Back to Journals » Neuropsychiatric Disease and Treatment » Volume 15
The role of long noncoding RNA in traumatic brain injury
Authors Li Z, Han K, Zhang D , Chen J, Xu Z , Hou L
Received 24 February 2019
Accepted for publication 8 May 2019
Published 28 June 2019 Volume 2019:15 Pages 1671—1677
DOI https://doi.org/10.2147/NDT.S206624
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yuping Ning
Video abstract presented by Lijun Hou.
Views: 157
Zhenxing Li,* Kaiwei Han,* Danfeng Zhang,* Jigang Chen, Zheng Xu, Lijun Hou
Department of Neurosurgery, Changzheng Hospital, Second Military Medical University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Abstract: Traumatic brain injury (TBI), a mainly lethal and highly debilitating condition, is increasing worldwide. However, the underlying mechanism has not been fully elucidated and effective therapy is needed. Long noncoding RNAs (lncRNAs), which form a major class of noncoding RNAs, have emerged as novel targets for regulating physiological functions and mediating numerous neurological diseases. Notably, gene expression profile analyses have demonstrated aberrant changes in lncRNA expression in the cerebral cortex and hippocampus of rats, mice and human after TBI. lncRNAs may be associated with multiple pathophysiological processes following TBI and might play a crucial role in complications of TBI, such as traumatic optic neuropathy due to the regulation of specific signaling pathways. Some lncRNAs have also been found to be therapeutic targets for motor and cognitive recovery after TBI. lncRNAs may be promising biomarkers for TBI diagnosis, treatment, and prognosis prediction. However, further research isneeded to clarify the underlying mechanisms and therapeutic effects of lncRNAs on TBI. We review the current progress of studies on lncRNAs in TBI to draw more attention to their roles in this debilitating condition.
Keywords: long non-coding RNA, traumatic brain injury, neuropathy
Introduction
In recent years, the incidence of traumatic brain injury (TBI) is increasing as a result of general use of motor vehicles, sports and falls, making it the leading cause of death and disability worldwide.1,2 It brings heavy emotional and economic burdens to individuals, families, and society. TBI survivors usually face a series of problems, such as epilepsy, motor and sensory disturbances, cognitive and neuropsychological dysfunction.3 Although the pathophysiological mechanism of TBI is known to be associated with primary damage of shearing and tearing forces, and secondary damage, the exact mechanism is still unclear.4,5 Secondary damage frequently occurs several minutes or months after TBI, and results in persistent neurological impairment. Generally, secondary damage is regulated by related pathways of glutamate excitotoxicity,6,7 free radical generation,8 and neuroinflammation,9 which contribute to oxidative stress, inflammation, neuronal apoptosis and other biochemical changes. Current treatment consists mainly of early detection, symptoms control, supportive therapy, and functional rehabilitation. However, the prompt prevention of secondary injury and effective treatment measures for satisfactory recovery are still lacking.10
A variety of noncoding RNA genes exists in the human and rodent genomes, such as ribosomal RNAs, transfer RNAs, small nuclear RNAs, microRNAs (miRNAs), and long noncoding RNAs (lncRNAs). It was gradually recognized that miRNAs were differentially expressed after TBI and would be novel biomarkers for the diagnosis, treatment, and prognosis prediction of such injury.11 For example, Di Pietro et al12 analyzed the expression miRNAs in serum of five mild TBI with extracranial injury patients, five severe TBI with extracranial injury patients and five healthy volunteers at different times post injury. They identified two miRNAs (miR-425-5p, miR-502) as ideal candidates for diagnosis of mild TBI and two miRNAs (miR-21 and miR-335) as valid biomarkers for diagnosis of severe TBI. As a large type of noncoding RNAs, lncRNAs have been emerging as important regulators of multiple disease processes.13 However, the functional significance of lncRNAs in TBI and their underlying mechanisms are less known compared with those of miRNAs. In this review, we summarize current knowledge about the roles of lncRNAs associated with TBI, which would help in the search for new therapeutic strategies.
Overview of lncRNAs
Biology of lncRNAs
As nonprotein-coding transcripts, noncoding RNAs account for at least 98% of the total human genome.14 They can be classified into miRNAs, P-element-induced wimpy testis (PIWI)-interacting RNAs, small nucleolar RNAs, lncRNAs and other types of ncRNAs.15 lncRNAs constitute the largest portion of the mammalian noncoding RNAs.16 Once regarded as transcriptional noise, increasing evidence has demonstrated that lncRNAs have multiple domains that can bind to DNA, RNA, and protein, modify the chromatin status, regulate transcription and translation, and ultimately affect gene expression.17 Although the functions of lncRNAs have not been completely decoded, studies have shown that they play different roles in cell proliferation, survival, and migration, as well as in maintaining genomic stability through guidance, bait, scaffolds and signaling molecules.18,19
lncRNAs and neurological diseases
lncRNAs are highly expressed in the central nervous system (CNS).20 Their aberrant expression has been confirmed to be related to neurodegenerative diseases, ischemic cerebrovascular diseases, gliomas and other neurological diseases. Using a triple transgenic mouse model of Alzheimer’s disease, Lee et al21 identified 205 dysregulated lncRNAs from 4622 lncRNAs that would facilitate the process of molecule-targeted therapy. Kraus et al22 studied 30 brain specimens derived from 20 patients with Parkinson’s disease (PD) and 10 controls and revealed that five lncRNAs were significantly differentially expressed. The five lncRNAs were H19 upstream conserved 1 and 2 (which was downregulated), and long intergenic noncoding RNA p21 (lincRNA-p21), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), small nucleolar RNA host gene 1 (SNHG1), and tiny noncoding RNA (which were all upregulated). Zhang et al23 found that the knockout of lncRNA MALAT1 in a mouse model of oxygen and glucose depravation (OGD) led to the significantly aggravated expression of proapoptotic factors and proinflammatory cytokines, which indicated that MALAT1 may have a protective role in ischemic stroke. MALAT1 was also thought to decrease the sensitivity of resistant glioblastoma.24 Although many studies have recently reported the potential value of lncRNAs in neurological diseases, the topic is still in its infancy, especially on their roles in TBI.25
lncRNAs in traumatic brain injury
lncRNA expression profiles in brain tissue of animal models and humans
lncRNA microarrays, RNA-sequencing (RNA-Seq), and bioinformatic analysis are commonly used to explore the expression of lncRNAs and delineate their molecular functions. To date, two studies in particular have reported the alteration of lncRNAs in mice or rats after TBI. Zhong et al26 detected alterations in lncRNA and messenger RNA (mRNA) expression levels by high throughput sequencing in the mouse cortex at 24 hours postTBI, using a controlled cortical impact model. In a total of 64,530 transcripts including 27,457 transcripts identified as mRNA and 37,073 transcripts identified as lncRNA, 1,580 mRNAs and 823 lncRNAs were found to be significantly altered. Of the abnormally changed lncRNAs, 667 were upregulated and 156 were downregulated (|log2 (fold change)| >1, P<0.05. The good reproducibility and reliability of the RNA-Seq were confirmed by using the reverse transcription polymerase chain reaction. Their bioinformatic analysis showed that the mRNAs were mainly associated with inflammatory and immunological activities, metabolism, neuronal, and vascular networks. They also demonstrated the cross association between lncRNAs and mRNAs. Wang et al27 performed a microarray analysis to profile lncRNAs that were altered in the rat hippocampus after TBI. A total of 271 lncRNAs and 1046 mRNAs were differentially expressed. Similarly, gene ontology (GO) and pathway analyses suggested that the alterations were related to inflammation, apoptosis, and necroptosis. The top three differentially expressed lncRNAs were: NR_002704, ENSRNOT00000062543, and Zfas. The study proposed that the aberrantly expressed lncRNAs might regulate secondary injury through the mitogen-activated protein kinase signaling pathway, p53 signaling pathway, or cytokine-cytokine receptor interaction. However, their deduction deserves further validation. Moreover, when considering that secondary injury after TBI is a dynamic pathophysiological process,28 the time-dependent changes of lncRNAs at different phases after TBI (eg, at 1 hour, 6 hours, 3 days, or 7 days) need to be clarified.
Yang et al29 investigated the expression profiles of lncRNAs in human TBI, where three patients (two females and one male) were enrolled in the study. The samples analyzed were derived from contusion tissue and tissue adjacent to the contusion, both in the temporal and frontal lobes of the brain. Arraystar Human lncRNA Microarray version 2.0 (Aksomics, Shanghai, China) was designed for global profiling of human lncRNAs and protein coding transcripts. They found 43 upregulated and 56 downregulated lncRNAs, and concluded that the lncRNA and mRNA expression levels were significantly different between the injured site and surrounding tissue. In terms of the differences according to gender and the brain lobe sampled, the analytical methods needed to be more standardized and validation studies should be carefully carried out. Nevertheless, it is known that the genes of mice are different from those of humans, and the research thus confirmed that the roles lncRNAs play in the pathogenesis of TBI still need to be elucidated.
lncRNAs in the pathophysiological process of traumatic brain injury
TBI triggers complex pathophysiological processes,30 where neuroinflammation is prominent in both the primary and secondary injury.31 As the immune cells of the brain, microglia actively respond to changes in the CNS environment. Sun et al32 observed that human lncRNA growth arrest-specific 5 (lncRNA GAS5) suppressed the transcription of terminal nucleotidyltransferase 4 (TRF4; a key factor that control M2 macrophage polarization), by recruiting polycomb repressive complex 2 (PRC2), thereby inhibiting microglial M2 polarization. Human lincRNA-p21 has also been reported to promotes microglial activation through a p53-dependent transcriptional pathway.33 These studies provide valuable insights into the role of lncRNAs in the neuroinflammatory process of TBI. In addition, astrocyte apoptosis often occurs following the secondary injury of TBI. Yu et al34 found that the mouse lncRNA Gm4419 could promote the trauma-induced apoptosis of astrocytes by upregulating the expression of the inflammatory cytokine tumor necrosis factor-alpha (TNF-α). In particular, the lncRNA Gm4419 transcript could play the role of a sponge for miR-466l (a miRNA that targets the 3′ untranslated region of TNF-αfor degradation and translation inhibition). However, the underlying mechanism by which lncRNA mediate the pathophysiological process of TBI requires confirmation by further research.
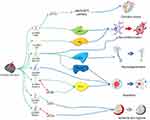
Ischemia generally proceeds after the occurrence of TBI.35 Neurons cultured underan OGD condition of are used as a common in vitro cell-damage model to simulate secondary pathophysiological changes after TBI.36,37 In the study by Yuan et al,38 PC-12 cells were subjected to an OGD environment and then stimulated with geniposide, an iridoid glycoside that has been reported to exert a neuroprotective effect on these cells.39 The treatment effect and the relative expression level of the lncRNA H19 were assessed. As a result, they found that geniposide protected PC-12 cells against OGD-induced injury via H19 upregulation. Human H19 has also been considered to alleviate hypoxia-induced injury by downregulating miR-28 expression in PC-12 cells.40 Moreover, Yang et al41 found that the lncRNA SNHG1 protected against OGD-induced injury in brain microvascular endothelial cells by sponging miR-338. These studies gives an insight into the role of lncRNA H19 and SNHG1 in ischemia and hypoxia injury after TBI. The different roles of lncRNAs in the pathophysiological process are summarized in Figure 1.
lncRNAs in the complication of traumatic brain injury
Traumatic optic neuropathy is one of the most feared and common complications of head and ocular trauma, for which a standard treatment has yet to be established.42 Yao et al.43 investigated the role of MALAT1 in the retinal neurodegeneration of optic nerve transection models of rats and mice. They found that MALAT1 might modulate retinal neurodegeneration through binding to the cAMP response element-binding protein (CREB), maintaining CREB phosphorylation by inhibiting protein phosphatase 2 (PP2A)-mediated dephosphorylation, and thus resulting in continuous CREB signaling activation. In addition to cell apoptosis and inflammatory responses, oxidative stress also plays an important role in secondary injury after TBI.44 Miao et al45 explored the role of the lncRNA GAS5 in mice retinal ganglion cells (RGCs) that were subjected to H2O2 injury in order to simulate an in vitro oxidation stress model of optic nerve injury. They found that lncRNA GAS5 knockdown would elicited competing endogenous RNAs (ceRNA) activity by upregulating miR-124 to alleviate the RGC injury. This process might be associated with activation of JAK/STAT3 signaling pathway and inhibition of the JNK signaling pathway. The above studies identified the roles of lncRNA MALAT1 and GAS5 in retinal neurodegeneration and optic nerve regeneration, which could be potential mechanisms underlying traumatic optic neuropathy.
TBI and post-traumatic stress disorder (PTSD) commonly co-occur in TBI patients such as veteran populations or individuals exposed to severe trauma, causing similar symptoms like sleep, concentration, memory, and mood disorders.46 PTSD and persistent sequelae due to TBI are thought to share common pathophysiological elements related to the CNS after acute injury or threat.47 PTSD affects a variety of physiological systems throughout the body, including the hypothalamus-pituitary-adrenal axis, cortical function, and immune system. Multiple biological studies on PTSD have been reported. As a novel research target, lncRNAs have also been explored with regard to their relationship with the PTSD model. Liu et al48 established a rat model of PTSD-like syndrome and conducted a microarray analysis of the hippocampal tissues. For microarray analysis, an Agilent Array platform was employed (Agilent Technologies, Santa Clara, CA, USA). They showed 293 lncRNAs and 476 mRNAs to be differentially expressed. GO and pathway analyses indicated the association of these lncRNAs with multiple biological processes, cellular components and molecular functions. Among the abnormally expressed lncRNAs, lncRNA MPAK159688_P1 (associated with the memory function-related Fos gene) was downregulated. Thus, lncRNAs would have promising value in discovering the mechanism of PTSD or sequelae due to TBI, and promoting effective rehabilitation.
lncRNAs in the treatment of traumatic brain injury
Inflammation commonly responds to TBI and leads to secondary brain edema, neuronal apoptosis, or cell death at different stages of the disease course. Patel et al10 isolated exosomes from human adipose-derived stem cells (hASCs) containing the lncRNA MALAT1, and then injected them intravenously into rats subjected to TBI. Exosomes depleted of MALAT1 or conditioned medium depleted of exosomes were also administered as control groups. They observed a significant recovery of motor function and amelioration of the cortical brain injury in the animals injected with the exosomes containing the lncRNA MALAT1. RNA-Seq suggested the MALAT1-dependent modulation of inflammation-related, cell cycle, and molecular regeneration pathways. The study demonstrated MALAT1 as a potential therapeutic approach for TBI. Similarly, MALAT1 from hASC exosomes was demonstrated to promote neuronal survival and cell proliferation after injury via alternative splicing of protein kinase C δII in a mouse hippocampal cell line.49 The upregulation of the lncRNA nuclear enriched abundant transcript 1 (NEAT1) has been shown to play a role in early apoptosis.50 Bexarotene, a selective agonist of retinoid X receptor, could exert a neuroprotective effect against cell apoptosis after TBI.51 Zhong et al52 studied the association between lncRNA Neat1 and bexarotene in the treatment of TBI in mice. They found that bexarotene promoted the level of NEAT1, which suppressed apoptosis and inflammation, thereby contributing to better motor and cognitive function recovery after TBI. Interestingly, Dai et al53 investigated the effect of the traditional Chinese medicine Changqin No. 1 on TBI, and found that lncRNA GAS5 could upregulate RAS p21 GTPase activating protein 1 (Rasa1) expression via sponging miR-335 in mouse neuronal cells. Moreover, lncRNA GAS5 exerted a neuroprotective effect by inhibiting neuronal apoptosis, which exploited a new field that explains the therapeutic mechanisms of some traditional Chinese medicines.
Oligodendrocyte death following TBI can induce pathological changes in white matter, which commonly results in network dysfunction and cognitive impairment.54 As a distinguishing biomarker of developing oligodendroglia progenitors, A2B5 from induced pluripotent stem cells has the potential to ameliorate TBI by inducing differentiation of the iPSCs into oligodendrocytes.55,56 Lyu et al57 performed a microarray analysis of lncRNAs and mRNAs in rats with TBI after A2B5-positive cell transplantation. The Agilent Array platform was employed for microarray analysis. They found 83 lncRNAs and 360 mRNAs to be differently expressed (P<0.05, fold change >2), with the crucial lncRNA and mRNA involved being ENSRNOT00000052577 and Kif2c, respectively. Therefore, the therapeutic effects of A2B5-positive iPSC transplantation could be related to the involvement of lncRNAs.
Future perspectives and conclusion
lncRNAs have received increasing attention for their ability to modulate transcriptional regulation in neurological diseases. The study on the roles of lncRNAs is currently at the infancy stage and their mechanisms remains be elucidated. First, although alterations in lncRNA expression have been reported in TBI, more research focused on the multidimensional interactions of lncRNAs studies not limited to their function as ceRNAs, and comprehensive analysis of their expression profiles are needed. Second, alterations in the expression levels of lncRNAs in blood samples after TBI has not been studied to date. Such lncRNAs with abnormal findings upon blood test would have the potential to be novel biomarkers. Third, despite the existence of various in vitro models of primary injury in TBI, such as transection, compression methods, and stretch-induced injury,35 most studies have only focused on cell models for secondary injury, such as the OGD and oxidative stress models. Therefore, in vitro experiments are relatively scarce and their validation function is not powerful enough.
In conclusion, lncRNAs have been recognized to play complex roles in TBI and may subsequently be promising biomarkers for diagnosis, evaluation, treatment, and prognosis prediction of this debilitating disease process. Future studies should focus on the detailed mechanisms and functional roles of lncRNAs so that targeted therapies for TBI can be developed.
Acknowledgment
This study was funded by the Shanghai Yang Fan Project (19YF1447900) to Danfeng Zhang and the Natural Science Foundation of China (81671206) to Lijun Hou.
Disclosure
The authors report no conflicts of interest in this work.
References
1.
2. James SL, Theadom A, Ellenbogen RG, et al. Global, regional, and national burden of traumatic brain injury and spinal cord injury, 1990–2016: a systematic analysis for the global burden of disease study 2016. Lancet Neurol. 2019;18:56–87. doi:10.1016/S1474-4422(18)30415-
3. Air M, Menon DK, Adelson PD, et al. Traumatic brain injury: integrated approaches to improve prevention, clinical care, and research. Lancet Neurol. 2017;16:987–7048.
4. Carbonara M, Fossi F, Zoerle T, et al. Neuroprotection in traumatic brain injury: mesenchymal stromal cells can potentially overcome some limitations of previous clinical trials. Front Neurol. 2018;9:885. doi:10.3389/fneur.2018.00885
5. Rana A, Singh S, Sharma R, Kumar A. Traumatic brain injury altered normal brain signaling pathways: implications for novel therapeutics approaches. Curr Neuropharmacol. 2018;16. doi:10.2174/1570159X16666180911121847
6. Dorsett CR, McGuire JL, DePasquale EAK, Gardner AE, Floyd CL, McCullumsmith RE. Glutamate neurotransmission in rodent models of traumatic brain injury. J Neurotrauma. 2017;34:263–272. doi:10.1089/neu.2015.4373
7. Faden AI, Demediuk P, Panter SS, Vink R. The role of excitatory amino acids and NMDA receptors in traumatic brain injury. Science. 1989;244:798–800.
8. Anthonymuthu TS, Kenny EM, Bayır H. Therapies targeting lipid peroxidation in traumatic brain injury. Brain Res. 2016;1640:57–76. doi:10.1016/j.brainres.2016.02.006
9. Simon DW, McGeachy M, Bayır H, Clark RSB, Loane DJ, Kochanek PM. The far-reaching scope of neuroinflammation after traumatic brain injury. Nat Rev Neurol. 2017;13:171–191. doi:10.1038/nrneurol.2017.13
10. Patel NA, Moss LD, Lee JY, et al. Long noncoding rna malat1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. J Neuroinflammation. 2018;15:204. doi:10.1186/s12974-018-1220-7
11. Pan Y-B, Sun Z-L, Feng D-F. The role of microRNA in traumatic brain injury. Neuroscience. 2017;367:189–199. doi:10.1016/j.neuroscience.2017.10.046
12. Di Pietro V, Ragusa M, Davies D, et al. MicroRNAs as novel biomarkers for the diagnosis and prognosis of mild and severe traumatic brain injury. J Neurotrauma. 2017;34:1948–1956. doi:10.1089/neu.2016.4857
13. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi:10.1038/ng.3192
14. Mattick JS. Non‐coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi:10.1093/embo-reports/kve230
15. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi:10.1038/nrg3074
16. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi:10.1038/nrg2521
17. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease.%a batista pj. Cell. 2013;152:1298–1307. doi:10.1016/j.cell.2013.02.012
18. Kung JTY, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi:10.1534/genetics.112.146704
19. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi:10.1146/annurev-biochem-051410-092902
20. Ng S-Y, Lin L, Soh S, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–468. doi:10.1016/j.tig.2013.03.002
21. Lee DY, Moon J, Lee S-T, et al. Distinct expression of long non-coding RNAs in an Alzheimer’s disease model. J Alzheimers Dis. 2015;45:837–849. doi:10.3233/JAD-142919
22. Kraus TFJ, Haider M, Spanner J, Steinmaurer M, Dietinger V, Kretzschmar HA. Altered long noncoding RNA expression precedes the course of Parkinson’s disease—a preliminary report. Mol Neurobiol. 2016;54:2869–2877. doi:10.1007/s12035-016-9854-x
23. Zhang X, Tang X, Liu K, Hamblin MH, Yin KJ. Long non-coding RNA MALAT1 regulates cerebrovascular pathologies in ischemic stroke. J Neurosci. 2017;37:1797. doi:10.1523/JNEUROSCI.3389-16.2017
24. Li H, Yuan X, Yan D, et al. Long non-coding RNA MALAT1 decreases the sensitivity of resistant glioblastoma cell lines to temozolomide. Cell Physiol Biochem. 2017;42:1192–1201. doi:10.1159/000478917
25. Chen Y, Zhou J. lncRNAs: macromolecules with big roles in neurobiology and neurological diseases. Metab Brain Dis. 2017;32:281–291. doi:10.1007/s11011-017-9965-8
26. Zhong J, Jiang L, Cheng C, et al. Altered expression of long non-coding RNA and mRNA in mouse cortex after traumatic brain injury. Brain Res. 2016;1646:589–600.
27. Wang CF, Zhao CC, Weng WJ, et al. Alteration in long non-coding RNA expression after traumatic brain injury in rats. J Neurotrauma. 2017;34:2100–2108. doi:10.1089/neu.2016.4642
28. Currie S, Saleem N, Straiton J, Macmullen-Price J, Warren DJ. Imaging assessment of traumatic brain injury. Postgrad Med J. 2016;92:41–50. doi:10.1136/postgradmedj-2014-133211
29. Yang L-X, Yang L-K, Zhu J, Chen J-H, Wang Y-H, Xiong K. Expression signatures of long non-coding RNA and mRNA in human traumatic brain injury. Neural Regen Res. 2019;14:632. doi:10.4103/1673-5374.247467
30. Chiu C-C, Liao Y-E, Yang L-Y, et al. Neuroinflammation in animal models of traumatic brain injury. J Neurosci Methods. 2016;272:38–49. doi:10.1016/j.jneumeth.2016.06.018
31. Lozano D, Gonzales-Portillo GS, Acosta S, et al. Neuroinflammatory responses to traumatic brain injury: etiology, clinical consequences, and therapeutic opportunities. Neuropsychiatr Dis Treat. 2015;11:97–106.
32. Sun D, Yu Z, Fang X, et al. lncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep. 2017;18:1801. doi:10.15252/embr.201643668
33. Ye Y, He X, Lu F, et al. A lincRNA-p21/miR-181 family feedback loop regulates microglial activation during systemic LPS-and MPTP-induced neuroinflammation. Cell Death Dis. 2018;9:803. doi:10.1038/s41419-018-0821-5
34. Yu Y, Cao F, Ran Q, Wang F. Long non-coding RNA Gm4419 promotes trauma-induced astrocyte apoptosis by targeting tumor necrosis factor α. Biochem Biophys Res Commun. 2017;491: 478–485.
35. Salvador E, Burek M, Förster CY. Stretch and/or oxygen glucose deprivation (OGD) in an in vitro traumatic brain injury (TBI) model induces calcium alteration and inflammatory cascade. Front Cell Neurosci. 2015;9:323. doi:10.3389/fncel.2015.00323
36. Choi YK, Maki T, Mandeville ET, et al. Dual effects of carbon monoxide on pericytes and neurogenesis in traumatic brain injury. Nat Med. 2016;22:1335. doi:10.1038/nm.4188
37. Park E, Park K, Liu E, Jiang R, Zhang J, Baker AJ. Bone-marrow–derived endothelial progenitor cell treatment in a model of lateral fluid percussion injury in rats: evaluation of acute and subacute outcome measures. J Neurotrauma. 2017;34:2801–2811. doi:10.1089/neu.2016.4560
38. Yuan Y, Zheng Z. Geniposide protects PC-12 cells against oxygen and glucose deprivation-induced injury by up-regulation of long-noncoding RNA H19. Life Sci. 2019;216:17–182. doi:10.1016/j.lfs.2018.11.047
39. Liu J, Zheng X, Yin F, et al. Neurotrophic property of geniposide for inducing the neuronal differentiation of PC12 cells. Int J Dev Neurosci. 2006;24:419–424. doi:10.1016/j.ijdevneu.2006.08.009
40. Chen Z, Chen X, Guo R, Meng J. Protective effects of lncRNA H19 silence against hypoxia-induced injury in PC-12 cells by regulating miR-28. Int J Biol Macromol. 2019;121:546–555. doi:10.1016/j.ijbiomac.2018.10.033
41. Yang X, Zi X-H. lncRNA SNHG1 alleviates OGD induced injury in BMEC via miR-338/HIF-1α axis. Brain Res. 2019;1714:174–181. doi:10.1016/j.brainres.2018.11.003
42. Ibrahim AS, Elmasry K, Wan M, et al. A controlled impact of optic nerve as a new model of traumatic optic neuropathy in mouse. Invest Ophthalmol Vis Sci. 2018;59:5548–5557. doi:10.1167/iovs.18-24773
43. Yao J, Wang XQ, Li YJ, et al. Long non‐coding RNA MALAT1 regulates retinal neurodegeneration through CREB signaling. EMBO Mol Med. 2016;8:e201505725.
44. Rodriguez-Rodriguez A, Jose Egea-Guerrero J, Murillo-Cabezas F, Carrillo-Vico A. Oxidative stress in traumatic brain injury. Curr Med Chem. 2014;21:1201–1211.
45. Miao X, Liang A. Knockdown of long noncoding RNA GAS5 attenuates H2O2‐induced damage in retinal ganglion cells through upregulating mir‐124: potential role in traumatic brain injury. J Cell Biochem. 2018;1–10. doi:10.1002/jcb.27560
46. Avallone KM, Smith ER, Ma S, et al. PTSD as a mediator in the relationship between post-concussive symptoms and pain among OEF/OIF/OND veterans. Mil Med. 2019;184:e118-e123. doi:10.1093/milmed/usy225
47. Hendrickson RC, Schindler AG, Pagulayan KF. Untangling PTSD and TBI: challenges and strategies in clinical care and research. Curr Neurol Neurosci Rep. 2018;18:106. doi:10.1007/s11910-018-0908-5
48. Liu Q, Ma J, Yu Z, Liu H, Chen C, Li W. Distinct hippocampal expression profiles of lncRNAs in rats exhibiting a PTSD-like syndrome. Mol Neurobiol. 2016;53:2161–2168. doi:10.1007/s12035-015-9180-8
49. El Bassit G, Patel RS, Carter G, et al. MALAT1 in human adipose stem cells modulates survival and alternative splicing of PKCδII in HT22 cells. Endocrinology. 2016;158:183–195.
50. Hirose T, Virnicchi G, Tanigawa A, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. doi:10.1091/mbc.E13-09-0558
51. Zhong J, Cheng C, Liu H, et al. Bexarotene protects against traumatic brain injury in mice partially through apolipoprotein E. Neuroscience. 2017;343:434–448. doi:10.1016/j.neuroscience.2016.05.033
52. Zhong J, Li J, Huang Z, et al. The long non-coding RNA NEAT1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav Immun. 2017;65:183–194.
53. Dai X, Yi M, Wang D, Chen Y, Xu X. Changqin no. 1 inhibits neuronal apoptosis via suppressing GAS5 expression in traumatic brain injury mice model. Biol Chem. 2019;400:753–763. doi:10.1515/hsz-2018-0340
54. Flygt J, Gumucio A, Ingelsson M, et al. Human traumatic brain injury results in oligodendrocyte death and increases the number of oligodendrocyte progenitor cells. J Neuropathol Exp Neurol. 2016;75:503–515. doi:10.1093/jnen/nlw025
55. Baracskay KL, Kidd GJ, Miller RH, Trapp BD. NG2‐positive cells generate A2B5‐positive oligodendrocyte precursor cells. Glia. 2007;55:1001–1010. doi:10.1002/glia.20519
56. Ogawa S-I, Tokumoto Y, Miyake J, Nagamune T. Immunopanning selection of A2B5-positive cells increased the differentiation efficiency of induced pluripotent stem cells into oligodendrocytes. Neurosci Lett. 2011;489:79–83. doi:10.1016/j.neulet.2010.11.070
57. Lyu Q, Zhang Z-B, Fu S-J, Xiong L-L, Liu J, Wang T-H. Microarray expression profile of lncRNAs and mRNAs in rats with traumatic brain injury after A2B5+ cell transplantation. Cell Transplant. 2017;26:1622–1635. doi:10.1177/0963689717723014
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.