAF and MEK inhibitors in BRAF (V600E) melanoma. Sci Transl Med. 2015;18(279):41–279.
17. Yao Y, Chen S, Zhou X, Xie L, Chen A. 5-FU and ixabepilone modify the microRNA expression profiles in MDA-MB-453 triple-negative breast cancer cells. Oncol Lett. 2014;7:541–547. doi:10.3892/ol.2013.1697
18. Altmann KH. Epothilone B and its analogs - a new family of anticancer agents. Mini Rev Med Chem. 2003;3:149–158.
19. Sharma PC, Bansal KK, Sharma A, Sharma D, Deep A. Thiazole-containing compounds as therapeutic targets for cancer therapy. Eur J Med Chem. 2020;188:112016.
20. Cascioferro S, Parrino B, Carbone D, et al. Thiazoles, their benzofused systems, and thiazolidinone derivatives: versatile and promising tools to combat antibiotic resistance. J Med Chem. 2020;63:7923–7956. doi:10.1021/acs.jmedchem.9b01245
21. Ayati A, Emami S, Asadipour A, Shafiee A, Foroumadi A. Recent applications of 1,3-thiazole core structure in the identification of new lead compounds and drug discovery. Eur J Med Chem. 2015;97:699–718. doi:10.1016/j.ejmech.2015.04.015
22. Sayed AR, Gomha SM, Taher EA, et al. Synthesis of novel thiazoles as potential anti- cancer agents. Drug Des Devel Ther. 2020;14:1363–1375. doi:10.2147/DDDT.S221263
23. Das D, Sikdar P, Bairagi M. Recent developments of 2-aminothiazoles in medicinal chemistry. Eur J Med Chem. 2016;109:89–98. doi:10.1016/j.ejmech.2015.12.022
24. Lu Y, Li CM, Wang Z, et al. Design, synthesis, and SAR studies of 4-substituted methoxylbenzoyl-arylthiazoles analogues as potent and orally bioavailable anticancer agents. J Med Chem. 2011;54:4678–4693. doi:10.1021/jm2003427
25. Chowdhury A, Patel S, Sharma A, Das A, Meshram P, Shard A. A perspective on environmentally benign protocols of thiazole synthesis. Chem Heterocycl Comp. 2020;56:455–463. doi:10.1007/s10593-020-02680-x
26. Braga SFP, Fonseca NC, Ramos JP, Fagundes EMS, Oliveira RB. Synthesis and cytotoxicity evaluation of thiosemicarbazones and their thiazole derivatives. Braz J Pharm Sci. 2016;52:299–308. doi:10.1590/S1984-82502016000200008
27. Shaik SP, Nayak VL, Sultana F, et al. Design and synthesis of imidazo[2,1-b]thiazole linked triazole conjugates: microtubule-destabilizing agents. Eur J Med Chem. 2017;126:36–51. doi:10.1016/j.ejmech.2016.09.060
28. Cascioferro S, Petri GL, Parrino B, et al. Imidazo[2,1-b] [1,3,4]thiadiazoles with antiproliferative activity against primary and gemcitabine-resistant pancreatic cancer cells. Eur J Med Chem. 2020;189:112088. doi:10.1016/j.ejmech.2020.112088
29. Cascioferro S, Petri GL, Parrino B, et al. 3-(6-phenylimidazo[2,1-b][1,3,4]thiadiazol-2-yl)-1H-indole derivatives as new anticancer agents in the treatment of pancreatic ductal adenocarcinoma. Molecules. 2020;25:329. doi:10.3390/molecules25020329
30. Zhao MY, Yin Y, Yu X, et al. Synthesis, biological evaluation and 3D-QSAR study of novel 4,5-dihydro-1H-pyrazole thiazole derivatives as B-RAF (V600E) inhibitors. Bioorg Med Chem. 2015;23:46–54. doi:10.1016/j.bmc.2014.11.029
31. Andreani A, Burnelli S, Granaiola M, et al. New antitumour imidazo[2,1-b]thiazole guanylhydrazones and analogues. J Med Chem. 2008;51:809–816.
32. Gomha SM, Edrees MM, Altalbawy F. Synthesis and characterization of some new bis-pyrazolyl-thiazoles incorporating the thiophene moiety as potent antitumor agents. Int J Mol Sci. 2016;17:1499. doi:10.3390/ijms17091499
33. Mohareb RM, Abdallah AE. Synthesis and cytotoxicity evaluation of thiazole derivatives obtained from 2-amino-4, 5, 6, 7-tetrahydrobenzo [b] thiophene-3-carbonitrile. Acta Pharmaceutica. 2017;67:495–510. doi:10.1515/acph-2017-0040
34. Mohareb RM, Fleita DH, Sakka OK. Novel synthesis of hydrazidehydrazone derivatives and their utilization in the synthesis of coumarin, pyridine, thiazole and thiophene derivatives with antitumour activity. Molecules. 2011;16:16–27. doi:10.3390/molecules16010016
35. Zhen Y, Stenmark H. Cellular functions of Rab GTPases at a glance. J Cell Sci. 2015;128:3171–3176. doi:10.1242/jcs.166074
36. Pei G, Bronietzki M, Gutierrez MG. Immune regulation of Rab proteins expression and intracellular transport. J Leukoc Biol. 2012;92:41–50. doi:10.1189/jlb.0212076
37. Martinez O, Goud B. Rab proteins. Biochim Biophys Acta. 1998;1404:101–112. doi:10.1016/S0167-4889(98)00050-0
38. Progida C, Cogli L, Piro F, De Luca A, Bakke O. Rab7b controls trafficking from endosomes to the TGN. J Cell Sci. 2010;123:1480–1491. doi:10.1242/jcs.051474
39. Progida C, Nielsen MS, Koster G, Bucci C, Bakke O. Dynamics of Rab7b-dependent transport of sorting receptors. Traffic. 2012;13:1273–1285. doi:10.1111/j.1600-0854.2012.01388.x
40. BasuRay S. Rab7a: the master regulator of vesicular trafficking. Biomed Rev. 2014;25:67–81. doi:10.14748/bmr.v25.1049
41. Borg Distefano M, Hofstad HL, Wang Y. TBC1D5 controls the GTPase cycle of Rab7b. J of Cell Sci. 2018;131:jcs216630. doi:10.1242/jcs.216630
42. Guerra F, Bucci C. Multiple roles of the small GTPase Rab7. Cells. 2016;5:34. doi:10.3390/cells5030034
43. Abdelmonsef AH, Dulapalli R, Dasari T, Padmarao LS, Mukkera T, Vuruputuri U. Identification of novel antagonists for Rab38 protein by homology modeling and virtual screening. Comb Chem High Throughput Screen. 2016;19:875–892. doi:10.2174/1386207319666161026153237
44. Abdelmonsef AH. Computer-aided identification of lung cancer inhibitors through homology modeling and virtual screening. Egypt J Med Hum Genet. 2019;20:1–14.
45. Abdelmonsef AH, Mosallam AM. Synthesis, in vitro biological evaluation and in silico docking studies of new quinazolin‐2,4‐dione analogues as possible anticarcinoma agents. J Heterocycl Chem. 2020;57:1637–1654. doi:10.1002/jhet.3889
46. Xie J, Yan Y, Liu F, et al. Knockdown of Rab7a suppresses the proliferation, migration, and xenograft tumor growth of breast cancer cells. Biosci Rep. 2019;39:BSR20180480. doi:10.1042/BSR20180480
47. Rashdan HRM, Gomha SM, El-Gendey MS, El-Hashash MA, Soliman AMM. Eco-friendly one-pot Synthesis of some new pyrazolo[1,2-b]phthalazinediones with antiproliferative efficacy on human hepatic cancer cell lines. Green Chem Lett Rev. 2018;11:264–274. doi:10.1080/17518253.2018.1474270
48. Gomha SM, Riyadh SM, Mahmmoud EA, Elaasser MM. Synthesis and anticancer activities of thiazoles, 1,3-thiazines, and thiazolidine using chitosan-grafted-poly (vinylpyridine) as basic catalyst. Heterocycles. 2015;91:1227–1243. doi:10.3987/COM-15-13210
49. Dasari T, Kondagari B, Dulapalli R, et al. Design of novel lead molecules against RhoG protein as cancer target–a computational study. J Biomol Struct Dyn. 2017;35:3119–3139. doi:10.1080/07391102.2016.1244492
50. Hollingsworth SA, Karplus PA. A fresh look at the Ramachandran plot and the occurrence of standard structures in proteins. Biomol Concepts. 2010;1:271–283. doi:10.1515/bmc.2010.022
51. Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46(W1):W363–W367. doi:10.1093/nar/gky473
52. O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi:10.1186/1758-2946-3-33
53. Rappé AK, Casewit CJ, Colwell KS, Goddard WA, Skiff WM. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J Am Chem Soc. 1992;114:10024–10035. doi:10.1021/ja00051a040
54. Dallakyan S, Olson AJ. Small-Molecule Library Screening by Docking with PyRx. Chem Biol Springer. 2015;1263:243–250.
55. El-Naggar M, Mohamed ME, Mosallam AM, Salem W, Rashdan HR, Abdelmonsef AH. Synthesis, characterization, antibacterial activity, and computer-aided design of novel quinazolin-2,4-dione derivatives as potential inhibitors against vibrio cholerae. Evol Bioinform. 2020;16:1–13. doi:10.1177/1176934319897596
56. Aboubakr HA, Lavanya SP, Thirupathi M, Rohini R, Sarita RP, Uma V. Human Rab8b protein as a cancer target - An in silico study. J Comput Sci Syst Biol. 2016;9:132–149.
57. Sahu S, Ghosh SK, Gahtori P, Pratap SU, Bhattacharyya DR, Bhat HR. In silico ADMET study, docking, synthesis and antimalarial evaluation of thiazole-1,3,5-triazine derivatives as Pf-DHFR inhibitor. Pharmacol Rep. 2019;71:762–767. doi:10.1016/j.pharep.2019.04.006
58. Wang Y, Xing J, Xu Y, et al. In silico ADME/T modelling for rational drug design. Q Rev Biophys. 2015;48:488–515. doi:10.1017/S0033583515000190
59. Cheng F, Li W, Zhou Y, et al. A comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model. 2012;52:3099–3105. doi:10.1021/ci300367a
60. Makam P, Thakur PK, Kannan T. In vitro and in silico antimalarial activity of 2-(2-hydrazinyl) thiazole derivatives. Eur J Pharmaceut Sci. 2014;52:138–145. doi:10.1016/j.ejps.2013.11.001
61. Gomha SM, Riyadh SM, Mahmmoud EA, Elaasser MM. Chitosan-grafted-poly (4-vinylpyridine) as a novel copolymer basic catalyst for synthesis of arylazothiazoles and 1,3,4-thiadiazoles under microwave irradiation. Chem Heterocycl Compd. 2015;51:1030–1038. doi:10.1007/s10593-016-1815-9
62. Gomha SM, Abdelrazek FM, Abdelrahman AH, Metz P. Synthesis of some novel thiazole, thiadiazole and 1, 4-phenylene-bis-thiazole derivatives as potent antitumor agents. Heterocycles. 2016;92:954–967. doi:10.3987/COM-16-13443
63. Sayed AR, Gomha SM, Abdelrazek FM, Farghaly MS, Hassan SA, Peter M. Design, efficient synthesis and molecular docking of some novel thiazolyl-pyrazole derivatives as anticancer agents. BMC Chem. 2019;13:116. doi:10.1186/s13065-019-0632-5
64. Abdelhamid AO, Gomha SM, Abdelriheem NA, Kandeel SM. Synthesis of new 3-heteroarylindoles as potential anticancer agents. Molecule. 2016;21:929. doi:10.3390/molecules21070929
65. Gomha SM, Muhammad ZA, Abdel‐aziz MR, Abdel‐aziz HM, Gaber HM, Elaasser MM. One‐pot synthesis of new thiadiazolyl‐pyridines as anticancer and antioxidant agents. J Heterocycl Chem. 2018;55:530–536. doi:10.1002/jhet.3088
66. Gomha SM, Kheder NA, Abdelaziz MR, Mabkhot YN, Alhajoj AM. A facile synthesis and anticancer activity of some novel thiazoles carrying 1,3,4‑thiadiazole moiety. Chemistry Central J. 2017;11:25. doi:10.1186/s13065-017-0255-7
67. Gomha SM, Abdelaziz MR, Kheder NA, Abdel-aziz HM, Alterary S, Mabkhot YNA. Facile access and evaluation of some novel thiazole and 1,3,4-thiadiazole derivatives incorporating thiazole moiety as potent anticancer agents. Chemistry Central J. 2017;11:105. doi:10.1186/s13065-017-0335-8
68. Gomha SM, Edrees MM, Muhammad ZA, El-Reedy AAM. 5-(Thiophen-2-yl)-1,3,4- thiadiazole derivatives: synthesis, molecular docking and in-vitro cytotoxicity evaluation as potential anticancer agents. Drug Des Devel Ther. 2018;12:1511–1523. doi:10.2147/DDDT.S165276
69. Gomha SM, Abdelhamid AO, Kandil OM, Kandeel SM, Abdelrehem NA. Synthesis and molecular docking of some novel thiazoles and thiadiazoles incorporating pyranochromene moiety as potent anticancer agents. Mini-Rev Med Chem. 2018;18:1670–1682. doi:10.2174/1389557518666180424113819
70. Edrees MM, Abu-Melha S, Saad AM, Kheder NA, Gomha SM, Muhammad ZA. Eco-friendly synthesis, characterization and biological evaluation of some new pyrazolines containing thiazole moiety as potential anticancer and antimicrobial agents. Molecules. 2018;23:1970. doi:10.3390/molecules23112970
71. Abu-Melha S, Edrees MM, Salem HH, Kheder NA, Gomha SM, Abdelaziz MR. Synthesis and biological evaluation of some novel thiazole-based heterocycles as potential anticancer and antimicrobial agents. Molecules. 2019;24:539. doi:10.3390/molecules24030539
72. Shakya B, Yadav PN. Thiosemicarbazones as potent anticancer agents and their modes of action. Mini-Rev Med Chem. 2020;20:638–661.
73. Muhammed MT, Aki-Yalcin E. Homology modeling in drug discovery: overview, current applications, and future perspectives. Chem Biol Drug Des. 2019;93:12–20. doi:10.1111/cbdd.13388
74. Laskowski RA. PDB sum: summaries and analyses of PDB structures. Nucleic Acids Res. 2002;29:221–222. doi:10.1093/nar/29.1.221
75. Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi:10.1385/1-59259-584-7:531
76. Shehadi IA, Rashdan HRM, Abdelmonsef AH. Homology modeling and virtual screening studies of antigen MLAA-42Protein: identification of novel drug candidates against leukemia-an in silico approach. Comput Math Methods Med. 2020;12.
77. Yuriev E, Agostino M, Ramsland PA. Challenges and advances in computational docking: 2009 in review. J Mol Recognit. 2011;24:149–164. doi:10.1002/jmr.1077
78. Henrich S, Feierberg I, Wang T, Blomberg N, Wade RC. Comparative binding energy analysis for binding affinity and target selectivity prediction. Proteins. 2010;78:135–153. doi:10.1002/prot.22579
79. Haredi AA, Eldeeb MM, El-Naggar M, Temairk H, Mohamed MA. Novel quinazolin-2,4-dione hybrid molecules as possible inhibitors against malaria: synthesis and in silico molecular docking studies. Front Mol Biosci. 2020;7. doi: 10.3389/fmolb.2020.00105.
80. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and develop ment settings. Adv Drug Deliv Rev. 1997;23:3–25. doi:10.1016/S0169-409X(96)00423-1
81. Pajouhesh H, Lenz GR. Medicinal chemical properties of successful central nervous system drugs. NeuroRx. 2005;2:541–553. doi:10.1602/neurorx.2.4.541
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.