Back to Journals » OncoTargets and Therapy » Volume 8
The XRCC1 Arg194Trp polymorphism is significantly associated with lung adenocarcinoma: a case-control study in an Eastern European Caucasian group
Authors Cătană A, Pop M, Domokos Hincu B, Pop IV, Petrişor FM, Porojan MD, Popp R
Received 14 July 2015
Accepted for publication 22 September 2015
Published 27 November 2015 Volume 2015:8 Pages 3533—3538
DOI https://doi.org/10.2147/OTT.S92361
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Faris Farassati
Andreea Cătană,1 Monica Pop,2 Bianca Domokos Hincu,2 Ioan V Pop,1 Felicia M Petrişor,1 Mihai D Porojan,3 Radu A Popp1
1Department of Molecular Sciences, 2Department of Pneumology, 3Department of Internal Medicine, Iuliu Haţieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania
Abstract: DNA repair plays an important role in maintaining the integrity of the genome by repairing DNA damage induced by carcinogens. Certain genetic polymorphisms that occur in DNA-repair genes may affect the ability to repair DNA defects, and may represent a risk factor in carcinogenesis. The gene XRCC1 is involved in DNA repair. The purpose of our study was to investigate the association between XRCC1 Arg194Trp and Arg399Gln polymorphisms and the risk of lung cancer in a Romanian population. We recruited 222 healthy controls and 102 patients with lung cancer. Genotypes were determined by multiplex polymerase chain-reaction restriction fragment-length polymorphism. Statistical analysis (odds ratio, recessive model) revealed an increased risk for lung cancer for the homozygous 194Trp genotype (χ2=0.186, odds ratio 10.667, 95% confidence interval 1.309–86.933; P=0.007). Also, we found an association between the 194Trp allele and women with lung adenocarcinoma. In conclusion, the results of the study place the XRCC1 Arg194Trp polymorphism among independent risk factors for developing lung cancer.
Keywords: lung cancer, XRCC1 Arg194Trp, XRCC1 Arg399Gln
Introduction
Lung cancer, the most common type of cancer, is nowadays the main cause of cancer deaths for men and women worldwide.1 Over the last few years, the incidence of lung cancer has increased steadily.2 The risk of lung cancer has been associated with different kinds of environmental and genetic factors. Exposure to environmental carcinogens and cigarette smoking are considered major etiologic factors for lung cancer.1
It is well known that DNA repair plays an important role in ensuring the stability of the genome by repairing DNA damage induced by exogenous and endogenous carcinogens. It is accepted that polymorphisms that occur in DNA-repair genes may affect the ability to repair DNA defects, and may represent a risk factor in different malignancies, because of the change of base-excision repair (BER) functions.3 XRCC1 is a crucial gene involved in DNA repair, specifically in the BER pathway. BER, nucleotide-excision repair, and double-strand-break repair are the main DNA-repair pathways described. Previous reports have indicated that certain genetic polymorphisms, particularly the XRCC1-gene variants, were associated with a high risk of malignancy, such as primary lung cancer,3,4 hepatocellular carcinoma,5 cervical cancer,6 childhood acute lymphoblastic leukemia,7 and gastric cancer.8
A genome-wide association study of 1,154 ever-smoking non-small-cell lung cancer patients, with genotyping of 317,498 tagging single-nucleotide polymorphisms and 1,137 ever-smoking controls in a Texas population of self-reported European descent, identified new DNA-repair polymorphisms associated with lung cancer.9 A newer, more comprehensive analysis of genome-wide association-study data of the DNA repair-capacity phenotype and genotypes with a much large sample size has confirmed the important role of genetic variation of DNA-repair genes and their role in lung neoplasia.10 The most common polymorphisms of the XRCC1 gene are Arg194Trp (A194T, rs1799782), Arg280His (A280G, rs25489), and Arg399Gln (A399G, rs25487). DNA-repair capacity may be affected by these polymorphisms, and thus it may modulate cancer susceptibility.
Over the last few years, case-control studies have been performed to study the association between the XRCC1 Arg194Trp and Arg399Gln polymorphisms and the risk of lung cancer.1,11 The purpose of this study was to investigate whether the XRCC1 Arg194Trp and Arg399Gln polymorphisms were involved in susceptibility to lung cancer in a Romanian population.
Materials and methods
Patients and controls
The current study was approved by the ethics committee of the conducting institution (Iuliu Haţieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania). The study was conducted according to the Helsinki Declaration. Written informed consent was obtained from all subjects included in the present study.
A group of 324 individuals were included in the study (Caucasian subjects of Romanian and Hungarian ethnicity). The case group comprised 102 cases diagnosed with lung cancer. Lung cancer was confirmed by imaging (computed tomography scan), and histopathological examination and tumor-subtype classification were done according to World Health Organization criteria.12–18 The controls included 222 healthy volunteers with no history of any malignancy. All study participants were or had been active smokers, with an average tobacco consumption between 15 and 40 cigarettes a day for at least 10 years.
Genotypic analysis of XRCC1 gene
Genomic DNA was extracted from 300 μL venous blood samples using a Wizard® genomic DNA purification kit (Promega Corporation, Fitchburg, WI, USA) and ZymoBead™ genomic DNA kit (Zymo Research Corporation, Irvine, CA, USA). Arg194Trp and Arg399Gln polymorphisms of the XRCC1 gene were genotyped using the multiplex polymerase chain-reaction restriction fragment-length polymorphism technique. A total of 100 ng of genomic DNA was amplified in a total volume of 25 μL reaction mixture containing reaction buffer of 1.5 nM MgCl2, 10 pmol of each primer, and 200 μm of each deoxyribose nucleoside triphosphate, and 0.5 unit of Taq polymerase. Thermocycling conditions were carried out as follows: 94°C for 5 minutes and then 30 cycles of 94°C for 30 seconds, 62°C for 1 minute, 72°C for 45 seconds, and a final polymerization step at 72°C for 5 minutes (Mastercycler® Gradient; Eppendorf AG, Hamburg, Germany).
Genotyping analysis was interpreted according to the following criteria: the common allele corresponding with Arg at codon 194, resulting in a 293 bp fragment, and the variant allele corresponding to Trp in a fragment of 313 bp. For codon 399 of the XRCC1 gene, the presence of 375 and 240 bp fragments, respectively, characterized the common allele corresponding to Gln, while the variant allele corresponding to Arg was defined by the presence of the undigested 615 bp fragment. Genotypic analysis of the XRCC1 gene was adapted after a protocol performed by Abdel-Rahman et al in 2000.19
Statistical analysis
The distribution of genotype and allele frequency of each XRCC1 polymorphism between different groups was compared by Fisher’s exact test, followed by comparative analysis according to dominant and recessive models. For estimation of the relative risk and strength of association, we calculated odds ratios (ORs) at a 95% confidence interval (CI). We considered statistically significant a P-value <0.05. Statistical analysis was carried out using SPSS 18.0 for Windows (SPSS Inc, Chicago, Il, USA).
Results
The common tumor type identified for the lung cancer group was squamous cell carcinoma (69.6%), with a frequency of 12.7% in women and 87.3% in men, followed by adenocarcinoma (18.6%), at 26.3% in women and 73.7% in men. Small-cell squamous carcinoma was found in 11.7% of patients. The average age at diagnosis was 62.2 years for squamous carcinoma (CI 60.4–63.9 years) and 60.3 years (CI 53.8–66.9 years) for small-cell lung cancer. The lowest average age of onset of clinical symptoms was recorded in patients with adenocarcinoma: 58 years (CI 53.9–64.7 years). Genotype distribution and frequency of alleles of the Arg194Trp and Arg399Gln polymorphisms of XRCC1 in patients with lung cancer and controls are presented in Table 1.
Comparative analysis (Fisher’s exact test) of dominant and recessive models for variant carriers of lung cancer risk is presented in Table 2. Comparative analysis to assess the risk for lung cancer in the study group compared with the control group for variant allele XRCC1 194Trp carriers (Fisher test analysis, ORs, dominant model), identified a statistical significance risk for the study group (χ2=0.135, OR 2.675, CI 0.963–7.432; P=0.052). Also, statistical comparative analysis (odd ratios, Fisher test for recessive model) revealed an increased risk for lung cancer for the homozygous 194Trp genotype (χ2=0.186, OR 10.667, CI 1.309–86.933; P=0.007). In this context, we can say that the results of the study place the XRCC1 Arg194Trp polymorphisms among independent risk factors for developing lung cancer.
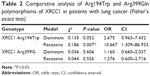  | Table 2 Comparative analysis of Arg194Trp and Arg399Gln polymorphisms of XRCC1 in patients with lung cancer (Fisher’s exact test) |
Comparative analysis to assess lung cancer risk in the study group compared with the control group for the variant Arg399Gln XRCC1 gene carriers (Fisher test analysis, ORs for dominant model) did not identify an increased risk for lung cancer (χ2=0.036, OR 1.160, CI 0.660–2.037; P=0.606). Statistical analysis according to Fisher’s exact test and ORs for the recessive model did not reveal any significant differences among patients and controls (χ2=0.044, OR 1.276, CI 0.600–2.716; P=0.526).
Comparative analysis of the Arg194Trp and Arg399Gln polymorphisms of the XRCC1 gene in patients with lung cancer according to sex and histopathological type revealed an increased frequency of the 194Trp allele in women with adenocarcinoma compared to males with adenocarcinoma (P=0.0003). There were no other statistically significant associations between the 194Trp and 399Gln alleles and different histopathological types of lung cancer (Table 3).
  | Table 3 Comparative analysis of Arg194Trp and Arg399Gln polymorphisms of XRCC1 gene in patients with lung cancer according to sex and histopathological type (Fisher’s exact test) |
Discussion
To our knowledge, this is the first study to perform a comparative analysis of the Arg194Trp and Arg399Gln polymorphisms of the XRCC1 gene in patients with lung cancer in Eastern Europe. Overall, our study provides evidence that the Arg194Trp polymorphism is associated with a high risk of lung cancer.
When we performed subgroup analyses, we observed that the XRCC1 Arg194Trp polymorphism was significantly associated with the risk of lung cancer. According to the literature, adenocarcinoma is most commonly found in women and young smokers, whereas in men and older smokers, first place is represented by small-cell lung cancer.20,21 David-Beabes and London observed that the 194Trp allele was associated with a significantly decreased risk of lung cancer among African–Americans.11
Statistical analysis revealed a frequency of 5.5% for the variant 194Trp allele of the XRCC1 gene, below those reported for the European and North American populations (11%–15%) and a frequency of 65% for the variant 399Arg allele codon for the XRCC1 gene, above the statistical limits reported in the literature (40%–65%).22 The results of the present research place our country among populations with the lowest frequency for the Arg194Trp variant allele of the XRCC1 gene.11,23
Our study showed that there was no association between the XRCC1 Arg399Gln polymorphism and the risk of lung cancer. Park et al found an association between the XRCC1 399Gln allele and increased risk of squamous cell carcinoma of the lung in male Korean patients (OR 1.66, 95% CI 0.99–2.79).3 Divine et al observed that the XRCC1 399Gln/Gln genotype was associated with a high risk of adenocarcinoma of the lung (OR 2.45, 95% CI 1.1–5.8).12 An association between the XRCC1 Arg399Gln polymorphism and risk for lung cancer in Asians has been found by Kiyohara et al; in contrast, no increase in this risk has been observed in Caucasians, highlighting that there is a particular distribution of allele variants in the European population.15
No relationship between lung cancer and the XRCC1 Arg399Gln polymorphism was observed by Hu et al or Hung et al.13,14 In a meta-analysis, Wang et al reported no association between the XRCC1 Arg399Trp polymorphism and lung cancer risk in the studied population.1 In the same meta-analysis, when studies were stratified by control source, they reported that the variant XRCC1 399Gln/Gln and Arg/Gln or Gln/Gln genotypes had a protective effect for lung cancer (OR 0.73, 95% CI 0.58–0.92, and OR 0.86, 95% CI 0.77–0.97, respectively). Another meta-analysis conducted by Kiyohara et al found no association between XRCC1 polymorphisms and an increased risk of lung cancer among Caucasians and Asians.15 No association was observed between an increased risk for the development of lung cancer in Turkish patients and the XRCC1 Arg399Gln polymorphism.17
Another meta-analysis from eight eligible studies in Chinese populations reported that variant genotypes of XRCC1 Arg399Gln might alter interindividual susceptibility to lung cancer.16 On the basis of the same meta-analysis, Zheng et al did not observe an association between the 194Trp and 280His alleles and lung cancer risk.16
For the XRCC1 Arg399Gln polymorphism, David-Beabes and London11 observed some evidence of a reduced risk for the homozygous variant genotype among heavier smokers (African–Americans, OR 0.3, 95% CI 0.0–2.9; Caucasians, OR 0.4, 95% CI 0.2–1.0).
In a recent meta-analysis of 44 case-control studies, the variant homozygous Trp/Trp genotype of codon 194 showed an increased risk for developing lung cancer (OR 1.19, 95% CI 1.01–1.39), especially in Asians (OR 1.21, 95% CI 1.02–1.43).24 Similar observations were reported by Jiang et al (OR 1.22, 95% CI 1.04–1.44).25 In contrast, another meta-analysis found a decreased lung cancer risk among subjects with a variant heterozygous genotype (Arg/Trp) for the XRCC1 Arg194Trp polymorphism (OR 0.88, 95% CI 0.79–0.97).1 Dai et al did not find a similar association in the total population, but did in a population-based control. They also found the presence of the heterozygous genotype Arg/Trp was associated with a reduced risk of non-small-cell lung cancer (OR 0.69).24
On the basis of a meta-analysis using 22 studies including 7,515 cases and 9,560 controls, Jiang et al failed to show an association between the XRCC1 Arg194Trp polymorphism and the risk of squamous cell carcinoma, small cell lung cancer, adenocarcinoma, or other histologic types of lung cancer.25
Currently, there are no specialized data about the distribution of these genetic variants in the Romanian population, but the results of the current study highlight some particularities of the studied population compared with existing data in the literature. An interesting finding is that the frequency of the Trp variant allele of the 194 codon of the XRCC1 gene is lower common compared to the American, European, and Central Asian populations (~35%), and approaching values reported in the northern US and northern African populations (6%–11%).11,13,26–28
Studies have shown different susceptibilities for smokers to develop lung cancer according to sex, with the assumption of differences in ability to repair DNA lesions, with a deficiency of these mechanisms in women.29,30 The results of our analysis revealed no statistically significant general difference between the sexes in the XRCC1 194Trp and 399Arg variants. However, we cannot confirm this was because cancer susceptibility does not involve just particular molecular features of DNA-repair pathways but other immunological and hormonal variables.
One of the major findings in the present study is that the variant 194Trp allele of the XRCC1 gene was more common in women with adenocarcinoma compared to men with adenocarcinoma, which could mean that this genetic variant could be an important inborn risk factor in the etiology of women’s lung adenocarcinoma. Our results are in agreement with other studies in which this DNA-repair genetic variant was associated with lung adenocarcinoma in women, although most of these studies focused on nonsmoking women.31,32
One of the limits of the present study is the small number of subjects and the fact that they were not followed thereafter to assess the association with different demographic characteristics in relation to other individual or pathological factors (disease stage and treatment). Because the study included Caucasian subjects with Balkan, Slavonic, Romanian, and Asian ancestry33 as part of the Eastern European population, genetic characteristics may have varied due to demographic particularities; therefore, it is imperative to conduct large-scale studies to determine the allele frequency and appropriate connections of these genetic variants in lung cancer in Eastern European patients.
Conclusion
We can say that our statistical results place our study group among the populations with the lowest frequency for the Arg194Trp variant allele of the XRCC1 gene. The findings of the study place the XRCC1 Arg194Trp polymorphism among independent risk factors for developing lung cancer, particularly lung adenocarcinoma in women.
Acknowledgment
This paper was published under the framework of the European Social Fund, Human Resources Development Operational Programme 2007–2013, project POSDRU/ 159/1.5/138776.
Disclosure
The authors report no conflicts of interest in this work.
References
Wang Y, Yang H, Li H, et al. Association between X-ray repair cross complementing group 1 codon 399 and 194 polymorphisms and lung cancer risk: a meta-analysis. Cancer Lett. 2009;285(2):134–140. | ||
International Agency for Research and Cancer. World Health Organization. [webpage on the Internet]. GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. Available from: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx. Accessed November 17, 2015. | ||
Park JY, Lee SY, Jeon HS, et al. Polymorphism of the DNA repair gene XRCC1 and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2002;11(1):23–27. | ||
Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11(12):1513–1530. | ||
Li J, Li Z, Feng L, Guo W, Zhang S. Polymorphisms of DNA repair gene XRCC1 and hepatocellular carcinoma risk among East Asians: a meta-analysis. Tumour Biol. 2012;34(1):261–269. | ||
Li Y, Liu F, Tan SQ, Wang Y, Li SW. X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and cervical cancer risk: a huge systematic review and meta-analysis. PLoS One. 2012;7(9):e44441. | ||
Wang L, Yin F, Xu X, Hu X, Zhao D. X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and risk of childhood acute lymphoblastic leukemia: a meta-analysis. PLoS One. 2012;7(4):e34897. | ||
Chen B, Zhou Y, Yang P, Wu XT. Polymorphisms of XRCC1 and gastric cancer susceptibility: a meta-analysis. Mol Biol Rep. 2012;39(2):1305–1313. | ||
Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. | ||
Wang LE, Gorlova OY, Ying J, et al. Genome-wide association study reveals novel genetic determinants of DNA repair capacity in lung cancer. Cancer Res. 2013;73(1):256–264. | ||
David-Beabes GL, London SJ. Genetic polymorphism of XRCC1 and lung cancer risk among African-Americans and Caucasians. Lung Cancer. 2001;34(3):333–339. | ||
Divine KK, Gilliland FD, Crowell RE, et al. The XRCC1 399 glutamine allele is a risk factor for adenocarcinoma of the lung. Mutat Res. 2001;461(4):273–278. | ||
Hu Z, Ma H, Chen F, Wei Q, Shen H. XRCC1 polymorphisms and cancer risk: a meta-analysis of 38 case-control studies. Cancer Epidemiol Biomarkers Prev. 2005;14(7):1810–1818. | ||
Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162(10):925–942. | ||
Kiyohara C, Takayama K, Nakanishi Y. Lung cancer risk and genetic polymorphisms in DNA repair pathways: a meta-analysis. J Nucleic Acids. 2010;2010:701760. | ||
Zheng H, Wang Z, Shi X, Wang Z. XRCC1 polymorphisms and lung cancer risk in Chinese populations: a meta-analysis. Lung Cancer. 2009;65(3):268–273. | ||
Karkucak M, Yakut T, Evrensel T, et al. XRCC1 gene polymorphisms and risk of lung cancer in Turkish patients. Int J Hum Genet. 2012;12(2):113–117. | ||
Brambilla E, Travis WD. Lung cancer. In: Stewart BW, Wild CP, editors. World Cancer Report. Lyon: World Health Organization; 2014:350–361. | ||
Abdel-Rahman SZ, Soliman AS, Bondy ML, et al. Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett. 2000;159(1):79–86. | ||
Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics 2000. CA Cancer J Clin. 2000;50(1):7–33. | ||
Charloux A, Hedelin G, Dietemann A, et al. Prognostic value of histology in patients with non-small cell lung cancer. Lung Cancer. 1997;17(1):123–134. | ||
Duarte MC, Colombo J, Rossit AR, et al. Polymorphisms of DNA repair genes XRCC1 and XRCC3, interaction with environmental exposure and risk of chronic gastritis and gastric cancer. World J Gastroenterol. 2005;11(42):6593–6600. | ||
Freedman ML, Reich D, Penney KL, et al. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36(4):388–393. | ||
Dai L, Duan F, Wang P, Song C, Wang K, Zhang J. XRCC1 gene polymorphisms and lung cancer susceptibility: a meta-analysis of 44 case-control studies. Mol Biol Rep. 2012;39(10):9535–9547. | ||
Jiang J, Liang X, Zhou X, et al. A meta-analysis of randomized controlled trials comparing irinotecan/platinum with etoposide/platinum in patients with previously untreated extensive-stage small cell lung cancer. J Thorac Oncol. 2010;5(6):867–873. | ||
Abdel-Rahman SZ, Soliman AS, Bondy ML, et al. Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett. 2000;159(1):79–86. | ||
Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58(4):604–608. | ||
Ratnasinghe D, Yao SX, Tangrea JA, et al. Polymorphisms of the DNA repair gene XRCC1 and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10(2):119–123. | ||
Improta G, Sgambato A, Bianchino G, et al. Polymorphisms of the DNA repair genes XRCC1 and XRCC3 and risk of lung and colorectal cancer: a case-control study in a southern Italian population. Anticancer Res. 2008;28(5B):2941–2946. | ||
Letkova L, Matakova T, Musak L, et al. DNA repair genes polymorphism and lung cancer risk with the emphasis to sex differences. Mol Biol Rep. 2013;40(9):5261–5273. | ||
Li MC, Cui ZS, He QC, Zhou BS. [Association of genetic polymorphism in the DNA repair gene XRCC1 with susceptibility to lung cancer in non-smoking women]. Zhonghua Zhong Liu Za Zhi. 2005;27(12):713–716. Chinese. | ||
Wei Q, Cheng L, Amos CI, et al. Repair of tobacco carcinogen-induced DNA adducts and lung cancer risk: a molecular epidemiologic study. J Natl Cancer Inst. 2000;92(21):1764–1772. | ||
Derenko M, Malyarchuk B, Denisova G, et al. Complete mitochondrial DNA analysis of eastern Eurasian haplogroups rarely found in populations of Northern Asia and Eastern Europe. PLoS One. 2012;7(2):e32179. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

