Back to Journals » Infection and Drug Resistance » Volume 14
The Potential of Digital Polymerase Chain Reaction for Improving Diagnostic Yield of Nontuberculous Mycobacteria Pulmonary Disease
Authors Nishii Y, Furuhashi K, Nakamura S, Nishio M, Nakamura Y, Ushiro K, Ito K, Sakaguchi T, Suzuki Y, Fujiwara K, Yasuma T, Kobayashi T , D'Alessandro-Gabazza C, Gabazza EC , Taguchi O, Hataji O
Received 7 September 2021
Accepted for publication 30 October 2021
Published 1 December 2021 Volume 2021:14 Pages 5079—5087
DOI https://doi.org/10.2147/IDR.S338165
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Yoichi Nishii,1 Kazuki Furuhashi,1 Saki Nakamura,1 Miho Nishio,1 Yuki Nakamura,1 Kengo Ushiro,1 Kentaro Ito,1 Tadashi Sakaguchi,1 Yuta Suzuki,1 Kentaro Fujiwara,1 Taro Yasuma,2 Tetsu Kobayashi,3 Corina D’Alessandro-Gabazza,2 Esteban C Gabazza,2 Osamu Taguchi,1 Osamu Hataji1
1Respiratory Center, Matsusaka Municipal Hospital, Matsusaka, Mie, Japan; 2Department of Immunology, Faculty and Graduate School of Medicine, Mie University, Tsu-city, Mie, Japan; 3Department of Pulmonary and Critical Care Medicine, Faculty and Graduate School of Medicine, Mie University, Tsu-city, Mie, Japan
Correspondence: Esteban C Gabazza
Department of Immunology, Mie University Graduate School of Medicine, Edobashi 2-174, Tsu-city, Mie, 514-8507, Japan
Tel +81 59 231 5017
Fax +81 59 231 5225
Email [email protected]
Introduction: Many patients with nontuberculous mycobacteria pulmonary disease are asymptomatic. The disease diagnosis is confirmed in only a small proportion of patients with radiological findings suspicious for nontuberculous mycobacteria pulmonary disease. Thus, many patients remained undiagnosed. Here, we evaluated the diagnostic value of digital polymerase chain reaction (PCR) in nontuberculous mycobacteria pulmonary disease.
Methods: We prospectively evaluated 123 patients with radiological findings suspicious for nontuberculous mycobacteria pulmonary disease. Digital PCR was performed using bronchial lavage fluid, sputum, saliva, blood, and urine.
Results: The culture of bronchial washing fluid was positive for nontuberculous mycobacteria in 53 patients and negative in 70. The positive detection rate of nontuberculous mycobacteria by digital PCR in patients with positive culture (n = 53) was as follows: bronchial lavage fluid 100%, sputum 62.9%, saliva 41.5%, blood 7.5%, and urine 3.8%. All patients with two or more positive partitions for nontuberculous mycobacteria in the digital PCR of bronchial lavage fluid showed nontuberculous mycobacteria growth in the bronchial lavage fluid culture. The digital PCR analysis of the bronchial lavage fluid showed a high sensitivity (100%), specificity (85.7%), positive predictive value (84.1%), negative predictive value (100%), and a high concordance rate (91.9%) with the bronchial lavage fluid culture results. In addition, the culture of bronchial lavage fluid was positive for nontuberculous mycobacteria in patients with two or more positive partitions in the digital PCR of sputum and saliva with a combined positive predictive value of 81.1%.
Conclusion: Digital PCR analysis of nontuberculous mycobacteria in bronchial lavage fluid shows a high concordance rate with the bronchial lavage fluid culture results and a high positive predictive value using both sputum and saliva, suggesting the potential usefulness of dPCR for diagnosis of nontuberculous mycobacteria pulmonary disease in clinical practice.
Keywords: digital PCR, nontuberculous mycobacteria pulmonary disease, diagnosis, bronchial lavage fluid culture, bronchoscopy
Plain Language Summary
The number of patients with nontuberculous mycobacteria pulmonary disease is globally increasing. Many patients with this infectious disease present no symptoms. Approximately one-third of patients with radiological findings suspicious of the nontuberculous mycobacteria pulmonary disease remain undiagnosed. Therefore, there is a compelling need for more accurate diagnostic techniques. The present study shows that the digital PCR analysis of nontuberculous Mycobacteria in bronchial lavage fluid shows a high concordance rate with results of the bronchial lavage fluid culture and a high positive predictive value using both sputum and saliva, suggesting the potential usefulness of digital PCR for diagnosis of nontuberculous mycobacteria pulmonary disease in clinical practice.
Introduction
Although the global number of patients with pulmonary tuberculosis is steadily decreasing, reports of nontuberculous mycobacteria (NTM) pulmonary disease are rising.1 NTM pulmonary disease is caused by NTM that belong to the genus Mycobacterium.2 The name of NTM pulmonary disease is used to differentiate it from diseases caused by Mycobacterium tuberculosis and Mycobacterium leprae that are included in the same genus Mycobacterium. NTM are common and free-living inhabitants of the natural environments, including soil, vegetation, and water systems (tap water).1 There are nearly 200 identified NTM species, and the number of species increases yearly.2 Identifying the NTM species is important because the course and therapy response depends on the bacterial species. About 80% of NTM pulmonary disease is caused by the Mycobacterium avium complex (MAC), which includes the microaerobic and slow-growing Mycobacterium avium and Mycobacterium intracellulare.3,4
The diagnosis and treatment guidelines of NTM pulmonary disease jointly established by the American Thoracic Society (ATS), European Respiratory Society (ERS), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), and Infectious Diseases Society of America (IDSA) were recently updated on 2020.5 There are clinical, imaging, and microbiological criteria. However, the diagnostic yield is only 34%, even in patients with suspicious chest radiological findings.6 The statement “presence or absence of subjective symptoms” has been deleted from the 2008 Japanese guidelines of NTM pulmonary disease because many patients are asymptomatic at presentation (https://www.kekkaku.gr.jp/medical_staff/#no4).
Diagnosis of NTM pulmonary disease with correct species identification is challenging in clinical practice. The 2020-year ATS/ERS/ESCMID/IDSA guidelines mentioned the use of nucleic acid amplification techniques, mass spectrometry, and other diagnostic methods.5 However, the international guidelines do not include the results of these advanced methods in the diagnostic criteria of NTM pulmonary disease.5 Nucleic acid amplification techniques are simple, highly sensitive, and less time-consuming tests than the canonical microbial culture methods, which require 1 or 2 months to provide a conclusive microbial diagnosis. Several nucleic acid amplification techniques, including real-time polymerase chain reaction (PCR), loop-mediated isothermal amplification (LAMP), transcription-reverse transcription concerted reaction (TRC), and digital PCR, have been developed.7,8 They have been applied for microbial identification (eg, virus and tuberculosis).7,8 Digital PCR (dPCR) is the most recently developed method. The principle of dPCR consists in separating a single sample into a large number of small partitions, performing PCR in each partition, and determining the number of copies of the target sequence from the number of positive and negative reactions in the partitions, and applying a Poisson correction. Apart from detecting MAC with high sensitivity, dPCR also allows comparing the number of copies or bacterial burden among different samples.
In the present study, we conducted a prospective study to evaluate the diagnostic value of dPCR in patients with NTM pulmonary disease.
Materials and Methods
Subjects and Study Design
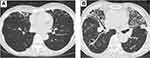
This study prospectively evaluated 123 patients with chest radiological findings suspicious for NTM pulmonary disease seen at the Matsusaka Municipal Hospital between March 2019 and January 2021 (Figure 1). Patients that fulfilled the criteria were consecutively enrolled in the study. The inclusion criteria were the presence of chest computed tomography (CT) findings suspicious for NTM pulmonary disease, history of negative sputum examination, or lack of expectoration. CT findings suspicious for NTM pulmonary disease were the presence of nodular and bronchiectatic lesions or fibrocavitary lesions (Figure 2A and B). The median time interval between CT findings suspicious for the disease to enrollment in the study (collection of bronchial lavage fluid for bacterial culture) was 9 days (range 1 ~ 120 days). Patients with a history of sputum culture-positive were excluded from the study. All patients had no history of previous treatment, including antituberculosis therapy. The human immunodeficiency virus test was negative in 95 cases but not performed in 28 cases. Digital PCR was performed using bronchial lavage fluid collected during bronchoscopy. We used samples of bronchial lavage fluid collected during bronchoscopy and sputum, saliva, blood, urine to perform dPCR in all cases.
Ethical Statement
The Ethics Committee for Clinical Investigation of Matsusaka Municipal Hospital approved the study protocol (Approval No J-42-190517-2-2), and the study was performed following the Principles of the Helsinki Declaration. Informed consent was obtained from all patients before the sample collection and all diagnostic procedures.
Procedure
Bronchial lavage fluid was collected during bronchoscopy. Before the bronchoscopic procedure, we anesthetized the pharyngeal area by spraying lidocaine (Spray Catheter, Olympus, Tokyo, Japan) and treated the patients with pentazocine hydrochloride (15 mg) and hydroxyzine hydrochloride (25 mg).9 During the bronchoscopic procedure, local anesthesia was performed by instilling lidocaine solution through the bronchoscope channels. The bronchoscope (Olympus Medical Systems, Tokyo, Japan) was advanced into the airway and inserted into the target lesion-related third bronchial segment previously determined by chest computed tomography study. The target lesion-related bronchial branch was wedged to wash with 20 mL of physiological saline and collect the washing sample. A suction kit (Muranaka Medical Instrument Co., Ltd.) was used for fluid recovery, and additional washing was performed when the recovered washing fluid was less than 10 mL. Five hundred microliters were separated from the total recovered bronchial lavage fluid and stored at −20 °C until use in the dPCR reaction, and the remaining lavage fluid was used for bacterial culture. Culture of bronchial lavage fluid was performed simultaneously in both liquid and solid media using a Mycobacteria Identification culture kit from Kyokuto Pharmaceutical Industrial Corporation (Tokyo, Japan), following the manufacturer’s instructions.
Sputum was collected from patients with expectoration (n = 76), whereas saliva, urine, and blood were collected from all patients (n = 123) (Table 1). The patients were asked to collect the sputum and saliva into sterile tubes that were immediately stored at −20 °C until use. Blood samples were drawn into tubes containing ethylenedinitrilotetraacetic acid disodium salt, centrifuged (3000 rpm) for 15 min at 4 °C, and the serum fraction was then separated and stored at −20 °C until use. Urine samples were collected into sterile tubes that were then centrifuged (3000 rpm) for 5 min at 4 °C, and the sediment was stored at −20 °C until use.
 |
Table 1 Patients’ Profile and Positive Partitions During Digital PCR |
Digital PCR
Digital PCR was performed using the collected samples. The cultured samples were not used for dPCR. DNA was extracted from the samples using the MagMAXTM Microbiome Ultra Nucleic Acid Isolation Kit (ThermoFisher Scientific). This DNA isolation kit includes a buffer to lyse bacteria and tubes with magnetic beads to collect nucleic acid. The sample DNA (3 µL) is then added to the dPCR reaction mixture containing the 1× QuantStudioTM 3D Digital PCR Master Mix v2, 100 nM of primers and probe and sterile distilled water in a total reaction volume per sample of 14.50 µL. The sequences of the primers and probe were previously validated: forward primer, 5ʹ-TTGGGCCCTGAGACAACACT-3ʹ, reverse primer: 5ʹ-GCAACCACTATCCAATACTCAAACAC-3ʹ, and the probe: ROX-CCGTGTGGAGTCCCTCCATCTTGG-BHQ1.10 The target of the primers is the ribosomal RNA internal transcribed spacer sequence, which is identical in both Mycobacterium avium-intracellulare complex (MAC).10
The Digital PCR was performed using the ProFlexTM PCR system and QuantStudioTM3D digital PCR system (ThermoFisher Scientific, Waltham, MA, USA). The QuantStudioTM3D digital PCR system has a silicon chip that contains a single array of individual reaction wells onto which the sample is dispensed and distributed over multiple separate reaction chambers or partitions. Therefore, once the dPCR reaction mixture (14.50 µL) is loaded onto the dPCR chip using the chip loader of the QuantStudioTM3D digital PCR system, the sample is automatically dispensed into 20,000 individual partitions (reaction volume per partition: 0.755 nL).11 PCR was run in the ProFlexTM PCR system with initial DNA denaturation at 96 °C for 10 minutes, followed by 35 cycles at 98 °C and annealing at 60 °C for 30 seconds and holding at 10 °C.
Every dPCR reaction included three chips with negative template control and positive template control. The chips were analyzed using the Quantstudio™ 3D Digital PCR Instrument and the Quantstudio® AnalysisSuite v3.0. Thresholds were set for each sample based on the fluorescence of positive and negative controls using the QuanStudiio 3DAnalysis SuiteTM Software (ThermoFisher Scientific) and following the manufacturer’s instructions. The software counts partitions with positive or negative reactions, calculates the ratio of positive-partition to negative-partition, and provides an absolute number of NTM copies after a Poisson correction.
Statistical Analysis
Data are expressed as in percentage, median, and range. The difference between variables was calculated using the two-tailed Mann–Whitney U-test. All statistical analysis was performed using the commercially available statistical software SPSS statistics, version 23 (SPSS Inc., Chicago, IL, USA). A p < 0.05 was considered statistically significant.
Results
Many Patients with CT Findings Suspicious for NTM Pulmonary Disease are Asymptomatic
The results of the bacterial culture of the bronchial lavage fluid from all 129 patients with radiological findings suspicious for NTM pulmonary disease were as follows: M. avium 33 cases, M. intracellulare 20, M. tuberculosis 4, NTM subspecies 2, and negative culture 70 cases. The significant geographic variation reported in the distribution of NTM species may explain the absence of infection by other mycobacteria species such as M. kansasii or M. abscessus.12 Patients with culture positive for Mycobacterium tuberculosis (n=4) and NTM subspecies (n=2) were excluded. Therefore the further study was performed only in the remaining 123 cases (Figure 1). The median age of the 123 patients (males 27, females 96) was 71 years old. The pulmonary lesions were nodular or bronchiectatic in 114 cases and fibrocavitary in 9 cases. Thirty-nice patients (31.7%) complained of cough, whereas 84 (68.3%) were asymptomatic (Table 1).
Digital PCR Analysis Showed a High Positive Rate in Patients with NTM Positive Culture
Samples of blood, bronchial lavage fluid, saliva, and urine were available for dPCR analysis in all 123 patients. However, sputum was available only in 76 patients (Table 1). The culture of bronchial lavage fluid was positive for NTM in 53 patients and negative in 70. The positive detection rate of NTM by dPCR analysis in all subjects (n = 123) was relatively high using bronchial lavage fluid, sputum, and saliva but low using blood and urine (Table 2). The positive detection rate of NTM by dPCR analysis in patients with positive culture for NTM was high using bronchial lavage fluid (100%), sputum (62.9%), and saliva (41.5%). However, in patients with negative culture for NTM, the detection rate of NTM by dPCR was relatively low using bronchial lavage fluid (22.8%), sputum (17.1%), and saliva (20.0%) (Table 2). The positive rate by dPCR was low using blood or urine in patients with either positive or negative culture.
 |
Table 2 Positive Detection Rate by Digital PCR Test in Patients with Positive or Negative Culture of Nontuberculous Mycobacteria Using Bronchial Lavage Fluid |
High Positive Predictive Value of dPCR in Patients with Positive NTM Culture
Patients with culture positive for NTM showed more dPCR-positive partitions for NTM using bronchial lavage fluid. All patients with two or more dPCR-positive partitions for NTM using bronchial lavage fluid showed NTM positive culture. However, the number of dPCR-positive partitions was low in patients with negative NTM culture (n = 70) (Table 3). Combining the dPCR results of sputum and saliva revealed that patients with 2 or more dPCR-positive partitions had positive NTM culture with a high positive predictive value (81.1%). The positive predictive value of dPCR using sputum or saliva was 100% when the number of positive partitions was more than 114 (Table 3).
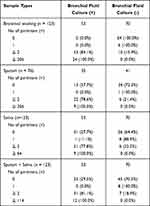 |
Table 3 Positive Predictive Values by Digital PCR |
Positive Partitions Using Blood and Urine Samples
Five cases showed dPCR-positive partitions using blood samples, and the number of positive partitions was 1 in all of them. The bronchial fluid culture was positive for NTM in 4 of these 5 cases; however, none showed disseminated disease. Six patients showed dPCR-positive partition using urine samples, and the number of positive partitions was 1 in all of them. Two of the 6 cases were positive for NTM culture.
High Sensitivity and Specificity of dPCR for NTM Detection
We calculated the sensitivity and specificity of dPCR for detecting NTM compared to the culture study by considering a positive NTM-dPCR analysis when there are 2 or more positive partitions (Table 4). The results showed that dPCR of NTM using bronchial lavage fluid has high sensitivity (100%), specificity (85.7%), positive predictive value (84.1%), and negative predictive value (100%). In addition, the concordance rate with the culture technique was high as 91.9%. The dPCR of NTM using saliva also showed high sensitivity (39.6%), specificity (91.4%), positive predictive value (77.8%), and negative predictive value (66.7%) with a high concordance rate with the culture technique (69.1%).
 |
Table 4 Sensitivity, Specificity, and Predictive Values of Digital PCR Test with Two or More Positive Partitions for Detecting Nontuberculous Mycobacteria |
Increased Number of Positive Partitions in Patients Showing Early Growth of NTM in the Culture Study
We found that the number of dPCR-positive partitions using bronchial lavage fluid or saliva was significantly increased in patients showing positive culture on the 4th week of microbial culture compared to the number of positive partitions in patients showing positive culture on the 8th week of microbial culture (Table 5).
 |
Table 5 Relationship Between Bronchial Fluid Culture Positivity and Number of Positive Partitions in the dPCR Test |
Discussion
The global number of patients with NTM pulmonary disease is increasing.4 The causative pathogen is MAC in approximately 80% of the patients.13 MAC infection that causes progressive destruction of the airways has a mortality rate as high as 25%.13 Many patients with positive NTM positive have few or no symptoms at presentation. According to the 2007 ATS/IDSA guidelines, only 62% of patients diagnosed with NTM pulmonary disease have productive expectoration or sputum.14 The diagnosis of NTM pulmonary disease is not considered by International Guidelines when there are no symptoms. However, many asymptomatic patients in Japan have chest radiological findings highly suspicious for NTM pulmonary disease with a positive bacterial culture. Therefore, the Guidelines of the Japanese Society for NTM Disease have removed “the presence of symptoms” as a requirement to diagnose NTM pulmonary disease from 2008 (https://www.kekkaku.gr.jp/medical_staff/#no4). The population enrolled in the present study is an illustrative example of NTM pulmonary disease cases experienced in Japan. Our study population included 40 asymptomatic patients (40/123=32.5%) with positive NTM culture and radiological suspicion of NTM pulmonary disease. To date, there is no reported longitudinal study describing the clinical outcome of asymptomatic patients with radiological findings suspicious for NTM pulmonary disease with positive bacterial culture. Therefore, it is unknown whether these asymptomatic patients with positive NTM culture will not become symptomatic overtime. On this basis, we believe it is urgent to consider using additional techniques to help reach diagnosis and decision-making in highly suspicious cases of NTM pulmonary disease. Here, we evaluated the usefulness of dPCR and found that dPCR analysis of NTM using bronchial lavage fluid was positive in 40 asymptomatic patients with suspicion of NTM pulmonary disease and positive NTM culture. Interestingly, the dPCR analysis of NTM using sputum and saliva was also positive in 28 of these asymptomatic patients. These observations suggest the potential of dPCR as an ancillary diagnostic tool in asymptomatic patients highly suspicious for NTM pulmonary disease.
Difficult-to-diagnose cases of NTM pulmonary disease with clinical symptoms are also common.6 Differential diagnosis of radiological findings suspicious for NTM is generally challenging and includes diffuse panbronchiolitis, pulmonary tuberculosis, primary ciliary dyskinesia, and immunodeficiency-associated bronchiectasis.6 Positive bacterial culture is a sine qua non condition to confirm the diagnosis of NTM pulmonary disease in difficult cases. The 2020 international guidelines advocate two separate positive sputum cultures or one positive bronchial lavage fluid culture to diagnose NTM pulmonary disease in suspected cases.5 However, a previous study reporting a positive NTM culture in only one-third (34%) of patients with radiological findings highly suspicious for NTM pulmonary disease casts doubts on the infallibility of culture techniques.6 Indeed, bacterial excretion may spontaneously stop during the natural course of NTM pulmonary disease despite clinical progression.15 In addition, NTM-culture techniques generally require a long time (weeks) to provide a positive or negative result. Therefore, the use of other technologies, including nucleic acid amplification methods (eg, dPCR, canonical quantitative PCR, and transcription-reverse transcription concerted reaction), 16S rRNA, or mass spectrometry (MALDI-TOF MS), should be considered to improve the diagnostic yield using culture techniques.11,16,17 Here, we evaluated the potential usefulness of dPCR for NTM identification. We found that a dPCR analysis positive for NTM (2 or more dPCR-positive partitions) has a high concordance rate (91.9%) with a positive NTM culture using bronchial lavage fluid. In addition, comparing the diagnostic yields of dPCR and culture using bronchial lavage fluid showed that dPCR has high sensitivity and negative predictive value. Interestingly, the number of dPCR-positive partitions was significantly higher in patients with 4-week positive culture than in cases with an 8-week positive culture. This observation suggests that a dPCR test, which requires only 6 hours to perform, may also indicate bacterial burden because samples showing earlier bacterial growth generally contain more microorganisms. Overall, these findings support the use of dPCR in combination with culture techniques for early NTM identification.
The lack of diagnostic materials or the invasiveness of procedures to collect specimens is another difficult problem in managing patients with NTM pulmonary disease. Collection of sputum is easy, but not all patients can expectorate. Bronchial lavage fluid and gastric juice are collected by endoscopy, but the procedures’ invasiveness restricts their general application in clinical practice.18,19 Collection of saliva, blood, and urine is also easy, but the value of these samples for NTM identification has not been validated. Here, we evaluated the usefulness of the dPCR test using saliva alone or in combination with sputum and found that the test can identify NTM in both samples. Two or more dPCR-positive partitions using sputum (78.6%) or saliva (77.8%) predicted positive culture of NTM in bronchial lavage fluid in a high proportion of patients. This predictive value was even higher (81.1%) when both sputum and saliva results were combined. Two or more dPCR-positive partitions using saliva alone or combined with sputum also showed high sensitivity and specificity. Overall, these results highlight the potential of dPCR analysis using easy-to-collect samples (sputum, saliva) to improve the diagnostic yield of NTM pulmonary disease.
Limitations
The failure to assess all NTM species and the small sample size are limitations of this study. The inherent limitations of the dPCR test include the inability to distinguish between living and dead bacteria and between pathogen and contaminant bacteria. In addition, dPCR cannot completely replace culture techniques in infectious diseases because a drug sensitivity test is generally required to select the most effective antibiotic. Patients with a positive partition in the dPCR analysis but negative for NTM in the culture study need clinical follow-up for diagnostic confirmation. However, some patients with NTM pulmonary disease (nodular or bronchiectatic lesion) show slow clinical progression, requiring long-term follow-up.
Conclusion
The number of patients with NTM pulmonary disease increases worldwide, and many cases remain undiagnosed because of a lack of symptoms. Therefore, there is a compelling need to improve the diagnostic yield of NTM pulmonary disease. The present study shows that the dPCR test has a high concordance rate with a positive NTM culture using bronchial lavage fluid and a high positive predictive value using both sputum and saliva, suggesting the potential usefulness of dPCR for the diagnosis of NTM pulmonary disease in clinical practice.
Abbreviations
NTM, nontuberculous mycobacteria; dPCR, digital polymerase chain reaction; MAC, Mycobacterium avium complex; LAMP, loop-mediated isothermal amplification; TRC, transcription-reverse transcription concerted reaction.
Acknowledgment
This research received no funding.
Disclosure
Dr Yoichi Nishii reports grants from AstraZeneca, Daiichi Sankyo, Takeda Pharmaceutical, Novartis Pharma, Boehringer Ingelheim, Kyowa Hakko Kirin, Sumitomo Dainippon Pharma, MSD, GlaxoSmithKline, Olympus, Bristol-Myers Squibb, Ono Pharmaceutical, Chugai Pharmaceutical, Eli Lilly, AbbVie, Taisho Pharmaceutical, and Celgene, outside the submitted work. Dr Kazuki Furuhashi reports grants from AstraZeneca, Daiichi Sankyo, Takeda Pharmaceutical, Novartis Pharma, Boehringer Ingelheim, Kyowa Hakko Kirin, Sumitomo Dainippon Pharma, MSD, GlaxoSmithKline, Olympus, Bristol-Myers Squibb, Ono Pharmaceutical, Chugai Pharmaceutical, Eli Lilly, AbbVie, Taisho Pharmaceutical, and Celgene, outside the submitted work. Dr Yuki Nakamura reports grants from AstraZeneca, Daiichi Sankyo, Takeda Pharmaceutical, Novartis Pharma, Boehringer Ingelheim, Kyowa Hakko Kirin, Sumitomo Dainippon Pharma, MSD, GlaxoSmithKline, Olympus, Bristol-Myers Squibb, Ono Pharmaceutical, Chugai Pharmaceutical, Eli Lilly, AbbVie, Taisho Pharmaceutical, and Celgene, outside the submitted work. Dr Tadashi Sakaguchi reports grants from AstraZeneca, Daiichi Sankyo, Takeda Pharmaceutical, Novartis Pharma, Boehringer Ingelheim, Kyowa Hakko Kirin, Sumitomo Dainippon Pharma, MSD, GlaxoSmithKline, Olympus, Bristol-Myers Squibb, Ono Pharmaceutical, Chugai Pharmaceutical, Eli Lilly, AbbVie, Taisho Pharmaceutical, and Celgene, outside the submitted work. Dr Yuta Suzuki reports grants from AstraZeneca, Daiichi Sankyo, Takeda Pharmaceutical, Novartis Pharma, Boehringer Ingelheim, Kyowa Hakko Kirin, Sumitomo Dainippon Pharma, MSD, GlaxoSmithKline, Olympus, Bristol-Myers Squibb, Ono Pharmaceutical, Chugai Pharmaceutical, Eli Lilly, AbbVie, Taisho Pharmaceutical, and Celgene, outside the submitted work. Dr Kentaro Fujiwara reports grants from AstraZeneca, Daiichi Sankyo, Takeda Pharmaceutical, Novartis Pharma, Boehringer Ingelheim, Kyowa Hakko Kirin, Sumitomo Dainippon Pharma, MSD, Glaxo Smith Kline, Olympus, Bristol-Myers Squibb, Ono Pharmaceutical, Chugai Pharmaceutical, Eli Lilly, AbbVie, Taisho Pharmaceutical, and Celgene, outside the submitted work. Dr Esteban C Gabazza reports grants from Shionogi pharmaceuticals and Astellas Pharmaceuticals, outside the submitted work. Dr Osamu Hataji reports grants, personal fees from AstraZeneca, Daiichi Sankyo, Takeda Pharmaceutical, Novartis Pharma, Boehringer Ingelheim, MSD, Glaxo Smith Kline, Olympus, Bristol-Myers Squibb, Ono Pharmaceutical, Chugai Pharmaceutical, Eli Lilly; personal fees from Pfizer, Kyorin Pharmaceutical, Taiho Pharmaceutical, Astellas Pharmaceutical, Meiji, Mitsubishi Tanabe Pharma, Sanofi; grants from Kyowa Hakko Kirin, Sumitomo Dainippon Pharma, AbbVie, Taisho Pharmaceutical, and Celgene, outside the submitted work. The authors report no other conflicts of interest in this work.
References
1. Ratnatunga CN, Lutzky VP, Kupz A, et al. The rise of non-tuberculosis mycobacterial lung disease. Front Immunol. 2020;11:303. doi:10.3389/fimmu.2020.00303
2. Gupta RS, Lo B, Son J. Phylogenomics and comparative genomic studies robustly support division of the genus mycobacterium into an emended genus mycobacterium and four novel genera. Front Microbiol. 2018;9:67. doi:10.3389/fmicb.2018.00067
3. Prevots DR, Shaw PA, Strickland D, et al. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med. 2010;182(7):970–976. doi:10.1164/rccm.201002-0310OC
4. Namkoong H, Kurashima A, Morimoto K, et al. Epidemiology of pulmonary nontuberculous mycobacterial disease, Japan. Emerg Infect Dis. 2016;22(6):1116–1117. doi:10.3201/eid2206.151086
5. Daley CL, Iaccarino JM, Lange C, et al. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Clin Infect Dis. 2020;71(4):e1–e36. doi:10.1093/cid/ciaa241
6. Koh W-J, Lee KS, Kwon OJ, et al. Bilateral bronchiectasis and bronchiolitis at thin-section CT: diagnostic implications in nontuberculous mycobacterial pulmonary infection. Radiology. 2005;235(1):282–288. doi:10.1148/radiol.2351040371
7. Pavsic J, Devonshire A, Blejec A, et al. Inter-laboratory assessment of different digital PCR platforms for quantification of human cytomegalovirus DNA. Anal Bioanal Chem. 2017;409(10):2601–2614. doi:10.1007/s00216-017-0206-0
8. Nyaruaba R, Mwaliko C, Kering KK, Wei H. Droplet digital PCR applications in the tuberculosis world. Tuberculosis. 2019;117:85–92. doi:10.1016/j.tube.2019.07.001
9. Nishii Y, Nakamura Y, Fujiwara K, et al. Use of ultrathin bronchoscope on a need basis improves diagnostic yield of difficult-to-approach pulmonary lesions. Front Med. 2020;7:588048. doi:10.3389/fmed.2020.588048
10. Rocchetti TT, Silbert S, Gostnell A, et al. Validation of a multiplex real-time PCR assay for detection of Mycobacterium spp., Mycobacterium tuberculosis complex, and Mycobacterium avium complex directly from clinical samples by use of the BD max open system. J Clin Microbiol. 2016;54(6):1644–1647. doi:10.1128/JCM.00241-16
11. Rutsaert S, Bosman K, Trypsteen W, Nijhuis M, Vandekerckhove L. Digital PCR as a tool to measure HIV persistence. Retrovirology. 2018;15(1):16. doi:10.1186/s12977-018-0399-0
12. Spaulding AB, Lai YL, Zelazny AM, et al. Geographic distribution of nontuberculous mycobacterial species identified among clinical isolates in the United States, 2009–2013. Ann Am Thorac Soc. 2017;14(11):1655–1661. doi:10.1513/AnnalsATS.201611-860OC
13. Diel R, Lipman M, Hoefsloot W. High mortality in patients with Mycobacterium avium complex lung disease: a systematic review. BMC Infect Dis. 2018;18(1):206. doi:10.1186/s12879-018-3113-x
14. Griffith DE, Aksamit T, Brown-Elliott BA, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367–416. doi:10.1164/rccm.200604-571ST
15. Moon SM, Jhun BW, Baek SY, et al. Long-term natural history of non-cavitary nodular bronchiectatic nontuberculous mycobacterial pulmonary disease. Respir Med. 2019;151:1–7. doi:10.1016/j.rmed.2019.03.014
16. Roth A, Fischer M, Hamid ME, et al. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol. 1998;36(1):139–147. doi:10.1128/JCM.36.1.139-147.1998
17. Alcaide F, Amlerova J, Bou G, et al. How to: identify non-tuberculous Mycobacterium species using MALDI-TOF mass spectrometry. Clin Microbiol Infect. 2018;24(6):599–603. doi:10.1016/j.cmi.2017.11.012
18. Ikedo Y. The significance of bronchoscopy for the diagnosis of Mycobacterium avium complex (MAC) pulmonary disease. Kurume Med J. 2001;48(1):15–19. doi:10.2739/kurumemedj.48.15
19. Takasa A, Hagiwara E, Sekine A, et al. [Clinical features of pulmonary Mycobacterium avium complex disease diagnosed through gastric aspirate culture]. Kekkaku. 2014;89(4):489–493. Japanese.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.