Back to Archived Journals » ChronoPhysiology and Therapy » Volume 7
The retinal clock in mammals: role in health and disease
Authors Felder-Schmittbuhl MP, Calligaro H , Dkhissi-Benyahya O
Received 10 March 2017
Accepted for publication 19 April 2017
Published 22 May 2017 Volume 2017:7 Pages 33—45
DOI https://doi.org/10.2147/CPT.S115251
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Marc Hebert
Marie-Paule Felder-Schmittbuhl,1,* Hugo Calligaro,2 Ouria Dkhissi-Benyahya2,*
1Institute of Cellular and Integratives Neurosciences, UPR3212, CNRS, Université de Strasbourg, Strasbourg, 2University of Lyon, Stem Cell and Brain Research Institute, INSERM U1208, Bron, France
*These authors contributed equally to this work
Abstract: The mammalian retina contains an extraordinary diversity of cell types that are highly organized into precise circuits to perceive and process visual information in a dynamic manner and transmit it to the brain. Above this builds up another level of complex dynamic, orchestrated by a circadian clock located within the retina, which allows retinal physiology, and hence visual function, to adapt to daily changes in light intensity. The mammalian retina is a remarkable model of circadian clock because it harbors photoreception, self-sustained oscillator function, and physiological outputs within the same tissue. However, the location of the retinal clock in mammals has been a matter of long debate. Current data have shown that clock properties are widely distributed among retinal cells and that the retina is composed of a network of circadian clocks located within distinct cellular layers. Nevertheless, the identity of the major pacemaker, if any, still warrants identification. In addition, the retina coordinates rhythmic behavior by providing visual input to the master hypothalamic circadian clock in the suprachiasmatic nuclei (SCN). This light entrainment of the SCN to the light/dark cycle involves a network of retinal photoreceptor cells: rods, cones, and intrinsically photosensitive retinal ganglion cells (ipRGCs). Although it was considered that these photoreceptors synchronized both retinal and SCN clocks, new data challenge this view, suggesting that none of these photoreceptors is involved in photic entrainment of the retinal clock. Because circadian organization is a ubiquitous feature of the retina and controls fundamental processes, the coherence from cell to tissue is critical for circadian functions, and disruption of retinal clock organization or its response to light can potentially have a major impact on retinal pathophysiology and vision.
Keywords: retina, clock gene, circadian, ipRGC, photoreceptor, light
Introduction
From algae to mammals, nearly all organisms have adapted their lifestyle to the environmental 24 h light/dark (LD) cycle. Daily rhythms in behavior and physiology are programmed by a group of biological clocks widely present in a variety of tissues and cells, including the retina, and organized in a hierarchical manner. The central clock in the suprachiasmatic nucleus (SCN) lies at the top of the network and coordinates the phases of many peripheral clocks throughout the body. The temporal phase of the SCN is adjusted daily by light, which is captured by the retina, in order to synchronize organism-wide behavioral and physiological rhythms with the LD cycle. One of the first rhythms described in mammalian retina was the finding that rod outer segment disk shedding cycles in relationship with lighting conditions.1 Furthermore, experiments performed in amphibians and birds demonstrated that the retina contains an endogenous circadian clock, able to oscillate in constant condition in a culture dish.2,3 However, it was not until 1996 that cultured hamster retina was reported to maintain autonomous circadian rhythms for melatonin secretion, providing evidence that an endogenous clock is also present in the mammalian retina,4,5 regardless of the absence of the SCN.6,7 Progress toward understanding the mechanisms of circadian rhythm generation and light entrainment of the retinal clock has been hampered by technical limitations. The successful development of bioluminescence recording using luciferase reporter coupled to clock genes fundamentally revolutionized studies on circadian oscillators.8,9 This technique provides the advantage of detecting and quantifying circadian rhythms in retinal living cells or explants, as well as revealing retinal circadian oscillations in vitro in long-term studies in mammals.10 Many reports have now clearly demonstrated that several molecular, cellular, and physiological retinal rhythms are under the control of a retinal circadian clock or, more likely, a network of clocks localized in multiple types of neurons and glial Müller cells (Figure 1).11
  | Figure 1 The circadian clock system in the mammalian retina. Notes: (A) Clock machinery has been described in all retinal neurons, besides rods, as well as in glial Müller cells (black oscillation). Light purple: SW cone; green: MW cone; gray: rod; purple: horizontal cell; red: bipolar cell; orange: amacrine cell; blue: ganglion cell; black: ipRGCs; yellow: Müller cell; white: retinal pigmented epithelium.124 (Adapted from Servier Medical Art [homepage on the Internet]. Available from: http://www.servier.com/Powerpoint-image-bank. Accessed January 1, 2017.x (https://creativecommons.org/licenses/by/3.0/). (B) The endogenous molecular mechanism generating circadian rhythms is based on transcriptional–translational feedback loops involving clock genes. The CLOCK/BMAL1 heterodimer binds to an enhancer box (E-box) and drives the transcription of the clock genes Period (Per1-2–3), Cryptochrome (Cry1-2), Rorα-β, and Rev-erbα-β. PER and CRY proteins in turn inhibit CLOCK/BMAL1 transcriptional activity. By competitive binding to retinoid-related orphan receptors response elements (ROR-E) sites, the REVERB and ROR proteins repress or activate Bmal1 transcription, respectively. Abbreviations: MW, middle-wavelength sensitive; SW, short-sensitive. |
Molecular organization and light entrainment of the retinal clock
Molecular basis of retinal circadian rhythms
The endogenous functioning of circadian clocks, including the retina, involves an integrated network of clock genes interacting in transcriptional–translational feedback loops.12 Complexes of BMAL1 and CLOCK proteins bind to E-box enhancer elements in the promoters of Period 1–3 (Per1–3), Cryptochrome 1–2 (Cry1–2) clock genes, and “clock-controlled genes” and thereafter drive their expression. PER and CRY proteins form heterodimers that translocate back into the nucleus to inhibit CLOCK/BMAL1 transcriptional activity and therefore their own expression, forming the core negative feedback loop (Figure 1). Fine-tuning and reinforcement of this loop are ensured by another loop involving activation by CLOCK/BMAL1 of the Ror and RevErb nuclear hormone receptor–encoding genes. By competitive binding to ROR-E sites, the REVERB and ROR proteins, respectively, repress or activate Bmal1 gene transcription. Importantly, additional posttranslational events, such as proteasomal degradation, phosphorylation, acetylation, ubiquitination, and sumoylation, are also key components in the generation and timing of circadian rhythms. The culmination of activities of these transcription factors regulates circadian biology, from molecular mechanisms to overall behavior, thus constituting a time code for the cell and the entire organism.
Impact of clock gene deletion on retinal functions
Mutant and transgenic rodent models have provided a useful tool to evaluate a specific clock gene’s function in circadian rhythms. However, the majority of clock genes have functionally redundant paralogs that are able to perform effectively as the primary gene. Thus, deleting a specific clock gene does not necessarily disrupt the clock mechanism within a cell but may change circadian parameters at the level of the tissue or the organism. Per2-knockout mice exhibit retinal vascular damage, reduction in retinal thickness, and a decrease in the electroretinogram (ERG) amplitude, similar to those found in type 2 diabetes.13–15 A double-clock gene mutant, the Per1Brdm1/Per2Brdm1 mouse, exhibits delayed photoreceptor differentiation and alterations in short-wavelength cone distribution.16 Cry1-/-/Cry2-/- and Bmal1 retina-specific knockout mice show a loss of ERG circadian rhythm, suggesting that functional Bmal1 and Cry1-Cry2 genes are required for rhythmic retinal processing of visual stimuli, independently from the central clock.17,18 In addition, Bmal1–/– mice present abnormal circadian gene expression profiles in LD cycles, suggesting that Bmal1 is involved in the light regulation of gene expression. Silencing of RevErbα using small interfering RNA induces retinal abnormalities, such as pan-retinal spotting and decreased response to light.19 By contrast, Ait-Hmyed et al20 reported a hypersensitivity to light in RevErbα–/– mice, concomitant with an increased number of melanopsin-containing ganglion cells, suggesting that this clock gene modulates light sensitivity and visual processing. Only one study has extensively explored the rhythmic phenotype of single clock gene knockout in retina. Using several clock-gene-deficient mice, Ruan et al21 reported that the retinal clock is more vulnerable to clock gene disruption than the SCN. The authors showed that Per1, Cry1, and Clock are each necessary for the expression of molecular circadian rhythms by the retina. By contrast, knocking out Per2, Per3, or Cry2 individually does not blunt the rhythm but modulates its period or amplitude.21 For example, retinal explants from Per3–/– mice show a period-shortening effect, whereas the period of the SCN clock is not altered. This differential pattern of circadian rhythm alterations between the retina and the SCN was proposed to relate to the fact that cellular coupling among clock cells/layers is not uniform throughout the retina,22 whereas oscillators are strongly coupled in the SCN.23
Entrainment of the retinal clock by light
An especially important finding was that “blind” transgenic mice lacking functional rods and cones (rd/rd cl) exhibit near-normal photoentrainment of circadian rhythms.24,25 This subsequently led to the discovery of a novel opsin, melanopsin (OPN4), expressed in a subset of intrinsically photosensitive retinal ganglion cells (ipRGCs), with a peak of sensitivity at ~480 nm.26–28 It is now clear that ipRGCs combine their direct intrinsic responses with signals derived from rods and cones to regulate diverse non-image-forming responses,29 with each photoreceptor encoding distinct parameters of the light.30–34 However, we currently lack a comprehensive understanding of how light regulates the retinal clock in mammals and of the role of the different photoreceptors.
Nevertheless, several lines of evidence suggest that ipRGCs influence retinal network and physiology. Melanopsin was shown to be critical for diurnal and circadian regulation of the mouse photopic and scotopic ERG, suggesting influence of melanopsin up to the photoreceptor level.35–37 In agreement with this, we reported that circadian rhythms of clock gene expression are lost in the photoreceptor layer (PRL) of Opn4–/– mice.38 In addition, synaptic contact of ipRGCs with other retinal neurons has been reported in rodents, monkeys, and humans.39–42 Specifically, ipRGCs provide excitatory sustained light responses to dopaminergic (DAergic) amacrine cells, which are influential modulators of retinal circuitry.43 In the absence of melanopsin, this sustained response is lost,40 and blue light fails to activate the DAergic system and to induce Per1-Per2 mRNA expression in the PRL.38 The function of these retrograde connections is currently unknown, but they represent a potential mechanism for transmitting irradiance information to the outer retina.
In mammals, the first rhythm shown in vitro to be synchronized with LD cycles was melatonin secretion in the mouse retina.5 Ten years later, Ruan et al10 clearly established that a pulse of white light administered at circadian time (CT)13 or CT19, respectively, delays or advances the phase of PER2Luc, principally via the DA D1 receptor. Recent data suggest that the photoreceptors regulating light response of retinal rhythms differ from those used by the SCN.44,45 PER2Luc rhythm is still entrained to LD cycle in rd1/rd1;Opn4–/– mice, lacking rods, cones, and melanopsin.44 The authors propose that an ultraviolet (UV)-sensitive opsin, neuropsin (OPN5), is the sole opsin required to phase-shift the retinal clock in response to light.45 Neuropsin is expressed in many vertebrate species and is localized in a subset of mouse RGCs,45–48 but it is unclear whether it is expressed in ganglion cells distinct from ipRGCs. However, Hughes et al49 suggested a limited role for neuropsin in driving light response in the mouse retina because UV light stimulation failed to induce both c-FOS and excitatory responses in mice lacking the essential components of the phototransduction signaling pathways present in rods, cones, and ipRGCs.49 All these studies raise important questions about the relationship between neuropsin and melanopsin and their respective roles in the light response of the retinal clock.
Cellular and layer organization of the retinal clock
At the cellular level
In early studies, analysis of the expression pattern of core clock genes in the whole retina was the classic approach used to assess the functioning of the retinal clock.50 However, this methodology led to significant dissimilarities among the results observed in different species or even within the same species among different strains.50 Moreover, the cellular diversity of mammalian retina renders difficult the analysis of the circadian clock at the whole-tissue level because differences in phasing and period between cells/layers may result in low-amplitude rhythmicity or masking of coherent clock gene expression.22,38,51–55 It is therefore essential to determine which cell types possess clock properties to understand rhythm generation in the retina. Based on the molecular and physiological data from amphibians and birds, the initial prevailing model for circadian organization in vertebrate retinas proposed that photoreceptors contain self-sustained circadian clocks.56–58 By contrast, the cellular location of the retinal clock in mammals has been a matter of long debate. In early research, retinal degenerative animal models such as the rd mouse and the Royal College of Surgeons rat, both characterized by progressive degeneration of photoreceptors, showed arrhythmic melatonin release, whereas the daily rhythms of DA were retained,5,59 suggesting that photoreceptors are necessary to maintain rhythmic melatonin production, but their integrity was not required for DA signaling. With the discovery of clock genes, photoreceptors have been the site of the most intensive controversy because initial reports claimed that expression of Clock, Bmal1, Per1-2, or Cry2 takes place in the outer nuclear layer (ONL) of rodents,60–62 whereas others failed to detect them.17,52,63 More recently, by using ONL isolated by laser microdissection, vibratome sectioning, or following kainic acid treatment, several studies demonstrated clock property in rat photoreceptors under an LD cycle, but these rhythms were severely damped in constant darkness, indicating that the photoreceptor clock is not robust,64–66 although data obtained in the mouse support the opposite conclusion.38 This latter analysis was the first to compare the kinetics of clock gene expression in ONL and inner retina (ganglion cell layer [GCL] + inner nuclear layer [INL]) and concluded that both retinal compartments show clock gene oscillations, interestingly without any obvious phase relationship (Figure 2). However, only ONL displayed the complete set of oscillating clock genes, suggesting that the main clock is located within the photoreceptors.38 Conversely, several studies support the idea that the generation of rhythms takes place mainly in the inner retina.17,52,63,67 The strongest evidence is provided by the study of Per2Luc mice. When analyzed on an rd background, on which photoreceptors degenerate quite early, PER2Luc activity was shown to be rhythmic52 and generated in the inner retina specifically.10 Responsible cell types have not been identified yet, although DAergic amacrine cells, which have been shown to express the whole set of clock genes,68 are a good candidate. Although their contribution to clock gene oscillations in the retina has been poorly evaluated, ganglion cells have also been reported to harbor clock gene transcripts or products,52,61–63,69 sometimes with sparse distribution. The most extensive ex vivo analysis of the distribution of clock gene expression in the retina was conducted by immunohistochemistry in the mouse. Surprisingly, the authors came to the conclusion that, although clock proteins are present in most cell types besides rods, rhythmic expression is detected uniquely in cones.53 Müller cells, the main glial cell type of the retina, have been demonstrated to also express sustained rhythms in primary culture of rodent or human cells transduced by luciferase-expressing viruses.70 Taken together and in spite of some discrepancy, these data suggest that clock genes are expressed throughout the retina.
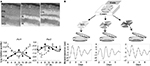  | Figure 2 The retinal clock contains a network of clocks. Notes: (A) Top panel: photomicrographs of a retinal section of wild-type mouse (a) before and after laser microdissection of (b) the IN of the retina, including the INL + GCL, and (c) the ON, which includes the PRL. Bottom panel: circadian expression of Per1 and Per2 clock genes in the inner (solid line) and the outer (dashed line) retinal layers of the wild-type mouse. Note that both compartments, the IN and the ON, show clock gene oscillations, interestingly without any obvious phase relationship. Cell Mol Life Sci, The absence of melanopsin alters retinal clock function and dopamine regulation by light, 70(18), 2013, 3435–3447, Dkhissi-Benyahya O, Coutanson C, Knoblauch K, et al. With permission of Springer.38 (B) Top panel: Vibratome-sectioning method for isolation of the PRL, the INL, and the GCL from freshly mounted retinas. Bottom panel: Representative bioluminescence recordings (detrended data following 24 h running average subtraction) of a GCL (left panel), an INL (middle panel), and a PRL (right panel) sample from PER2Luc mice during the first week of culture. Data from Jaeger et al and Sandu et al.22,66 Abbreviations: cps, counts per second; CT, circadian time; GCL, ganglion cell layer; IN, inner region; INL, inner nuclear layer; ON, outer region; PRL, photoreceptor layer. |
At the level of layers
Studying the specific clock properties and robustness of a retinal subregion by ex vivo analyses is limited by the existence of potential input received from neighboring cells. By using an original, vibratome-based procedure to isolate the retinal cellular layers (PRL, INL, and GCL) from Per2Luc mice, we established that these 3 layers can act as functional, self-sustained circadian pacemakers.22 However, explants of PRLs showed the lowest amplitude and robustness, which correlated with the absence of rhythms in PRLs isolated similarly from Per1-luciferase rats, in which GCL and INL showed clear rhythms (Figure 2).22 These results confirm that the retina is a network of circadian oscillators located in the different layers and in which the INL and GCL provide major contribution. In conclusion, expression of clock genes has been detected in most retinal neurons as well as in glial cells.70 The identity of the major pacemaker, if any, still warrants identification but it is likely that, as in the SCN, the global network plays a role in the sustainability of the retinal clock.
Intercellular/layer synchronization in the retina
Irrespective of the exact cellular location of the retinal clock in mammals, data from early studies indicate that the retinal oscillators are interconnected, notably through the DA/melatonin feedback loop.50 In the current view essentially derived from amphibian and avian models, the secretion of DA by a subset of amacrine cells in the light period and the synthesis of melatonin in PRLs during the night signal to the retina that it is day and night, respectively. Both molecules display reciprocal negative effects, involving, respectively, the melatonin receptors present in the INL, amacrine cells in particular,71,72 and the D2-like DA receptors located on photoreceptors.73,74 Taken together, these cells and neuromodulators constitute an intercellular feedback loop that regulates retinal rhythmic physiology (melatonin and DA displaying overt largely distributed receptors) and sustains the rhythmic activity of the clocks that have not been localized yet. In this respect, the case of most mouse lines is odd because they do not synthesize melatonin and are unable to maintain rhythmic DA release in constant dark conditions,59 although they otherwise display robust PER2Luc oscillations.10,22 This result indicates that DAergic cells, at least in melatonin-deficient mice, are not the site of rhythm generation and that photoreceptors in melatonin-proficient mice, contain a clock releasing melatonin as a synchronizing signal. The localization of the clock contained in the inner retina still awaits identification, but it has been suggested that it does not rely on any synchronizing signal from the remainder of the retina or within the clock population itself. Indeed, in the presence of agonists or antagonists of most common neurotransmitters, as well as upon blocking of neural transmission, the cycling bioluminescence from PER2Luc retinas was barely affected.10,22 Only treatment with gamma-aminobutyric acid (GABA) decreased the amplitude, indicating that it has a negative impact on the synchronization.10,22 We observed that, upon dividing retinas from the same strain of mice into distinct cellular layers or combinations thereof, the period of PER2Luc oscillations was strongly correlated with the degree of dissociation of the tissue. Moreover, communication between the INL and GCL appeared to rely on their synaptic connection because both layers sampled individually and cultured together exhibited periods similar to those of the independent layers and significantly higher than those of INL + GCL sampled together. In addition, the blocking of gap junctions, similar to dissociating the retinas, increased the period of the retinal clock, although this effect was only partial. These data support the idea that the retinal clock is rather composed of several oscillators, which are located in the distinct layers and strongly connected not only by gap junctions but also by other (notably synaptic) signals that remain to be identified.
Functions controlled by the retina clock
Many functions have been described to be rhythmic in the retina exposed to the LD cycle and are thought to optimize adaptation of vision to the daily changes in light intensity. Many of them have been shown to remain rhythmic in constant conditions and, hence, to be regulated by a circadian clock: this is the case for melatonin and DA synthesis,75 phagocytosis of photoreceptor outer segments,1 amplitude of the photopic ERG,35,76 contrast sensitivity,77 and rod–cone coupling.78 Since the demonstration of the existence of an autonomous clock in the mammalian retina,4 it has been essentially assumed that the rhythmic retinal functions are under the control of the retina clock seen as a whole. The ongoing paradigm about how circadian clocks control their outputs is based on the idea that clock transcription factors transfer their rhythmicity to their target genes. In this context, generation of tissue-specific cycling gene expression programs can result from either indirect transcriptional activation by transcription factors that are targets of the clock or direct effects from the combination of clock factors and tissue-specific regulators. By using high-throughput transcriptome analyses, it has been demonstrated that ~43% of protein-encoding genes in mammals are rhythmic.79 One groundbreaking study conducted on whole eyes sampled throughout the 24 h cycle nicely showed that, in LD conditions, the knockout of Bmal1 induces an alteration of rhythmic expression in 90% of the genes, with total loss of rhythmicity in 60% of them.17 This study hence places the circadian clock in the center of rhythmic functions in the retina, Bmal1 being required even to mediate the effects of the LD cycle on gene expression. However, analysis of the genes identified as rhythmic in this study did not reveal clear enrichment in specific functions, most major pathways, including photoreceptor signaling and synaptic transmission, being found in the gene list.17 A similar study was reported more recently, with whole eye globes sampled at 2 zeitgeber times (ZT): ZT1.5, coinciding with the peak of photoreceptor outer segment phagocytosis, and ZT9.80 The authors describe rhythmic expression of genes related to both the phosphoinositide metabolism and regulation of phagocytosis, with maxima correlating with the peak of phagocytosis in the retina. Concomitantly, another study was conducted on rat photoreceptors isolated by laser microdissection at ZT6 and ZT18.81 Forty genes displaying differential day/night expression in PRLs and confirmed in whole retina samples were reported, notably linked to transcriptional regulation, cell cycle and cell death, cell response to environmental signals, and retina-specific functions (Grk1, Rorb, Pax4, and Pgc1a), confirming that the clock controls many distinct processes in photoreceptors.81
More globally, insight into how the clock controls retinal function has been mainly obtained as a result of targeted analysis of gene expression, in most cases by Northern blotting and/or real-time quantitative polymerase chain reaction (qPCR), which has led to a wide list of cycling genes, either in LD or in LD and constant dark (DD) conditions (Table 1). Not surprisingly, the genes identified as rhythmic mainly refer to visual function, indicating that, even if there is a bias in the analysis, photoreceptors display rhythmic activity regulated at the gene expression level. They also indicate that part of this regulation involves cycling transcription factors and signaling proteins that are not retina specific.
  | Table 1 Genes whose expression (mRNA or protein) was reported to vary over the 24 h cycle in light/dark (LD), constant dark (DD), or both conditions (LD and DD) |
Regarding clock-controlled genes, the most investigated ones relate to the regulation of melatonin synthesis, described as a characteristic output from the retinal clock in most vertebrate species examined.2,82,83 In mammals, the site for melatonin synthesis has not been located strictly to the PRL because melatonin synthesis is retained upon photoreceptor degeneration84 and also because the enzymes involved in its synthesis are detected in the GCL.85 However, its regulation in the PRL has been the best characterized. Expression of several enzymes involved in the biochemical cascade generating melatonin was shown to be rhythmic in the rat retina: TPH, HIOMT, and AA-NAT,86 which catalyzes the penultimate, rate-limiting step. The promoter of the Aanat gene contains an E-box sequence, which was shown to be stimulated by BMAL1/CLOCK.87 This gene is likely regulated as well through its well-conserved CRE site, which mediates cyclic adenosine monophosphate (cAMP) activation potentially during the night, following photoreceptor depolarization. This CRE-mediated control probably works also in constant conditions because cAMP is rhythmic in constant darkness.88 Indeed, the BMAL1/CLOCK dimer directly stimulates transcription of Ac1 gene (encoding adenylate cyclase 1), potentially restricting its expression to the end of the day and night, thus gating cAMP increase and Aanat activation to the same temporal window.88
The potential role of the retinal clock in eye disease
A myriad of circadian rhythms has been described in the mammalian eye. Most of them, if not all, are regulated by the retinal clock.50 Furthermore, the mammalian retinal clock controls cell survival and growth processes in the eye, including the susceptibility of photoreceptors to degeneration from light damage or during aging.89,90 The widespread control of signaling, metabolism, and gene expression exerted by the retinal circadian clock suggests that its integrity is critical for maintaining normal rhythmic responses and retina homeostasis. Thus, this organ represents a major target for impairments induced by circadian misalignment or retinal diseases.
Several ocular pathologies, such as diabetes, glaucoma, or age-related macular degeneration, are associated with degeneration of retinal cells, including photoreceptors, which receive and integrate light signals from the environment. These pathologies represent the major causes of blindness that not only affect vision but also lead to a decrease or loss of the photic input to the circadian system, impairing proper synchronization to the environmental 24 h cycle and thus adding to vision loss another strong handicap.
Dysregulation of clock gene expression has been identified as a key factor in the pathogenesis of several diseases or retinal alterations. Bmal1–/– mice display ocular abnormalities, including corneal neovascularization, keratinization, and progressive inflammation,91 associated with reduced life span, as well as premature aging symptoms, such as cataracts, also reported in Clock-deficient mice.92 Clock mutant and Bmal1 conditional knockout mice develop type 1 diabetes,93 whereas Per2-knockout mice exhibit retinal phenotype similar to that found in type 2 diabetes.13–15 Conversely, diabetes is also associated with circadian disruption of clock gene expression. In rodent models of type 194,96 or type 295 diabetes, the expression of several clock genes, including Clock, Bmal1, Per1-Per2, and Cry1-2, was downregulated in the retina, whereas no alterations were observed in the SCN. We reported similar observations using a mouse model of type 1 diabetes induced by streptozotocin injection.55 In this model, the amplitude of Per1 and Cry1 gene expression is upregulated, whereas Per3 loses its circadian rhythmicity in the retina. In addition to the dysregulation of the retinal clock organization, we reported disturbances in one of its main outputs, the DAergic system, with 56% reduction in the total number of tyrosine hydroxylase (TH)-positive cells in diabetic mice,55 in accordance with previous studies performed both in mouse and rat models of diabetes.97–101 The diabetes-associated visual dysfunction reported in rodent models100,102,103 and in diabetic patients who exhibit defects in color perception and contrast sensitivity can be treated by DA precursors or DA agonists.100 However, the underlying mechanisms are still unclear. The decline in DA content during diabetes may be related to a reduced number of retinal DAergic neurons, low levels of DA synthesis, or an impairment of DA release from the cells. In addition, a deficit in light induction of Th, c-Fos, and Per1 in response to 480 nm light has been reported during the development of diabetic retinopathy, possibly through impaired ipRGCs (Figure 3).55,104 In patients with type 2 diabetes, ipRGC dysfunction has been proposed to be investigated for diagnostic purpose by using the postillumination pupil response (PIPR), a sustained constriction of the pupil after light offset.105
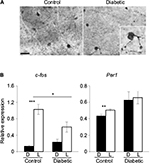  | Figure 3 ipRGCs and light-induced gene expression are altered in a mouse model of diabetic retinopathy. Notes: (A) Representative photomicrographs of flat-mounted retinas from control and diabetic mouse models immunostained by melanopsin antibody at 12 weeks postdiabetes. Morphological abnormalities of ipRGCs, such as soma swelling and increased varicosities in the dendrites, were observed in the peripheral retina of diabetic animals. Enlarged image of a melanopsin-positive cell (inset: magnification ×100). Scale bar =50 mm. (B) Relative light-induced c-Fos and Per1 mRNA levels in the retinas of control and diabetic mice at 12 weeks postdiabetes. Control and diabetic mice were exposed to a 480 nm monochromatic light pulse (1.17×1014 photons/cm2/s, 15 min duration) delivered at CT16. White bars represent animals that received the light pulse (L). The dark control mice (D, black bars) were handled the same way but did not receive any light stimulus. Data are presented as mean value ± SEM. Asterisk indicates a statistically significant difference: *p≤0.05; **p≤0.01; ***p≤0.001 (modified from55,104). Adapted from Lahouaoui H, Coutanson C, Cooper HM, Bennis M, Dkhissi-Benyahya O. Diabetic retinopathy alters light-induced clock gene expression and dopamine levels in the mouse retina. Mol Vis. 2016;22:959.55 Adapted from Lahouaoui H, Coutanson C, Cooper HM, Bennis M, Dkhissi-Benyahya O. Clock genes and behavioral responses to light are altered in a mouse model of diabetic retinopathy. PLoS One. 2014;9(7):e101584.104 Abbreviations: CT, circadian time; ipRGC, intrinsically photosensitive retinal ganglion cell; SEM, standard error of the mean. |
The classic hallmark of glaucomatous optic neuropathies is the progressive loss of RGCs and their axons, with concomitant insidious defects in the visual field. As for RGCs, ipRGC abnormalities have been reported in rodent models of experimental glaucoma106–109 and in glaucomatous patients.110,111 We reported an ∼50%–70% reduction of RGC axon terminals (including ipRGCs) in several visual and nonvisual structures, notably in the SCN, and an alteration of the expression of opsin genes in the retina in an experimental rat model of glaucoma,106 as observed by others.112 In addition, several nonvisual responses to light were affected, such as the pupillary light response, light-induced nocturnal pineal melatonin suppression, and locomotor activity rhythm.106,108 Dysfunction of ipRGCs in animal models of glaucoma have been corroborated by human studies because glaucoma patients with sleep disorders showed a reduction in the pupillary light response and suppression of nocturnal melatonin by light.113–118
The potential role of ipRGCs has been described in other pathological processes. Concurrent regression of embryonic hyaloid vasculature and formation of the retinal vasculature occur postnatally in the mouse,119 and the light response pathway that regulates both processes involves melanopsin.120 In neurodegenerative diseases such as Parkinson’s and Alzheimer’s, a contribution of retinal deficits to circadian abnormalities has been suggested because loss of ipRGCs has been found to be associated with β-amyloid deposits.121 Degeneration of cone and melanopsin pathways has also been reported in a mouse model of Huntington’s disease.122 All these data suggest alteration of both retinal photoreception and photic signal transmission necessary to entrain the retinal and the central clocks in several degenerative diseases. Using impaired circadian photoreception as a marker or biological indicator of retinal or brain pathologies will represent a valuable research tool for diagnostic purposes. ERG anomalies are, for instance, associated with several psychiatric disorders.123
Conclusion
The retina constitutes a fascinating clock model perfectly suitable both to understand the molecular mechanisms of circadian rhythm generation at the cellular and tissue levels and to characterize the synchronizing factors coordinating multiple oscillators at the tissue level. To elucidate these key unresolved questions, the development of in vitro culture of isolated cells or retinal explants from bioluminescent mice models, coupled with a spatial resolution of the signal, is crucial to monitor clock oscillations at the single-cell level in all retinal layers and over several days. Disruption of retinal clock organization or its entrainment by light may have major impacts on visual and nonvisual functions. In addition, given its highly rhythmic physiology, the retina potentially represents a major target for impairments induced by circadian misalignment. Thus, it will be important to assess the potential detrimental effects of repetitive clock phase shifts, such as those induced by shift work or repetitive jet lag, on retina function and health.
Acknowledgments
This research was supported by the University of Strasbourg Institute for Advanced Studies (USIAS) and Coopération et mobilité internationales Rhône-Alpes (Rhône-Alpes CMIRA).
Disclosure
The authors report no conflicts of interest in this work.
References
LaVail M. Rod outer segment disk shedding in rat retina: relationship to cyclic lighting. Science. 1976;194(4269):1071–1074. | ||
Besharse JC, Iuvone PM. Circadian clock in Xenopus eye controlling retinal serotonin N-acetyltransferase. Nature. 1983;305(5930):133–135. | ||
Pierce ME, Sheshberadaran H, Zhang Z, Fox LE, Applebury ML, Takahashi JS. Circadian regulation of iodopsin gene expression in embryonic photoreceptors in retinal cell culture. Neuron. 1993;10(4):579–584. | ||
Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272(5260):419–421. | ||
Tosini G, Menaker M. The clock in the mouse retina: melatonin synthesis and photoreceptor degeneration. Brain Res. 1998;789(2):221–228. | ||
Terman JS, Reme CE, Terman M. Rod outer segment disk shedding in rats with lesions of the suprachiasmatic nucleus. Brain Res. 1993;605(2):256–264. | ||
Sakamoto K, Oishi K, Shiraishi M, et al. Two circadian oscillatory mechanisms in the mammalian retina. Neuroreport. 2000;11(18):3995–3997. | ||
Yamazaki S, Numano R, Abe M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288(5466):682–685. | ||
Yoo S-H, Yamazaki S, Lowrey PL, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101(15):5339–5346. | ||
Ruan G-X, Allen GC, Yamazaki S, McMahon DG. An autonomous circadian clock in the inner mouse retina regulated by dopamine and GABA. PLoS Biol. 2008;6(10):e249. | ||
Besharse JC, McMahon DG. The retina and other light-sensitive ocular clocks. J Biol Rhythms. 2016;31(3):223–243. | ||
Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2016;18(3):164–179. | ||
Bhatwadekar AD, Glenn JV, Curtis TM, Grant MB, Stitt AW, Gardiner TA. Retinal endothelial cell apoptosis stimulates recruitment of endothelial progenitor cells. Invest Opthalmol Vis Sci. 2009;50(10):4967–4973. | ||
Bhatwadekar AD, Yan Y, Qi X, et al. Per2 mutation recapitulates the vascular phenotype of diabetes in the retina and bone marrow. Diabetes. 2013;62(1):273–282. | ||
Jadhav V, Luo Q, Dominguez M J 2nd, et al. Per2-mediated vascular dysfunction is caused by the upregulation of the connective tissue growth factor (CTGF). PLoS One. 2016;11(9):e0163367. | ||
Ait-Hmyed O, Felder-Schmittbuhl M-P, Garcia-Garrido M, et al. Mice lacking Period 1 and Period 2 circadian clock genes exhibit blue cone photoreceptor defects. Eur J Neurosci. 2013;37(7):1048–1060. | ||
Storch K-F, Paz C, Signorovitch J, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130(4):730–741. | ||
Cameron MA, Barnard AR, Hut RA, et al. Electroretinography of wild-type and cry mutant mice reveals circadian tuning of photopic and mesopic retinal responses. J Biol Rhythms. 2008;23(6):489–501. | ||
Mollema NJ, Yuan Y, Jelcick AS, et al. Nuclear receptor Rev-erb alpha (Nr1d1) functions in concert with Nr2e3 to regulate transcriptional networks in the retina. PLoS One. 2011;6(3):e17494. | ||
Ait-Hmyed Hakkari O, Acar N, Savier E, et al. Rev-Erbα modulates retinal visual processing and behavioral responses to light. FASEB J. 2016;30(11):3690–3701. | ||
Ruan G-X, Gamble KL, Risner ML, Young LA, McMahon DG. Divergent roles of clock genes in retinal and suprachiasmatic nucleus circadian oscillators. PLoS One. 2012;7(6):e38985. | ||
Jaeger C, Sandu C, Malan A, Mellac K, Hicks D, Felder-Schmittbuhl M-P. Circadian organization of the rodent retina involves strongly coupled, layer-specific oscillators. FASEB J. 2015;29(4):1493–1504. | ||
Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447(7143):477–481. | ||
Freedman MS, Lucas RJ, Soni B, et al. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science. 1999;284(5413):502–504. | ||
Lucas RJ, Freedman MS, Munoz M, Garcia-Fernadez J, Foster RG. Novel ocular photoreceptors regulate the mammalian circadian system: II. Acute inhibition of pineal melatonin. Science. 1999;284(5413):502–504. | ||
Provencio I, Jiang G, De Grip WJ, Hayes WP, Rollag MD. Novel skin and brain opsin, melanopsin is found in the chicken. Invest Ophthalmol Vis Sci. 1998;39(4):S236. | ||
Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295(5557):1070–1073. | ||
Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295(5557):1065–1070. | ||
Hughes S, Jagannath A, Rodgers J, Hankins MW, Peirson SN, Foster RG. Signalling by melanopsin (OPN4) expressing photosensitive retinal ganglion cells. Eye. 2016;30(2):247–254. | ||
Dkhissi-Benyahya O, Gronfier C, De Vanssay W, Flamant F, Cooper HM. Modeling the role of mid-wavelength cones in circadian responses to light. Neuron. 2007;53(5):677–687. | ||
Altimus CM, Güler AD, Alam NM, et al. Rod photoreceptors drive circadian photoentrainment across a wide range of light intensities. Nat Neurosci. 2010;13(9):1107–1112. | ||
Dollet A, Albrecht U, Cooper HM, Dkhissi-Benyahya O. Cones are required for normal temporal responses to light of phase shifts and clock gene expression. Chronobiol Int. 2010;27(4):768–781. | ||
Lall GS, Revell VL, Momiji H, et al. Distinct contributions of rod, cone, and melanopsin photoreceptors to encoding irradiance. Neuron. 2010;66(3):417–428. | ||
Lucas RJ, Lall GS, Allen AE, Brown TM. How rod, cone, and melanopsin photoreceptors come together to enlighten the mammalian circadian clock. Prog Brain Res. 2012;199:1–18. | ||
Barnard AR, Hattar S, Hankins MW, Lucas RJ. Melanopsin regulates visual processing in the mouse retina. Curr Biol. 2006;16(4):389–395. | ||
Allen AE, Storchi R, Martial FP, et al. Melanopsin-driven light adaptation in mouse vision. Curr Biol. 2014;24(21):2481–2490. | ||
Milosavljevic N, Allen AE, Cehajic-Kapetanovic J, Lucas RJ. Chemogenetic activation of ipRGCs drives changes in dark-adapted (scotopic) electroretinogram. Invest Opthalmol Vis Sci. 2016;57(14):6305. | ||
Dkhissi-Benyahya O, Coutanson C, Knoblauch K, et al. The absence of melanopsin alters retinal clock function and dopamine regulation by light. Cell Mol Life Sci. 2013;70(18):3435–3447. | ||
Wong KY, Dunn FA, Graham DM, Berson DM. Synaptic influences on rat ganglion-cell photoreceptors. J Physiol. 2007;582(pt 1):279–296. | ||
Zhang D-Q, Belenky MA, Sollars PJ, Pickard GE, McMahon DG. Melanopsin mediates retrograde visual signaling in the retina. PLoS One. 2012;7(8):e42647. | ||
Reifler AN, Chervenak AP, Dolikian ME, et al. All spiking, sustained ON displaced amacrine cells receive gap-junction input from melanopsin ganglion cells. Curr Biol. 2015;25(21):2763–2773 | ||
Prigge CL, Yeh P-T, Liou N-F, et al. M1 ipRGCs influence visual function through retrograde signaling in the retina. J Neurosci. 2016;36(27):7184–7197. | ||
Zhang D-QQ, Wong KY, Sollars PJ, Berson DM, Pickard GE, McMahon DG. Intraretinal signaling by ganglion cell photoreceptors to dopaminergic amacrine neurons. Proc Natl Acad Sci U S A. 2008;105(37):14181–14186. | ||
Buhr ED, Van Gelder RN. Local photic entrainment of the retinal circadian oscillator in the absence of rods, cones, and melanopsin. Proc Natl Acad Sci U S A. 2014;111(23):8625–8630. | ||
Buhr ED, Yue WWS, Ren X, et al. Neuropsin (OPN5)-mediated photoentrainment of local circadian oscillators in mammalian retina and cornea. Proc Natl Acad Sci U S A. 2015;112(42):13093–13098. | ||
Kojima D, Mori S, Torii M, Wada A, Morishita R, Fukada Y. UV-sensitive photoreceptor protein OPN5 in humans and mice. PLoS One. 2011;6(10):e26388. | ||
Yamashita T, Ono K, Ohuchi H, et al. Evolution of mammalian Opn5 as a specialized UV-absorbing pigment by a single amino acid mutation. J Biol Chem. 2014;289(7):3991–4000. | ||
Tarttelin EE, Bellingham J, Hankins MW, Foster RG, Lucas RJ. Neuropsin (Opn5): a novel opsin identified in mammalian neural tissue. FEBS Lett. 2003;554(3):410–416. | ||
Hughes S, Rodgers J, Hickey D, Foster RG, Peirson SN, Hankins MW. Characterisation of light responses in the retina of mice lacking principle components of rod, cone and melanopsin phototransduction signalling pathways. Sci Rep. 2016;6:28086. | ||
McMahon DG, Iuvone M, Tosini G. Circadian organization of the mammalian retina: from gene regulation to physiology and diseases. Prog Retin Eye Res. 2014;39:58–76. | ||
Peirson SN, Butler JN, Duffield GE, Takher S, Sharma P, Foster RG. Comparison of clock gene expression in SCN, retina, heart, and liver of mice. Biochem Biophys Res Commun. 2006;351(4):800–807. | ||
Ruan G-X, Zhang D-QQ, Zhou T-R, Yamazaki S, McMahon DG. Circadian organization of the mammalian retina. Proc Natl Acad Sci U S A. 2006;103(25):9703–9708. | ||
Liu X, Zhang Z, Ribelayga CP. Heterogeneous expression of the core circadian clock proteins among neuronal cell types in mouse retina. PLoS One. 2012;7(11):e50602. | ||
Buonfiglio DC, Malan A, Sandu C, et al. Rat retina shows robust circadian expression of clock and clock output genes in explant culture. Mol Vis. 2014;20:742–752. | ||
Lahouaoui H, Coutanson C, Cooper HM, Bennis M, Dkhissi-Benyahya O. Diabetic retinopathy alters light-induced clock gene expression and dopamine levels in the mouse retina. Mol Vis. 2016;22:959. | ||
Cahill GM, Besharse JC. Circadian clock functions localized in xenopus retinal photoreceptors. Neuron. 1993;10(4):573–577. | ||
Zhu H, LaRue S, Whiteley A, Steeves TD, Takahashi JS, Green CB. The Xenopus clock gene is constitutively expressed in retinal photoreceptors. Brain Res Mol Brain Res. 2000;75(2):303–308. | ||
Zhu H, Green CB. Three cryptochromes are rhythmically expressed in Xenopus laevis retinal photoreceptors. Mol Vis. 2001;7:210–215. | ||
Doyle SE, Grace MS, McIvor W, Menaker M. Circadian rhythms of dopamine in mouse retina: the role of melatonin. Vis Neurosci. 2002;19(5):593–601. | ||
Gekakis N, Staknis D, Nguyen HB, et al. Role of the clock protein in the mamalian circadian mechanism. Science. 1998;280:1564–1568. | ||
Namihira M, Honma S, Abe H, Masubuchi S, Ikeda M, Honma K. Circadian pattern, light responsiveness and localization of rPer1 and rPer2 gene expression in the rat retina. Neuroreport. 2001;12(3):471–475. | ||
Dinet V, Ansari N, Torres-Farfan C, Korf H-WW. Clock gene expression in the retina of melatonin-proficient (C3H) and melatonin-deficient (C57BL) mice. J Pineal Res. 2007;42(1):83–91. | ||
Witkovsky P, Veisenberger E, LeSauter J, et al. Cellular location and circadian rhythm of expression of the biological clock gene Period 1 in the mouse retina. J Neurosci. 2003;23(20):7670–7676. | ||
Tosini G, Davidson AJ, Fukuhara C, Kasamatsu M, Castanon-Cervantes O. Localization of a circadian clock in mammalian photoreceptors. FASEB J. 2007;21(14):3866–3871. | ||
Schneider K, Tippmann S, Spiwoks-Becker I, et al. Unique clockwork in photoreceptor of rat. J Neurochem. 2010;115(3):585–594. | ||
Sandu C, Hicks D, Felder-Schmittbuhl M-P. Rat photoreceptor circadian oscillator strongly relies on lighting conditions. Eur J Neurosci. 2011;34(3):507–516. | ||
Miyamoto Y, Sancar A. Vitamin B2-based blue light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc Natl Acad Sci U S A. 1998;95(11):6097–6102. | ||
Dorenbos R, Contini M, Hirasawa H, Gustincich S, Raviola E. Expression of circadian clock genes in retinal dopaminergic cells. Vis Neurosci. 2007;24(4):573–580. | ||
García-Fernández JM, Álvarez-López C, Cernuda-Cernuda R. Cytoplasmic localization of mPER1 clock protein isoforms in the mouse retina. Neurosci Lett. 2007;419(1):55–58. | ||
Xu L, Ruan G, Dai H, Liu AC, Penn J, Mcmahon DG. Mammalian retinal Müller cells have circadian clock function. Mol Vis. 2016;22:275–283. | ||
Dubocovich ML. Melatonin is a potent modulator of dopamine release in the retina. Nature. 1983;306(5945):782–784. | ||
Fujieda H, Hamadanizadeh S, Wankiewicz E, Pang SF, Brown GM. Expression of mt1 melatonin receptor in rat retina: evidence for multiple cell targets for melatonin. Neuroscience. 1999;93(2):793–799. | ||
Nguyen-Legros J, Chanut E, Versaux-Botteri C, Simon A, Trouvin J-H. Dopamine inhibits melatonin synthesis in photoreceptor cells through a D2-like receptor subtype in the rat retina: biochemical and histochemical evidence. J Neurochem. 1996;67(6):2514–2520. | ||
Tosini G, Dirden JC. Dopamine inhibits melatonin release in the mammalian retina: in vitro evidence. Neurosci Lett. 2000;286(2):119–122. | ||
Iuvone PM, Tosini G, Pozdeyev N, Haque R, Klein DC, Chaurasia SS. Circadian clocks, clock networks, arylalkylamine N-acetyltransferase, and melatonin in the retina. Prog Retin Eye Res. 2005;24(4):433–456. | ||
Danilenko KV, Plisov IL, Cooper HM, Wirz-Justice A, Hébert M. Human cone light sensitivity and melatonin rhythms following 24-hour continuous illumination. Chronobiol Int. 2011;28(5):407–414. | ||
Hwang CK, Chaurasia SS, Jackson CR, Chan GC-K, Storm DR, Iuvone PM. Circadian rhythm of contrast sensitivity is regulated by a dopamine–neuronal pas-domain protein 2–adenylyl cyclase 1 signaling pathway in retinal ganglion cells. J Neurosci. 2013;33(38):14989–14997. | ||
Ribelayga CP, Cao Y, Mangel SC. The circadian clock in the retina controls rod-cone coupling. Neuron. 2008;59(5):790–801. | ||
Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111(45):16219–16224. | ||
Mustafi D, Kevany BM, Genoud C, Bai X, Palczewski K. Photoreceptor phagocytosis is mediated by phosphoinositide signaling. FASEB J. 2013;27(11):4585–4595. | ||
Kunst S, Wolloscheck T, Hölter P, et al. Transcriptional analysis of rat photoreceptor cells reveals daily regulation of genes important for visual signaling and light damage susceptibility. J Neurochem. 2013;124(6):757–769. | ||
Hamm HE, Menaker M. Retinal rhythms in chicks: circadian variation in melantonin and serotonin N-acetyltransferase activity. Proc Natl Acad Sci U S A. 1980;77(8):4998–5002. | ||
Pang SF, Yu HS, Suen HC, Brown GM. Melatonin in the retina of rats: a diurnal rhythm. J Endocrinol. 1980;87(1):89–93. | ||
Sakamoto K, Liu C, Tosini G. Classical photoreceptors regulate melanopsin mRNA levels in the rat retina. J Neurosci. 2004;24(43):9693–9697. | ||
Gianesini C, Clesse D, Tosini G, Hicks D, Laurent V. Unique regulation of the melatonin synthetic pathway in the retina of diurnal female Arvicanthis ansorgei (Rodentia). Endocrinology. 2015;156(9):3292–3308. | ||
Tosini G, Baba K, Hwang CK, Iuvone PM. Melatonin: an underappreciated player in retinal physiology and pathophysiology. Exp Eye Res. 2012;103:82–89. | ||
Tosini G, Fukuhara C. The mammalian retina as a clock. Cell Tissue Res. 2002;309(1):119–126. | ||
Fukuhara C, Liu C, Ivanova TN, et al. Gating of the cAMP signaling cascade and melatonin synthesis by the circadian clock in mammalian retina. J Neurosci. 2004;24(8):1803–1811. | ||
Organisciak DT, Darrow RM, Barsalou L, Kutty RK, Wiggert B. Circadian-dependent retinal light damage in rats. Invest Opthalmol Vis Sci. 2000;41(12):3694–3701. | ||
Baba K, Pozdeyev N, Mazzoni F, et al. Melatonin modulates visual function and cell viability in the mouse retina via the MT1 melatonin receptor. Proc Natl Acad Sci U S A. 2009;106(35):15043–15048. | ||
Yang G, Chen L, Grant GR, et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med. 2016;8(324):324ra16. | ||
Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20(14):1868–1873. | ||
Marcheva B, Ramsey KM, Buhr ED, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627–631. | ||
Kuriyama K, Sasahara K, Kudo T, Shibata S. Daily injection of insulin attenuated impairment of liver circadian clock oscillation in the streptozotocin-treated diabetic mouse. FEBS Lett. 2004;572(1–3):206–210. | ||
Busik JV, Tikhonenko M, Bhatwadekar AD, et al. Diabetic retinopathy is associated with bone marrow neuropathy and a depressed peripheral clock. J Exp Med. 2009;206(13):2897–2906. | ||
Wang Q, Tikhonenko M, Bozack SN, et al. Changes in the daily rhythm of lipid metabolism in the diabetic retina. PLoS One. 2014;9(4):e95028. | ||
Nishimura C, Kuriyama K. Alterations in the retinal dopaminergic neuronal system in rats with streptozotocin-induced diabetes. J Neurochem. 1985;45(2):448–455. | ||
Seki M, Tanaka T, Nawa H, et al. Involvement of brain-derived neurotrophic factor in early retinal neuropathy of streptozotocin-induced diabetes in rats. Diabetes. 2004;53(9):2412–2419. | ||
Gastinger MJ, Singh RSJ, Barber AJ. Loss of cholinergic and dopaminergic amacrine cells in streptozotocin-diabetic rat and Ins2Akita-diabetic mouse retinas. Invest Opthalmol Vis Sci. 2006;47(7):3143. | ||
Aung MH, Park HN, Han MK, et al. Dopamine deficiency contributes to early visual dysfunction in a rodent model of type 1 diabetes. J Neurosci. 2014;34(3):726–736. | ||
Szabadfi K, Szabo A, Kiss P, et al. PACAP promotes neuron survival in early experimental diabetic retinopathy. Neurochem Int. 2014;64:84–91. | ||
Kirwin SJ, Kanaly ST, Hansen CR, Cairns BJ, Ren M, Edelman JL. Retinal gene expression and visually evoked behavior in diabetic long evans rats. Invest Opthalmol Vis Sci. 2011;52(10):7654. | ||
Akimov NP, Rentería RC. Spatial frequency threshold and contrast sensitivity of an optomotor behavior are impaired in the Ins2Akita mouse model of diabetes. Behav Brain Res. 2012;226(2):601–605. | ||
Lahouaoui H, Coutanson C, Cooper HM, Bennis M, Dkhissi-Benyahya O. Clock genes and behavioral responses to light are altered in a mouse model of diabetic retinopathy. PLoS One. 2014;9(7):e101584. | ||
Feigl B, Zele AJ, Fader SM, Howes AN, Hughes CE, Jones KA. The post-illumination pupil response of melanopsin-expressing intrinsically photosensitive retinal ganglion cells in diabetes. Acta Ophthalmol Scand. 2012;90(3):230–234. | ||
Drouyer E, Dkhissi-Benyahya O, Chiquet C, et al. Glaucoma alters the circadian timing system. PLoS One. 2008;3(12):e3931. | ||
Wang H, Lu Q, Wang N, Liu H, Zhang L, Zhan G. Loss of melanopsin-containing retinal ganglion cells in a rat glaucoma model. Chin Med J (Engl). 2008;121(11):1015–1019. | ||
de Zavalía N, Plano SA, Fernandez DC, et al. Effect of experimental glaucoma on the non-image forming visual system. J Neurochem. 2011;117(5):904–914. | ||
El-Danaf RN, Huberman AD. Characteristic patterns of dendritic remodeling in early-stage glaucoma: evidence from genetically identified retinal ganglion cell types. J Neurosci. 2015;35(6):2329–2343. | ||
Adhikari P, Zele AJ, Thomas R, Feigl B. Quadrant field pupillometry detects melanopsin dysfunction in glaucoma suspects and early glaucoma. Sci Rep. 2016;6:33373. | ||
Kelbsch C, Maeda F, Strasser T, et al. Pupillary responses driven by ipRGCs and classical photoreceptors are impaired in glaucoma. Graefes Arch Clin Exp Ophthalmol. 2016;254(7):1361–1370. | ||
Ortín-Martínez A, Salinas-Navarro M, Nadal-Nicolás FM, et al. Laser-induced ocular hypertension in adult rats does not affect non-RGC neurons in the ganglion cell layer but results in protracted severe loss of cone-photoreceptors. Exp Eye Res. 2015;132:17–33. | ||
Pérez-Rico C, de la Villa P, Arribas-Gómez I, Blanco R. Evaluation of functional integrity of the retinohypothalamic tract in advanced glaucoma using multifocal electroretinography and light-induced melatonin suppression. Exp Eye Res. 2010;91(5):578–583. | ||
Feigl B, Mattes D, Thomas R, Zele AJ. Intrinsically photosensitive (melanopsin) retinal ganglion cell function in glaucoma. Invest Opthalmol Vis Sci. 2011;52(7):4362. | ||
Kankipati L, Girkin CA, Gamlin PD. The post-illumination pupil response is reduced in glaucoma patients. Invest Opthalmol Vis Sci. 2011;52(5):2287–2292. | ||
Lanzani MF, de Zavalía N, Fontana H, Sarmiento MIK, Golombek D, Rosenstein RE. Alterations of locomotor activity rhythm and sleep parameters in patients with advanced glaucoma. Chronobiol Int. 2012;29(7):911–919. | ||
Münch M, Léon L, Collomb S, Kawasaki A. Comparison of acute non-visual bright light responses in patients with optic nerve disease, glaucoma and healthy controls. Sci Rep. 2015;5:15185. | ||
Obara EA, Hannibal J, Heegaard S, Fahrenkrug J. Loss of melanopsin-expressing retinal ganglion cells in severely staged glaucoma patients. Invest Opthalmol Vis Sci. 2016;57(11):4661. | ||
Fruttiger M. Development of the retinal vasculature. Angiogenesis. 2007;10(2):77–88. | ||
Rao S, Chun C, Fan J, et al. A direct and melanopsin-dependent fetal light response regulates mouse eye development. Nature. 2013;494(7436):243–246. | ||
La Morgia C, Ross-Cisneros FN, Koronyo Y, et al. Melanopsin retinal ganglion cell loss in Alzheimer disease. Ann Neurol. 016;79(1):90–109. | ||
Ouk K, Hughes S, Pothecary CA, Peirson SN, Jennifer Morton A. Attenuated pupillary light responses and downregulation of opsin expression parallel decline in circadian disruption in two different mouse models of Huntington’s disease. Hum Mol Genet. 2016;25(24):ddw359. | ||
Lavoie J, Maziade M, Hébert M. The brain through the retina: the flash electroretinogram as a tool to investigate psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2014;48:129–134. | ||
Servier Medical Art [homepage on the Internet]. Available from: http://www.servier.com/Powerpoint-image-bank. Accessed January 1, 2017. | ||
Li H, Zhang Z, Blackburn MR, Wang SW, Ribelayga CP, O’Brien J. Adenosine and dopamine receptors coregulate photoreceptor coupling via gap junction phosphorylation in mouse retina. J Neurosci. 2013;33(7):3135–3150. | ||
Roseboom PH, Coon SL, Baler R, McCune SK, Weller JL, Klein DC. Melatonin synthesis: analysis of the more than 150-fold nocturnal increase in serotonin N-acetyltransferase messenger ribonucleic acid in the rat pineal gland. Endocrinology. 1996;137(7):3033–3045. | ||
Sakamoto K, Ishida N. Circadian expression of serotonin N-acetyltransferase mRNA in the rat retina. Neurosci Lett. 1998;245(2):113–116. | ||
Bai L, Zimmer S, Rickes O, et al. Daily oscillation of gene expression in the retina is phase-advanced with respect to the pineal gland. Brain Res. 2008;1203:89–96. | ||
Bobu C, Sandu C, Laurent V, Felder-Schmittbuhl M-P, Hicks D. Prolonged light exposure induces widespread phase shifting in the circadian clock and visual pigment gene expression of the Arvicanthis ansorgei retina. Mol Vis. 2013;19:1060–1073. | ||
Vancura P, Wolloscheck T, Baba K, Tosini G, Iuvone PM, Spessert R. Circadian and Dopaminergic Regulation of Fatty Acid Oxidation Pathway Genes in Retina and Photoreceptor Cells. Bartell PA, ed. PLoS One. 2016;11(10):e0164665. | ||
Jackson CR, Chaurasia SS, Hwang CK, Iuvone PM. Dopamine D4 receptor activation controls circadian timing of the adenylyl cyclase 1/cyclic AMP signaling system in mouse retina. Eur J Neurosci. 2011;34(1):57–64. | ||
Katti C, Butler R, Sekaran S. Diurnal and circadian regulation of connexin 36 transcript and protein in the mammalian retina. Invest Ophthalmol Vis Sci. 2013;54(1):821–829. | ||
de Zavalía N, Fernandez DC, Sande PH, et al. Circadian variations of prostaglandin E2 and F2 α release in the golden hamster retina. J Neurochem. 2010;112(4):972–979. | ||
Yan Y, Salazar TE, Dominguez JM, et al. Dicer expression exhibits a tissue-specific diurnal pattern that is lost during aging and in diabetes. PLoS One. 2013;8(11):e80029. | ||
Klitten LL, Rath MF, Coon SL, Kim J-S, Klein DC, Møller M. Localization and regulation of dopamine receptor D4 expression in the adult and developing rat retina. Exp Eye Res. 2008;87(5):471–477. | ||
Humphries A, Carter DA. Circadian dependency of nocturnal immediate-early protein induction in rat retina. Biochem Biophys Res Commun. 2004;320(2):551–556. | ||
Man P-S, Evans T, Carter DA. Rhythmic expression of an egr-1 transgene in rats distinguishes two populations of photoreceptor cells in the retinal outer nuclear layer. Mol Vis. 2008;14:1176–1186. | ||
Kunst S, Wolloscheck T, Grether M, Trunsch P, Wolfrum U, Spessert R. Photoreceptor cells display a daily rhythm in the orphan receptor Esrrβ. Mol Vis. 2015;21:173–184. | ||
Yoshida K, Kawamura K, Imaki J. Differential expression of c-fos mRNA in rat retinal cells: Regulation by light/dark cycle. Neuron. 1993;10(6):1049–1054. | ||
Kamphuis W, Cailotto C, Dijk F, Bergen A, Buijs RM. Circadian expression of clock genes and clock-controlled genes in the rat retina. Biochem Biophys Res Commun. 2005;330(1):18–26. | ||
Brann M, Cohen L. Diurnal expression of transducin mRNA and translocation of transducin in rods of rat retina. Science 1987;235(4788):585–587. | ||
Gauer F, Craft CM. Circadian regulation of hydroxyindole-O-methyltransferase mRNA levels in rat pineal and retina. Brain Res. 1996;737(1):99–109. | ||
Hölter P, Kunst S, Wolloscheck T, et al. The retinal clock drives the expression of Kcnv2 , a channel essential for visual function and cone survival. Investig Opthalmology Vis Sci. 2012;53(11):6947. | ||
Law A-L, Parinot C, Chatagnon J, et al. Cleavage of Mer tyrosine kinase (MerTK) from the cell surface contributes to the regulation of retinal phagocytosis. J Biol Chem. 2015;290(8):4941–4952. | ||
Wang Y, Osterbur DL, Megaw PL, et al. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol. 2001;1(1):9. | ||
Kunst S, Wolloscheck T, Kelleher DK, et al. Pgc-1α and Nr4a1 Are Target Genes of Circadian Melatonin and Dopamine Release in Murine Retina. Invest Ophthalmol Vis Sci. 2015;56(10):6084–6094. | ||
Sakamoto K, Liu C, Kasamatsu M, Iuvone M, Tosini G. Intraocular injection of kainic acid does not abolish the circadian rhythm of arylalkylamine N-acetyltransferase mRNA in rat photoreceptors. Mol Vis. 2006;12:117–124. | ||
von Schantz M, Lucas RJ, Foster RG. Circadian oscillation of photopigment transcript levels in the mouse retina. Brain Res Mol Brain Res. 1999;72(1):108–114. | ||
Mathes A, Engel L, Holthues H, Wolloscheck T, Spessert R. Daily Profile in Melanopsin Transcripts Depends on Seasonal Lighting Conditions in the Rat Retina. J Neuroendocrinol. 2007;19(12):952–957. | ||
Sakamoto K, Liu C, Kasamatsu M, Pozdeyev N V, Iuvone PM, Tosini G. Dopamine regulates melanopsin mRNA expression in intrinsically photosensitive retinal ganglion cells. Eur J Neurosci. 2005;22(12):3129–3136. | ||
Hannibal J, Georg B, Fahrenkrug J. Differential expression of melanopsin mRNA and protein in Brown Norwegian rats. Exp Eye Res. 2013;106:55–63. | ||
Wolloscheck T, Spiwoks-Becker I, Rickes O, Holthues H, Spessert R. Phosphodiesterase10A: Abundance and circadian regulation in the retina and photoreceptor of the rat. Brain Res. 2011;1376:42–50. | ||
Wiechmann AF, Sinacola MK. Diurnal expression of recoverin in the rat retina. Mol Brain Res. 1997;45(2):321–324. | ||
Ban N, Ozawa Y, Inaba T, et al. Light–dark condition regulates sirtuin mRNA levels in the retina. Exp Gerontol. 2013;48(11):1212–1217. | ||
Liang J, Wessel JH, Iuvone PM, Tosini G, Fukuhara C. Diurnal rhythms of tryptophan hydroxylase 1 and 2 mRNA expression in the rat retina. Neuroreport. 2004;15(9):1497–1500. | ||
Scoma HD, Humby M, Yadav G, Zhang Q, Fogerty J, Besharse JC. The de-ubiquitinylating enzyme, USP2, is associated with the circadian clockwork and regulates its sensitivity to light. PLoS One. 2011;6(9):e25382. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
