Back to Journals » OncoTargets and Therapy » Volume 8
The relationship between UGT1A1 gene polymorphism and irinotecan effect on extensive-stage small-cell lung cancer
Authors Xiao X, Xia S, Zou M, Mei Q, Zhou L, Wang S, Chen Y
Received 26 August 2015
Accepted for publication 4 November 2015
Published 3 December 2015 Volume 2015:8 Pages 3575—3583
DOI https://doi.org/10.2147/OTT.S95149
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Xiao-guang Xiao, Shu Xia, Man Zou, Qi Mei, Lei Zhou, Shu-jing Wang, Yuan Chen
Department of Oncology, Tongji Hospital, Tongji Medical College of Huazhong University of Science and Technology, Wuhan, People’s Republic of China
Aims: To analyze the distribution of uridine diphosphate glucuronosyltransferase (UGT)1A1 gene polymorphisms in Chinese patients with extensive-stage small-cell lung cancer (E-SCLC), and to evaluate correlations between the UGT1A1 gene polymorphisms and toxicity, and efficacy of irinotecan (CPT-11) based regimen in the patients with E-SCLC.
Methods: The study analyzed the distribution of UGT1A1*28/*6 gene polymorphisms by polymerase chain reaction amplification and pyrosequencing. The analysis of UGT1A1*28 and UGT1A1*6 gene polymorphisms was performed in 67 patients with E-SCLC admitted to the clinic in the Department of Oncology from June 2011 to January 2013. A total of 67 cases with E-SCLC treated with irinotecan (CPT-11)-based regimen were enrolled to observe the adverse events and efficacy during the chemotherapy, including objective response rate, progression-free survival (PFS) and overall survival (OS). The correlation between UGT1A1 gene polymorphisms and severe adverse events was analyzed. The influences of UGT1A1*6/*28 polymorphisms on objective response rate, PFS, and OS were also analyzed.
Results: The distribution of UGT1A1 genotypes among 67 patients was as follows: UGT1A1*28 wild-type (WT) genotype TA6/6 (56, 83.6%), heterozygous mutant genotype TA6/7 (11, 16.4%); UGT1A1*6 WT genotype G/G (45, 67.2%), heterozygous mutant genotype G/A (22, 32.8%); no significant difference of PFS and OS was observed between different genotypes. The incidence of grade 3 and 4 delayed diarrhea and neutropenia in the patients carrying UGT1A1*6 G/A mutation was higher than that in the WT genotype (36.4% vs 6.6% P=0.034; 27.2% vs 4.4% P=0.026, respectively). The incidence of grade 3 and 4 thrombocytopenia in the patients carrying UGT1A1*28 TA6/7 mutation was higher than that in the WT genotype (27.2% vs 1.8% P=0.017). The patients simultaneously carrying UGT1A1*28 TA6/7 and UGT1A1*6 G/A mutations were prone to suffering grade 3 and 4 delayed diarrhea and neutropenia.
Conclusion: For irinotecan-based regimens in E-SCLC, the UGT1A1*28 and UGT1A1*6 locus mutations can be regarded as predictors for severe adverse events. We also found that neither clinical response nor prognosis was significantly associated with the UGT1A1 gene polymorphisms.
Keywords: small-cell lung cancer, irinotecan, uridine diphosphate glucuronosyltransferase 1A1, gene polymorphism
Introduction
As a type of lung cancer with special pathological features, small-cell lung cancer (SCLC) is highly malignant and has the characteristics of poor differentiation, short tumor cell doubling time, fast tumor progress, and being prone to metastases at an early stage. Furthermore, approximately two-thirds of SCLC cases clinically belong to extensive-stage (E-SCLC) when initially diagnosed.1 Many combinations have been evaluated in the patients with extensive-stage disease, with little consistent evidence of benefit when compared with etoposide and cisplatin (EP). The combination of irinotecan and a platinum agent has provided the greatest challenge to EP. Initially, in 2002, a small Phase III trial performed in Japan reported that patients with E-SCLC who were treated with irinotecan plus cisplatin (IP) experienced a median survival of 12.8 months compared with 9.4 months for patients treated with EP (P=0.002). In addition, the 2-year survival was 19.5% in the IP group vs 5.2% in the EP group, respectively.2 Compared to the EP regimen, IP program can significantly prolong the survival of the patients with tolerable adverse effect. However, two subsequent large Phase III trials performed in the United States comparing IP with EP failed to show a significant difference in response rate or overall survival (OS) between the regimens.3,4 A Phase III randomized trial found that median OS was slightly improved with irinotecan and carboplatin compared with carboplatin and oral etoposide (8.5 vs 7.1 months, P=0.04).5 Based on these findings, the carboplatin and irinotecan regimen has been added to the National Comprehensive Cancer Network Guidelines as an option for patients with extensive-stage disease.6 A meta-analysis suggests an improvement in progression-free survival (PFS) and OS with IP regimens compared with etoposide plus platinum regimens.7 However, the relatively small absolute survival benefit needs to be balanced against the toxicity profile of irinotecan-based regimens. Therefore, the National Comprehensive Cancer Network Panel continues to consider etoposide plus platinum as the standard regimen for patients with either limited-stage or E-SCLC.
Uridine diphosphate glucuronosyltransferase (UGT)1A1 plays an important role in the metabolism of irinotecan. Extensive research attention had been paid to the correlation between UGT1A1 gene polymorphism and irinotecan-related delayed diarrhea and neutropenia, but it is still controversial. Several clinical studies conducted in colorectal cancer showed that the polymorphism of UGT1A1*28 gene can be used to evaluate the risk of severe neutropenia and diarrhea occurring in patients receiving irinotecan chemotherapy. In 2005, the US Food and Drug Administration required that testing for UGT1A1*28 as a risk factor for severe adverse events should be included on the irinotecan label.8
The clinical studies conducted outside of the People’s Republic of China suggested the risk of severe granulocyte decrease associated with CPT-11 chemotherapy can be assessed by analyzing the polymorphism of UGT1A1*28. The risk of grade 3–4 neutropenia and diarrhea is significantly increased for the patient carrying the mutations of UGT1A1*28.9,10 However, there were significant racial differences in the distribution of UGT1A1*28 gene polymorphisms. For instance, the proportion for Asian populations with UGT1A1*28 homozygous mutant (TA7/7) is only from 0% to 5%, which was much lower than the frequency of 12%–27% in Africans and 5%–15% in Caucasians.9–11 Therefore, whether the UGT1A1*28 gene polymorphisms can be the predictors of adverse reactions in the same way for Chinese patients remain unclear. The study by Gao et al found that the Asian patients who received CPT-11 based treatment and carrying UGT1A1*6 mutations had the risk of the grade 4 neutropenia, thrombocytopenia and diarrhea significantly increased.12 Comparatively, the mutation of UGT1A1*6 gene was more common in Asian populations, which accounted for more than 20%.13 Furthermore, studies suggested that UGT1A1*6 and UGT1A1*28 were similar in function, and that both of them can lead to the reduction of metabolism of CPT-11 inactivity in vivo and increase the risk for adverse reactions. Therefore, when predicting the occurrence of relevant adverse events in the Chinese patient population, we should take UGT1A1*28 and UGT1A1*6 superior gene polymorphisms’ effect together into consideration.
This study examined UGT1A1*28 and UGT1A1*6 gene polymorphism in 67 patients with E-SCLC who were willing to receive irinotecan-based chemotherapy. The purpose is to explore the distribution frequency of UGT1A1 gene polymorphism in the Chinese patient population and to evaluate correlations between UGT1A1 gene polymorphism and toxicity and efficacy of irinotecan in patients with E-SCLC.
Patients and methods
Clinical data
A total of 67 patients with E-SCLC treated in the Department of Oncology (Tongji Hospital, Tongji Medical College of Huazhong University of Science and Technology) from June 2011 to January 2013 were retrospectively included in this study. The study was approved by the medical ethics committee of Tongji Hospital, Tongji Medical College of Huazhong University of Science and Technology. All the patients enrolled in this study provided written informed consent.
Inclusion criteria
- Diagnosed with SCLC confirmed by cytology or histology and proved to be E-SCLC by radiographic examination;
- Have complete medical and follow-up visit records; have neutrophil counts of 2×109/L or higher and platelets of 80×109/L or higher; have alanine aminotransferase and aspartate aminotransferase levels 2.5 times the upper limit of normal value; have a total bilirubin level 1.5 times the upper limit of normal value or higher; have serum creatinine levels at or above the upper limit of normal value; and have a normal electrocardiogram.
- In view of the high-sensitivity to chemotherapy of SCLC, Eastern Cooperative Oncology Group (ECOG) score is between 0 and 2 points;
- Ethnic Chinese, male or female, at least 18 years old;
- Have measurable lesion that can be evaluated by imageology and have not received chemotherapy;
- The expected survival time is 3 months or more.
Exclusion criteria
- Have severe infection, severe diarrhea, or any other serious systemic diseases;
- The presence of contraindications to chemotherapy;
- Have second primary tumors;
- History of allergies to biological products;
- Pregnant and lactating women.
Specimen extraction and determination of UGT1A1 gene polymorphism
The study subjects were selected from the patients with E-SCLC who attended the clinic in the Department of Oncology between June 2011 and January 2013. The Flinders Technology Associates card (Whatman plc, Maidstone, UK) was used to collect the finger blood sample from qualified patients before starting the treatment for the use in UGT1A1*28 and UGT1A1*6 gene polymorphism detection.
Below are the steps and methods used in the determination of the UGT1A1 gene polymorphism:
- Extract genomic DNA and polymerase chain reaction
The whole blood Genomic DNA was extracted with the QiaAmp kit (Qiagen NV, Venlo, the Netherlands), in accordance with the instructions. The condition of polymerase chain reaction (PCR) system is described next: extraction first started with 94°C 5 minutes degeneration, and then followed by 94°C 15 seconds, 55°C 25 seconds, 72°C 50 seconds for 40 cycles, and then 72°C 5 minutes extension.
UGTA1*28 primer: 5′-ATGGCACAGGGTACGTCTTC-3′
UGTA1*6 primer: 5′-ACCTCTGGCAGGAGCAAAG-3′ - Direct sequencing method
The PCR target products were identified by the high-resolution capillary electrophoresis. If the bands are clear and single, the specimen was then sent to Shanghai Yuanqi Bio-Pharmaceutical Company Limited (Shanghai, People’s Republic of China) for direct sequencing after conforming to the standard.
Treatment regimen
All the patients received irinotecan-based combination chemotherapy, which was administered in one of the following two ways: 1) CPT-11 60 mg/m2 intravenous infusion on days 1,8, and 15; repeated every 4 weeks; 2) CPT-11 85 mg/m2 intravenous infusion on days 1and 8; repeated every 3 weeks. Appropriate platinum drug (cisplatin, carboplatin, or lobaplatin) was selected based on the patient’s general physical condition, concurrent diseases and tolerance to the chemotherapy.
Evaluation of treatment effect and follow-up visit
The treatment effect was evaluated after two cycles using Response Evaluation Criteria In Solid Tumors 1.1 standard. The patients with partial response were required to have another radiographic examination to confirm the effect after 4 weeks. For the patients with progressive disease, the treatment was switched to another chemotherapy scheme or the best support treatment was given to them. The PFS is defined as the time from the start of treatment to tumor progression or death resulting from any cause, and the OS is defined as the time from the start of the treatment until death caused by any reason. Adverse events were evaluated all the time by using National Cancer Institute Common Toxicity Criteria Adverse Events (NCI-CTCAE) version 3.0. The follow-up visit was done every 3 months after the end of chemotherapy. The median follow-up period is 15 months (range from 8 to 28 months).
Statistical method
The data were analyzed using the Statistical Package for the Social Sciences version 18.0 software (SPSS Inc., Chicago, IL, USA). The OS and PFS curves were evaluated by the Kaplan–Meier method. The efficacy and adverse events between different genotypes were compared with chi-square test or Fisher’s exact test. The P-value of less than 0.05 is defined as having a statistically significant difference.
Results
All 67 patients who had efficacy and adverse event data completed genotype testing. The change of TA repeats in the TATA box of the UGT1A1 promoter caused UGT1A1*28 polymorphism, resulting in three genotypes: wild-type (WT) TA6/6, heterozygous mutation TA6/7, and homozygous mutation TA7/7. The UGT1A1*6 polymorphism, characterized by a single-nucleotide substitution in exon 1 of UGT1A1, resulting in three genotypes: WT G/G, heterozygous mutation G/A, and homozygous mutation A/A. The combination of UGT1A1*28 and UGT1A1*6 genotype was divided into WT, 1 site mutation, and 2 site mutation. We analyzed not only the effect of each individual UGT1A1*28 and UGT1A1*6 genotypes separately, but also the combination of these two different genotypes on the treatment efficacy and adverse events.
Distribution of UGT1A1*28 gene polymorphism
Among 67 SCLC patients enrolled in this study, 56 patients (83.6%) have the genotype of homozygous WT TA6/6 with UGT1A1 gene promoter region TA series repeating six times; eleven patients (16.4%) have the genotype of heterozygous mutant TA6/7 with TA series repeating six and seven times; no homozygous mutations TA7/7 with TA series repeating seven times was observed.
Distribution of UGT1A1*6 gene polymorphism
Among 67 SCLC patients enrolled in this study, 45 patients (67.2%) have WT G/G; 22 patients (32.8%) have heterozygous mutation type G/A. No A/A homozygous mutations were observed.
Distribution of combined genotypes
We looked into the combination of genotypes of these two genes (Table 1) and found that 38 patients have the combined genotype of TA6/6 and G/G (WT); 18 patients have TA6/6 and G/A (heterozygous variant) type; seven patients have TA6/7 and G/G (heterozygous variant) type; four patients have TA6/7 and G/A (two sites heterozygous variants) type. The double homozygous mutation was not observed.
  | Table 1 Patient’s combination genotype distribution |
Relationship between UGT1A1 gene polymorphism and treatment efficacy
There was no significant difference on treatment efficacy for the patients with different UGT1A1 genotypes. We also found no statistical significant difference on short-term efficacy in the patients with UGT1A1 genotype combination (Tables 2 and 3, respectively).
Relationship between UGT1A1 gene polymorphism and PFS
The median PFS of UGT1A1*28 WT (TA6/6) and mutant type (TA6/7) was 9.9 months and 10 months, respectively. There is no statistically significant difference (P=0.589) (Figure 1). The median PFS of UGT1A1*6 WT (G/G) and mutant type (G/A) was 9.7 months and 9.9 months, respectively. There is also no statistical difference between them (P=0.408) (Figure 2). There is no significant difference between different combination genotype and PFS either (P=0.491) (Figure 3).
  | Figure 1 PFS curve of UGT1A1*28 different genotypes. |
  | Figure 2 PFS curve of UGT1A1*6 different genotypes. |
  | Figure 3 PFS curve of UGT1A1*28 and UGT1A1*6 different combination genotypes. |
Relationship between UGT1A1 gene polymorphism and survival time
The median OS of the patients with UGT1A1*28 WT (TA6/6) was 13.9 months, and the median OS of the patients with UGT1A1*28 mutant type (TA6/7) was 14.5 months, there was no statistical difference (P=0.816) (Figure 4). The median OS of the patients with UGT1A1*6 WT (G/G) was 13.8 months, and the median OS of the patients with UGT1A1*6 mutant type (G/A) was 14.1 months, there was also no significant statistical difference (P=0.607) (Figure 5). The OS between different combination genotype had no significant statistical difference either (P=0.258) (Figure 6).
  | Figure 4 OS curve of UGT1A1*28 different genotypes. |
  | Figure 5 OS curve of UGT1A1*6 different genotypes. |
  | Figure 6 OS curve of UGT1A1*28 and UGT1A1*6 different combination genotypes. |
Relationship between UGT1A1 gene polymorphism and adverse reactions
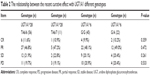
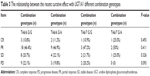
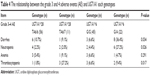
Statistical analysis found the risk of grade 3–4 thrombocytopenia for the patients carrying mutant type UGT1A1*28 (TA6/7) is significantly increased and the risk of grade 3–4 diarrhea and neutropenia for the patients carrying UGT1A1*6 heterozygous mutant (G/A) is significantly increased (P<0.05) (Table 4). The comparison between adverse events and different combination found that patients simultaneously carrying TA6/7 and G/A 2 sites heterozygous mutation are prone to have grade 3–4 degree diarrhea and neutropenia (P<0.05) (Table 5).
  | Table 4 The relationship between the grade 3 and 4 adverse events (AE) and UGT1A1 each genotypes |
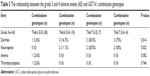  | Table 5 The relationship between the grade 3 and 4 adverse events (AE) and UGT1A1 combination genotypes |
Discussion
The occurrence of adverse events for CPT-11 has obvious individual differences. The CPT-11 is one of the S-phase cell-cycle specific drugs. The CPT-11 is converted to its biologically active form 7-ethyl-10-hydroxycamptothecin (SN-38) under the effect of carboxylesterase in the liver and gastrointestinal tract in vivo. The SN-38 mainly inhibits DNA topoisomerase, preventing the repair of DNA chain scission, interfering with the DNA replication and RNA transcription, leading to cell death. A key enzyme in the intestine UGT1A1 converts SN-38 to inactive SN-38G through glucuronidation.14 The SN-38 leads to intestinal mucosa injury and delayed diarrhea and can also be catalyzed by the UGT enzyme into SN-38G in the intestine. The UGT1A1 gene mutation can lower the activity of UGT1A1 and therefore reduces its capability in UGT1A1 inactivating SN-38. This results in the accumulation of SN-38 in the intestine causing damage to the intestinal mucosa and therefore severe delayed diarrhea.15 Therefore, the expression and activity of the UGTs enzymes are closely related to the efficacy and adverse reaction of CPT-11, of which UGT1A1 plays a vital role. Studies showed that the activity of the UGT1A1 enzyme was closely related to the UGT1A1 gene polymorphisms, especially UGT1A1*6 and UGT1A1*28.16
The distribution of UGT1A1*28 polymorphism of 68 ethnic Chinese SCLC patients is: 56 cases carrying UGT1A1*28 WT (TA6/6), accounted for 83.6%; eleven cases carrying UGT1A1*28 heterozygous mutation type (TA6/7), accounted for 16.4% correspondingly; no UGT1A1*28 homozygous mutant (TA7/7) was found. This result is consistent with the study reported on the distribution of UGT1A1*28 gene variation in Chinese conducted by Li et al.17 The results of the study also show that the frequency of carrying heterozygous mutant TA6/7 and homozygous mutant TA7/7 is significantly lower than that of Caucasian and African populations. For Caucasian population, the TA6/7 heterozygous mutation frequency is high at 35%–50%, TA7/7 homozygous mutation frequency is 10%–15%, while for the African population, the TA6/7 heterozygous mutation frequency is 44%–53% and the TA7/7 homozygous mutation frequency is 12%–17%.
The distribution of gene polymorphism of UGT1A1*6 described as follows: 45 cases were UGT1A1*6 WT (G/G) and accounted for 67.2%; 22 cases were UGT1A1*6 heterozygous mutation type (G/A) and accounted for 32.8%; no homozygous mutant (A/A) was observed. The result is similar to UGT1A1 gene mutation frequency in Japanese population reported by Nakamura et al18 and Minami et al19, and in Asian population reported by Jada et al.20 Contrary to UGT1A1*28 and UGT1A1*6 mutation frequencies in the Caucasus and African populations are relatively lower.16 This study also found that UGT1A1*6 gene mutation frequency is higher than the UGT1A1*28 in Chinese population. The rate of the UGT1A1*28 gene mutations (TA6/7) was only 16.4%, while UGT1A1*6 mutation type (G/A) was 32.8%.
Because UGT1A1 gene polymorphism has the closest relationship to irinotecan adverse event, further analysis was performed and the result shows that only 1.8% patients with UGT1A1*28 WT (TA6/6) had grade 3–4 thrombocytopenia, while the percentage of patients with UGT1A1*28 heterozygous mutation type (TA6/7) who had the same adverse event is as high as 27.2%. There is a significant difference between two different genotypes (P=0.017). The percentage of the patients having grade 3–4 neutropenia and anemia is similar between these two genotypes. So the relationship between UGT1A1*28 gene polymorphism and diarrhea was more pronounced in Asia, but the relationship with neutropenia required further study.21
For gastrointestinal toxicity, 9.1% UGT1A1*28 heterozygous mutation type (TA6/7) patients had grade 3–4 diarrhea, while it is 10.7% for WT UGT1A1*28 (TA6/6) patients. There is no statistically significant difference between the two genotypes. Similar to other study results, it is rare to have grade 3–4 diarrhea for Chinese patients having UGT1A1*28 mutation.17,22 Compared to the patients with WT (G/G), the patients with UGT1A1*6 heterozygous mutation type (G/A) are more likely to have grade 3–4 neutropenia (27.2% vs 4.4%, P=0.026) and diarrhea (36.4% vs 6.6%, P=0.034). This result is similar to several clinical studies conducted in Japan.18,19 This may be caused by the inactivation of SN-38. As an active metabolite of CPT-11, SN-38 prevents the repair of the DNA chain and interferes with the replication and transcription of nucleic acid through inhibiting DNA topoisomerase. After being metabolized by UGT1A1, SN-38 is converted into the inactive SN-38G through glucuronidation by the enzyme in the gut. When UGT1A1*6 heterozygous mutation occurs, it reduces UGT1A1 activity, so that the metabolization of SN-38 to SN-38G is decreased. This causes a large amount of SN-38 accumulated in the intestine, which continues to deliver cytotoxicity and directly damages the intestinal mucosal barrier, leading to severe diarrhea.23
Studies have found that when CPT-11 was administered in low dose (50–180 mg/m2), UGT1A1*28 cannot prompt the increase in hematologic toxicity, and the hematologic toxicity of the patients with UGT1A1*28 homozygous mutant type will significantly increase only in the medium or high dose (200–350 mg/m2).24 Other investigators also proposed that the toxicities of low-dose CPT-11 were not affected by the status of the UGT1A1 gene.25 A meta-analysis that included 1,998 patients indicated that when CPT-11 was used in low dose, the UTG1A1*28 genotype was also linked to neutropenia, but the risk for neutropenia was significantly increased after the application of high-dose CPT-11.26 The result of another meta-analysis that contained 1,760 patients and was related to CPT-11 induced diarrhea showed that both UGT1A1*28 homozygous mutant-type and heterozygous mutant-type will increase the risk for delayed diarrhea, but this trend is not significant when the dose of CPT-11 was lower than 125 mg/m2.27 The dose of CPT-11 in this study was not high, and with the limitation of sample size, we did not detect hematologic toxicity being significantly increased in UGT1A1*6 and UGT1A1*28 homozygous mutant-type patients.
The relationship between UGT1A1 gene polymorphisms and the clinical response of CPT-11-based regimens was one of the hotspots. There are few studies on the impact of UGT1A1*28 gene polymorphism on the efficacy of CPT-11.28 The clinical studies demonstrated that irinotecan-based chemotherapy can improve patients’ efficacy, PFS, and OS.29 Toffoli et al did a multi-center clinical study with 250 colorectal cancer patients with the chemotherapy of CPT-11 combined with 5-FU/CF.30 The study assessed the relationship between the UGT1A1*28 polymorphism and tumor response rate and OS. The result showed that the patients with purebred mutation TA7/7 had the highest objective response (95%) and the lowest risk of tumor progression. Probably it is because TA7/7 type UGT1A1 genotype can downregulate the transcription of UGT1A1 and therefore decrease its expression and is unable to inactivate SN-38, resulting in high blood concentrations of CPT-11 and enhanced cell toxicity thus improving the objective response rate. Although these studies have proposed the clinical response of homozygous mutant-type was better, so far there has been no consistent conclusion. The two major studies found no difference in efficacy between different genotypes of UGT1A1.31,32
This study found that treatment efficacy and long-term survival are not related to UGT1A1 gene polymorphism. The median PFS for the patients without UGT1A1*28 mutations was 9.9 months, while the median PFS for the patients with UGT1A1*28 mutant (TA6/7) was 10 months. There is no significant difference (P=0.589). The median PFS of UGT1A1*6 WT (G/G) patients was 9.7 months, while the median PFS of UGT1A1*6 mutant (G/A) patients was 9.9 months. There is no statistically significant difference (P=0.408). The UGT1A1*28 WT (TA6/6) and mutant (TA6/7) had a median OS of 13.9 months and 14.5 months, respectively, with no significant difference (P=0.816) between the two groups. For UGT1A1*6 WT (G/G), the median OS was 13.8 months, while for UGT1A1*6 mutant (G/A), the median OS was 14.1 months, with no statistically significant difference (P=0.607). The result of our study is consistent with results reported by another researcher, which shows no correlation between UGT1A1*28 and UGT1A1*6 two allele gene polymorphisms and the patient treatment outcomes.33
Whether the polymorphism of UGT1A1 gene can predict the efficacy of CPT-11 has no uniform conclusion. The mutation of UGT1A1 gene can increase the level of SN-38, which is the active metabolite of CPT-11. So the efficacy was likely to increase. However, the majority of the clinical trials showed no significant difference between the efficacy of CPT-11 and UGT1A1 genotypes. The reason that chemotherapy did not differ between the various genotypes may be this study is a retrospective study with small samples, high proportion of patients in extensive stage, and irinotecan dosage is not uniform. Therefore, we should consider not only the UGT1A1 gene polymorphism but also the status of the patients, the intensity of drug dose, the course of treatment, the combination of medication, and the other factors when we predict the clinical effect.
Disclosure
The authors report no conflicts of interest in this work.
References
Xiao X, Wang S, Xia S, et al. Retrospective study of irinotecan/cisplatin followed by etoposide/cisplatin or the reverse sequence in extensive-stage small cell lung cancer. Onco Targets Ther. 2015;21(8):2209–2214. | ||
Noda K, Nishiwaki Y, Kawahara M, et al. Irinotecan plus cisplatin compared with etoposide plus cisplatin for extensive small-cell lung cancer. N Engl J Med. 2002;346(2):85–91. | ||
Lara PN Jr, Natale R, Crowley J, et al. Phase III trial of irinotecan/cisplatin compared with etoposide/cisplatin in extensive-stage small-cell lung cancer: clinical and pharmacogenomic results from SWOG S0124. J Clin Oncol. 2009;27(15):2530–2535. | ||
Hanna N, Bunn PA Jr, Langer C, et al. Randomized phase III trial comparing irinotecan/cisplatin with etoposide/cisplatin in patients with previously untreated extensive-stage disease small-cell lung cancer. J Clin Oncol. 2006;24(13):2038–2043. | ||
Hermes A, Bergman B, Bremnes R, et al. Irinotecan plus carboplatin versus oral etoposide plus carboplatin in extensive small-cell lung cancer: a randomized phase III trial. J Clin Oncol. 2008;26(26): 4261–4267. | ||
NCCN.org [homepage on the Internet]. Houston: National Comprehensive Cancer Network Online Resources, Inc.; c2012-02 [updated July 25, 2011; cited October 25, 2015]. Available from: http://www.nccn.org/. Accessed February 26, 2012. | ||
Lima JP, dos Santos LV, Sasse EC, et al. Camptothecins compared with etoposide in combination with platinum analog in extensive stage small cell lung cancer: systematic review with meta-analysis. J Thorac Oncol. 2010;5(12):1986–1993. | ||
Cecchin E, Innocenti F, D’Andrea M, et al. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol. 2009;27(15):2457–2465. | ||
Stewart CF, Panetta JC, O’Shaughnessy MA, et al. UGT1A1 promoter genotype correlates with SN-38 pharmacokinetics, but not severe toxicity in patients receiving low-dose irinotecan. J Clin Oncol. 2007;25(18):2594–2600. | ||
Kaniwa N, Kurose K, Jinno H, et al. Racial variability in haplotype frequencies of UGT1A1 and glucuronidation activity of a novel single nucleotide polymorphism 686C> T (P229L) found in an African-American. Drug Metab Dispos. 2005;33(3):458–465. | ||
Sai K, Sawada J, Minami H. Irinotecan pharmacogenetics in Japanese cancer patients: roles of UGT1A1*6 and *28. Yakugaku Zasshi. 2008;128(4):575–584. | ||
Gao J, Zhou J, Li Y, Lu M, Jia R, Shen L. UGT1A1 6/28 polymorphisms could predict irinotecan-induced severe neutropenia not diarrhea in Chinese colorectal cancer patients. Med Oncol. 2013;30(3):604. | ||
Fujita K, Ando Y, Nagashima F, et al. Genetic linkage of UGT1A7 and UGT1A9 polymorphisms to UGT1A1*6 is associated with reduced activity for SN-38 in Japanese patients with cancer. Cancer Chemother Pharmacol. 2007;60(4):515–522. | ||
Di Paolo A, Bocci G, Polillo M, et al. Pharmacokinetic and pharmacogenetic predictive markers of irinotecan activity and toxicity. Curr Drug Metab. 2011;12(10):932–943. | ||
Hirose K, Kozu C, Yamashita K, et al. Correlation between plasma concentration ratios of SN-38 glucuronide and SN-38 and neutropenia induction in patients with colorectal cancer and wild-type UGT1A1 gene. Oncol Lett. 2012;3(3):694–698. | ||
Park SR, Kong SY, Rhee J, et al. Phase II study of a triplet regimen of S-1 combined with irinotecan and oxaliplatin in patients with metastatic gastric cancer: clinical and pharmacogenetic results. Ann Oncol. 2011;22(4):890–896. | ||
Li M, Wang Z, Guo J, et al. Clinical significance of UGT1A1 gene polymorphisms on irinotecan-based regimens as the treatment in metastatic colorectal cancer. Onco Targets Ther. 2014;23(7):1653–1661. | ||
Nakamura Y, Soda H, Oka M, et al. Randomized phase II trial of irinotecan with paclitaxel or gemcitabine for non-small cell lung cancer: association of UGT1A1*6 and UGT1A1*27 with severe neutropenia. J Thorac Oncol. 2011;6(1):121–127. | ||
Minami H, Sai K, Saeki M, et al. Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet Genomics. 2007;17(7):497–504. | ||
Jada SR, Lim R, Wong CI, et al. Role of UGT1A1*6, UGT1A1*28 and ABCG2 c.421C>A polymorphisms in irinotecan-induced neutropenia in Asian cancer patients. Cancer Sci. 2007;98(9):1461–1467. | ||
Palomaki GE, Bradley LA, Douglas MP, et al. Can UGT1A1 genotyping reduce morbidity and mortality in patients with metastatic colorectal cancer treated with irinotecan? An evidence-based review. Genet Med. 2009;11(1):21–34. | ||
Zhou CF, Ma T, Su Y, et al. UGT1A1 gene polymorphisms and the toxicities of FOLFIRI in Chinese Han patients with gastrointestinal cancer. Anticancer Agents Med Chem. 2013;13(2):235–241. | ||
Schulz C, Heinemann V, Schalhorn A, et al. UGT1A1 gene polymorphism: impact on toxicity and efficacy of irinotecan-based regimens metastatic colorectal cancer. World J Gastroenterol. 2009;15(40):5058–5066. | ||
Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99(17):1290–1295. | ||
Sugiyama T, Hirose T, Kusumoto S, et al. The UGT1A1*28 genotype and the toxicity of low-dose irinotecan in patients with advanced lung cancer. Oncol Res. 2010;18(7):337–342. | ||
Hu ZY, Yu Q, Pei Q, Guo C. Dose-dependent association between UGT1A1*28 genotype and irinotecan-induced neutropenia: low doses also increase risk. Clin Cancer Res. 2010;16(15):3832–3842. | ||
Hu ZY, Yu Q, Zhao YS. Dose-dependent association between UGT1A1*28 polymorphism and irinotecan-induced diarrhoea: a meta-analysis. Eur J Cancer. 2010;46(10):1856–1865. | ||
Douillard JY, Cunningham D, Roth AD, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355(9209):1041–1047. | ||
Saltz LB, Cox JV, Blanke C, et al; Irinotecan Study Group. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. N Engl J Med. 2000;343(13):905–914. | ||
Toffoli G, Cecchin E, Corona G, et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24(19):3061–3068. | ||
McLeod HL, Sargent DJ, Marsh S, et al. Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. J Clin Oncol. 2010;28(20):3227–3233. | ||
Glimelius B, Garmo H, Berglund A, et al. Prediction of irinotecan and 5-fluorouracil toxicity and response in patients with advanced colorectal cancer. Pharmacogenomics J. 2011;11(1):61–71. | ||
Rouits E, Boisdron Celle M, Dumont A, et al. Relevance of different UGTlAl polymorphisms in irinotecan induced toxicity: a molecular and clinical study of 75 patients. Clin Cancer Res. 2011;10(1):5151–5159. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.