Back to Journals » Infection and Drug Resistance » Volume 15
The Performance of Metagenomic Next-Generation Sequence in the Diagnosis of Suspected Opportunistic Infections in Patients with Acquired Immunodeficiency Syndrome
Authors Liu L, Yuan M, Sun S, Wang J, Shi Y, Yu Y, Su X
Received 12 June 2022
Accepted for publication 29 August 2022
Published 24 September 2022 Volume 2022:15 Pages 5645—5653
DOI https://doi.org/10.2147/IDR.S378249
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Li Liu,1,2 Mingjuan Yuan,3 Siqing Sun,4 Jinrong Wang,5 Yi Shi,6 Yamin Yu,7 Xin Su1,6
1Department of Respiratory and Critical Care Medicine, Jinling Hospital, The First School of Clinical Medicine, Southern Medical University, Nanjing, People’s Republic of China; 2Department of Infectious Disease, Hunan Provincial People’s Hospital (The First Affiliated Hospital of Hunan Normal University), Changsha, Hunan Province, People’s Republic of China; 3Department of Infectious Disease, The Central Hospital of Yueyang, Yueyang, Hunan Province, People’s Republic of China; 4Department of General Medicine, The Second Hospital of Nanjing, Nanjing Hospital of Nanjing University of Chinese Medicine, Nanjing, People’s Republic of China; 5Department of Respiratory and Critical Care Medicine, Hengshui People’s hospital, Hengshui, Hebei Province, People’s Republic of China; 6Department of Respiratory and Critical Care Medicine, Jinling Hospital, Medical School of Nanjing University, Nanjing, People’s Republic of China; 7Department of Nephrology, Ningxiang People’s Hospital, Changsha, Hunan Province, People’s Republic of China
Correspondence: Xin Su, Email [email protected]
Background: For acquired immunodeficiency syndrome (AIDS) patients with suspected opportunistic infections, the rapid and accurate identification of pathogens remains a challenge. Metagenomic next-generation sequencing (mNGS) has emerged as a pan-pathogen assay for infectious diseases diagnosis, but its guiding significance for diagnosis and antimicrobials treatment in AIDS patients with suspected opportunistic infections is still not well established. In this study, we compared the microbiological diagnostic value of mNGS with that of conventional microbiological tests (CMTs) in AIDS patients with suspected opportunistic infections.
Methods: From January 2018 to February 2021, a retrospective study was performed at four tertiary teaching hospitals in China and data of 86 AIDS patients with suspected opportunistic infections were collected. The pathogen detection performance of mNGS and CMTs were compared.
Results: Positive agreement between mNGS and clinical diagnosis was significantly higher than that of CMTs (65/86 (75.6%) vs 37/86 (43.0%)). In addition, mNGS identified more bacterial (25 vs 2), fungal (5 vs 3), viral (9 vs 2) organisms compared with CMTs. Mixed infection were detected in 34 patients by mNGS combined with CMTs. Viruses (94.1%, 32/34) and fungi (94.1%, 32/34) were commonly seen in the mixed infection cases. mNGS helped identify the pathogen or guide appropriate treatment in 49/86 (57%) patients. Meanwhile, CMTs also contributed in the decision of appropriate treatment in 28 patients. The successful de-escalation or discontinuation of treatment was supported in 37 patients with the help of mNGS. We observed a significant reduction in the number of patients being prescribed foscarnet (52.3% vs 23.26%, p < 0.001), moxifloxacin (34.9% vs 10.5%, p = 0.005), and levofloxacin (32.6% vs 14%, p = 0.001) before and after mNGS.
Conclusion: For AIDS patients with suspected opportunistic infections, mNGS can provide early, noninvasive, and rapid microbiological diagnosis. mNGS may lead to a more precise antimicrobial treatment and reduced the unreasonable use of antimicrobials.
Keywords: mNGS, AIDS, opportunistic infections
Corrigendum for this paper has been published.
Introduction
Human Immunodeficiency Virus (HIV) infection is still a great public health threat around the world. Opportunistic infections often occur in AIDS patients even under the antiretroviral therapy (ART). It has a significant influence on patients’ life quality, health-care costs and the survival.1–3 Opportunistic infections have become the major cause of morbidity and mortality among AIDS patients.4,5
The majority of HIV-related opportunistic infections include Candida esophagitis, Pneumocystis jirovecii pneumonia (PJP), active pulmonary tuberculosis, Mycobacterium avium complex infection, cytomegalovirus and Cryptococcal meningitis.6 The spectrum of opportunistic infections differs among various countries in part due to different climates and socio-economic conditions. Fithamlak did a retrospective cross-sectional study on opportunistic infection in AIDS patients in Dawro Zone Hospital in Ethiopia. They found that pulmonary tuberculosis, severe community acquired pneumonia and oral candidiasis were the most common opportunistic infections.7 There are few studies about spectrum of opportunistic infections in AIDS patients in China. Wenwen Pang indicated that bacterial pneumonia was the most common opportunistic infections, followed by candida infection, Pneumocystis jiroveci pneumonia, tuberculosis and infectious diarrhoea in Sichuan Province, China.8 In Shanghai, Pneumocystis pneumonia and bacterial coinfection were the most common opportunistic infections, followed by tuberculosis, Cytomegalovirus, Cryptococcosis, and Mycobacterium avium complex infection.9
However, the diagnosis of opportunistic infections in AIDS patients is a great challenge to clinicians because of its broad spectrum of potential pathogens. Physicians often initiate non-specific empirical antibiotic therapy on undiagnosed cases.10,11 In actual clinical practice, up to 46% of empirical antibiotic treatments were shown to be inappropriate and approximately 50% were either unnecessary or broad-spectrum antibiotics.12,13 Therefore, prompt identification of causative agents is critical for appropriate treatment.
Metagenomic next-generation sequence (mNGS) has been addressed in detection the causative pathogens and guided targeted antimicrobial therapy quickly.14–19 However, its guiding significance for diagnosis and antimicrobial treatment in AIDS patients with suspected opportunistic infections is still not well established. Therefore, we conducted a retrospective study in the central area of China to analyze the diagnostic value of mNGS for AIDS patients with suspected opportunistic infections and optimization of early antimicrobial strategies in adult AIDS patients.
Methods
Study Design and Participants
The retrospective multicenter study was enrolled at Hunan Provincial People’s Hospital (Hunan province, South Central Region of China), The Central Hospital of Yueyang (Hunan province, South Central Region of China), Second Hospital of Nanjing (Jiangsu province, East Central Region of China) and Hengshui People’s Hospital (Hebei province, North Central Region of China). To explore the value of metagenomic next-generation sequence in Acquired Immunodeficiency Syndrome with suspected opportunistic infections, we evaluated AIDS patients with suspected opportunistic infections and had underwent mNGS between January 2018 and February 2021. Patients were included if all the following criteria were met: (1) age between 18 and 80 years; (2) patients with AIDS in whom opportunistic infections was suspected either at admission or during hospitalization; (3) patients underwent mNGS. Ethical approval for this study was obtained from the institutional ethics committee at each study site (Hunan Provincial People’s Hospital, The Central Hospital of Yueyang, Second Hospital of Nanjing and Hengshui People’s Hospital).
For each case, clinical informations and the results of other conventional tests were reviewed to collect from electronic patient records, including demographics, whole blood cell count, procalcitonin level, C-reactive protein, CD4+ T lymphocyte cell count, smear test results, culture, and PCR results. The patients’ treatment regimen was also collected. Antimicrobial treatment before mNGS, initial antimicrobial treatment at admission and adjustment later based on the results of mNGS were also collected. The final diagnosis was made by two independent clinicians based on mNGS results, CMTs, chest imaging, clinical manifestations and patients’ responses to the antimicrobial therapy.
In our clinical practice, patients suspected opportunistic infections underwent three times blood culture, Human gammaherpesvirus 4 DNA, cytomegalovirus DNA, serum (1,3)-β-D Glucan (BDG), serum galactomannan testing, Tuberculosis spot test, Mycoplasma pneumoniae antibody, Chlamydophila pneumoniae antibody, Syphilis antibody, toxoplasma Gondii antibody, Human herpesvirus 1 antibody, chest computer tomography (CT) after informed consent. Sputum/bronchoalveolar lavage fluid (BALF)/pleural samples were also sent for conventional testing, including bacterial and fungal smear; acid-fast stain; (1,3)-β-D Glucan; galactomannan testing; culture of bacteria, fungal organisms and mycobacterium species in all patients. When patients were suspected to have central nervous system infection, cerebrospinal fluid (CSF) samples were sent for bacterial and fungal smear; acid-fast stain; culture of bacteria, fungal organisms, mycobacterium species and cryptococcus antigen.
Metagenomic Next Generation Sequencing and Data Analysis
Samples (blood, BALF, bone marrow, CSF, pleural) from patients were collected according to standard operating procedures, and then sent to BGI-Huada Genomics Institute (Wuhan, China) for pathogens detection as described previously.19 Each sample was mixed and shaken with glass beads, and then the mixture was attached to a horizontal platform on a vortex mixer and agitated vigorously at 2800–3200 rpm for 30 min. The DNA was extracted using the TIANamp MicroDNA kit (Tiangen Biotech) according to standard procedures. The total DNA or cDNA was subjected to library construction through DNA-fragmentation (150bp), end-repair, adapter-ligation, and unbiased PCR amplification. An Agilent 2100 Bioanalyzer was used for quality control of the library and a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) was used for quality control of the DNA library concentration. After low-quality and short (length <35bp) reads were removed, high-quality sequences were generated. Then the remaining sequence were aligned to the current virus, bacterial, fungal, and protozoan databases (https://www.ncbi.nlm.nih.gov/genomes), which is comprised of whole genome sequences of 4061 viral taxa, 2473 bacterial genomes or scaffolds, and genomic sequences for 199 fungi related to human infection and 135 parasites associated with human diseases.
Microbiologic Methods
Using conventional microbiologic methods, samples (blood, BALF, bone marrow, CSF, pleural effusion) were examined by microscopy with routine laboratory staining and cultures of bacteria, fungi, and mycobacteria. Regarding the serology methods, peripheral blood samples were submitted for the detection of Mycoplasma pneumoniae antibody, Chlamydophila pneumoniae antibody, Syphilis antibody, toxoplasma Gondii antibody, Human herpesvirus 1 antibody using commercial enzyme-linked immunosorbent assays according to the manufacturer’s instructions. Both serum and CSF cryptococcus antigen were detected using gold immunochromatographic assay according to the manufacturer’s instructions. Human gammaherpes virus 4 DNA and cytomegalovirus DNA were tested by a real-time PCR assay. Serum BDG was detected according to the manufacturer’s instructions. Both BALF and serum galactomannan detection were performed using a double-sandwich ELISA according to the manufacturer’s instructions.
Criteria for a Positive mNGS Result
For bacteria (excluding mycobacteria), viruses, fungi, and parasites, whose reads number was in the top 10 in the complete belonging list, mNGS identified a microbe (on species level).20 The result was considered Mycobacterium tuberculosis-positive when at least one specific read was mapped to the species or genus level.21
For cases who were detected Cytomegalovirus22 and Human gammaherpesvirus 423 by mNGS, if serum PCR indicated positive at the same time, we identified that they were infectious pathogens. Furthermore, the mixed infection was defined as the isolation of more than one pathogenic species. After obtaining laboratory tests, chest imaging and mNGS results, clinical features were taken into consideration by two senior chief physicians to identify the pathogens and reached a consensus.24
Statistical Analysis
Continuous variables were expressed as the medians. Categorical variables were summarized as the counts and percentages in each category. Statistical analyses were performed using SPSS version 20.0 (IBM Corporation, Armonk, NY, USA). Figures were conducted using GraphPad Prism 5 software. A p value less than 0.05 was considered statistically significant.
Results
Population Characteristics
Eighty-six patients, who were presenting with acquired immunodeficiency syndrome with suspected opportunistic infections, underwent mNGS between January 2018 and February 2021. Among 86 patients, the majority of patients were diagnosed with pneumonia (56/86 [65.1%]), followed by blood stream infection (`18/86 [20.9%]), and central nervous system infection (12/86 [14%]) (Table 1). The proportion of samples is shown in Figure 1.
 |
Table 1 The Detailed Information Related to 86 Patients of AIDS Patients with Suspected Opportunistic Infections |
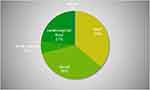 |
Figure 1 The proportion of samples in this study. |
There are 17 female and 69 male were included. Their median age was 42.8years (range 27–77). All (100%) patients had advanced clinical symptoms. The median CD4 T cell count was 56.9 cells/mm3 (range from 7 to 283) (Table 2).
 |
Table 2 Clinical Characteristics of AIDS Patients with Suspected Opportunistic Infections |
Summary of Pathogens Detected by mNGS
In this study, the mNGS sequencing results for 76 of 86 (88.4%) patients were positive for microbial pathogens. A comparison of the diagnostic results from mNGS with conventional microbiological tests (CMTs) is shown in Figure 2. CMTs and mNGS results were concordant for 37 of 86 patients (43%). Positive agreement between mNGS and clinical diagnosis was significantly higher than that of CMTs (65/86 (75.6%) vs 37/86 (43.0%)). Overall, mNGS identified more bacterial (25 vs 2), fungal (5 vs 3), viral (9 vs 2) organisms compared with CMTs. Cytomegalovirus (CMV) (n = 35, 40.7%), Human gammaherpesvirus 4 (EBV) (n = 35, 40.7%), Pneumocystis jirovecii (n = 27, 31.4%), were the most common pathogens detected, followed by Talaromyces marneffei (n = 16, 18.6%), Torque tenovirus (n = 16, 18.6%) and Mycobacterium tuberculosis (n = 11, 12.8%).
 |
Figure 2 Breakdown of organisms identified by both metagenomics next-generation sequencing (mNGS) and conventional microbiological tests (CMTs), mNGS only, and CMTs only. |
Mixed infection was defined when two or more infectious pathogens detected. Mixed infection were detected in 34 patients by mNGS combined with CMTs. The most frequent pattern of multiple pathogens was virus and fungi mixed infection (17 patients, 17/86=19.8%), followed by bacteria, virus and fungi mixed infection (7 patients, 7/86=8.1%). Six patients were diagnosed as bacteria and fungi mixed infection (6/86=7%); two patients were diagnosed as bacteria and virus mixed infection (2/86=1.2%); two patients were diagnosed as bacteria, pneumocystis jirovecii and mycoplasma mixed infection (2/86=1.2%). Notably, virus (94.1%, 32/34) and fungi (94.1%, 32/34) were commonly seen in the mixed infection cases.
When examining the cases in which mNGS was positive and CMTs were negative, the most commonly identified organisms were P. jirovecii (n = 27 cases), Streptococcus pneumoniae (n = 6 cases) and Haemophilus influenzae (n = 3). However, mNGS missed detecting pathogens in 16.3% (14/86) patients whose diagnosis based on clinical features, CMTs and the response to antimicrobial therapy. The pathogens missed by mNGS were Nontuberculosis mycobacteria, Mycobacterium tuberculosis, followed by Syphilis, Toxoplasma gondii (Table 3).
 |
Table 3 Pathogens Missed by mNGS |
Impact on Treatment Decisions by mNGS and CMTs
We finally attempted to assess the clinical impact of the mNGS findings. Empiric antimicrobial treatment of 86 cases were prescribed for initial consideration of possible infections. We compared the use proportion of broad-spectrum antimicrobials before and after mNGS. 78/86 (90.7%) patients received broad-spectrum antimicrobials (antimicrobials often cover Gram-negative, Gram-positive bacteria, atypic pathogens, virus and fungi) therapy before performing mNGS, while only 34/86 (39.5%) after mNGS (P < 0.0001).
mNGS helped identify the etiology or confirm appropriate treatment in 49/86 (57%) patients. Antiviral treatment (acyclovir) was added as Varicella zoster virus encephalitis in two cases was established based on mNGS results. For patients mNGS provided positive identification of mycobacterium avium infection, leading to precise changes in the anti-infection treatment. For patients with PJP confirmed by mNGS, sulphanilamide dose increased from prophylactic medication to therapeutic medication, for 20 patients with PJP we combined caspofungin and sulphanilamide. For patients with talaromyces marneffei infection, we added amphotericin B, discontinued antibacterial treatment.
Meanwhile, CMTs also helped give appropriate treatment in 28 patients. PCR confirmed EBV infection, and then we discontinued foscarnet and initiated acyclovir antiviral therapy. Blood cultures showed talaromyces marneffei positive, which supported the mNGS result, we continued amphotericin B therapy. Syphilis-specific antibody was positive, we administrated penicillin treatment. Toxoplasma antibody was positive and we gave lincomycin plus sulfadiazine.
The successful de-escalation or discontinuation of treatment was supported by mNGS in 37 patients. We observed a significant reduction in the number of patients being prescribed foscarnet (52.3% vs 23.26%, p < 0.001), moxifloxacin (34.9% vs 10.5%, p = 0.005), and levofloxacin (32.6% vs 14%, p = 0.001) before and after mNGS (Table 4).
 |
Table 4 Comparison of the Use Proportion of Antimicrobials Between Pre-mNGS and After-mNGS in Patients |
Discussion
We retrospectively enrolled patients diagnosed acquired immunodeficiency syndrome with suspected opportunistic infections during the period from January 2018 to February 2021, and attempted to identify the causal pathogens. Our study showed that AIDS patients with suspected opportunistic infections had a more complex microbial etiology spectrum, with higher detection rates of fungal, viral and coexisting infectious agents. Positive agreement between mNGS and clinical diagnosis was significantly higher than that of CMTs (75.6% vs 43.0% respectively). We accordingly found that CMV, EBV, Pneumocystis jirovecii were the most causal common pathogens detected by mNGS. All PJP cases were detected by mNGS in this study. Traditionally, the detection of P. jirovecii only be based on methods such as immunofluorescence staining and PCR assay. However, immunofluorescence shows low positivity, and immunofluorescence staining of P. jirovecii is not routinely performed in many hospitals. This may be the reason why CMTs reported few PJP cases. P. jirovecii PCR is very sensitive but it is not available in many hospitals in China. mNGS provide a new sensitive approach for detection of P. jirovecii. Our study showed early application of mNGS can facilitate the rapid and accurate diagnosis of PJP no matter BALF or blood. Jiang et al reported that mNGS showed a good specificity of 96.3% when compared with Gomori’s methenamine silver staining and serum (1,3)-β-D glucan.25 Meanwhile, mNGS not only identify the species of microorganism, but also it can identify to the level of genus. For example, different non-tuberculous mycobacterium genera has different treatment, the same therapy will lead to the failure of treatment, and induce the emergence of drug resistance. In this study, for patients mNGS provided positive identification of mycobacterium avium infection, leading to precise changes in the anti-infection treatment.
Although mNGS is sensitive, rapid and unbiased, there are still many problems, such as how to regulate its indications, the timing of its intervention, its diagnostic threshold of various pathogens, and lack of availability in underdeveloped countries. The Consensus of Chinese experts proposed that mNGS is the first-line choice for critical, difficult and immunodeficient patients.26 Meanwhile, mNGS could be performed for chronic infection without waiting for the results of conventional microbiological tests. Our team has discussed the threshold of mNGS for the diagnosis of PJP last year,27 and mNGS diagnostic threshold of bacteria, tuberculosis, virus, fungi and other pathogens needs further research and discussion in future. Meanwhile, we believe that the diagnostic threshold of mNGS should be different between the immunodeficient and non-immunodeficient patients.
CMTs and mNGS results reached an agreement in 37 out of 86 patients (43%) in this study. As we know, in conventional microbiological tests, culture is less positive and longer, smear is rapid but less sensitive. However, PCR and antigen/antibody detection are more sensitive and less time-consuming, and they are obviously superior to mNGS in speed, efficiency and economic cost. Regretly, we carry out too less CMTs in reality, which reduce the sensitivity and diagnostic ability of CMTs greatly. A report from Peking Union Medical College Hospital showed they did not observe improved diagnostic accuracy of mNGS in BALF compared with that of CMTs. It indicated CMTs is still valuable if CMTs methods are comprehensively and carefully carried out.28 However, CMTs is well performed only in 30% hospitals in China. So, there is a large room for us to improve the quality of CMTs. There were two Varicella zoster virus (VZV) encephalitis cases detected by mNGS in this study. It would be detected quickly and accurately if VZV PCR was available. The same for mycobacterium tuberculosis infection, the rate of detection would be greatly improved if the hospital carry out Gene Xpert.
Parize et al reported that mNGS had a high negative predictive value and detected more clinically relevant viruses and bacteria than CMTs. Reliable negative mNGS results may help support a conclusion that infection is not present, which will help reduce physicians’ concerns about active infections.29 At this point, it is advocated that mNGS results can help to narrow the diagnosis and de-escalation or discontinuation unnecessary treatment. In the present study, initial antimicrobial treatments were modified for 51.2% of patients after the report of mNGS results. More importantly, we observed an absolute reduction in antimicrobial prescribing. The reduction of therapy was most pronounced with antimicrobials targeted at virus, bacteria and atypical pathogens such as foscarnet, moxifloxacin and levofloxacin. Implementation of mNGS as a rule-out strategy may reduce the abuse and use proportion of antimicrobials, as well as the duration of antimicrobials. Meanwhile, the unbiased broad-spectrum detection of mNGS would further provide guidance for effective antimicrobial treatment. 11.6% patients did not receive caspofungin and 9.3% patients did not receive amphotericin B until the report of mNGS results. The median turnaround time for mNGS results from samples submission to pathogen identification decreased from 2 days at the beginning to within 24 hours later. This significantly helped clinicians in the decision of initial antimicrobial treatment. Of course, an important limitation of use of mNGS is lack of reliable drug susceptibility data.
In this study, we did not do complex PCR assays for patients in CSF and BALF, it decreased the positive rate of CMTs. Second, we enrolled a relatively small observational cohort. It needs more observation to investigate the diagnostic value of mNGS in AIDS patients with suspected opportunistic infections.
In this study, we evaluated the performance of mNGS in the diagnosis of acquired immunodeficiency syndrome patients with suspected opportunistic infections. These findings showed mNGS improved yield of pathogens and lead to personalized targeted antimicrobial treatment.
Data Sharing Statement
All data generated or analysed during this study are included in the published article.
Ethics Approval and Consent to Participate
All procedures performed in this study involving human participants were conducted in accordance with the ethical standards of the Ethics Committee of each study site (Hunan provincial People’s Hospital, The Central Hospital of Yueyang, Second Hospital of Nanjing and Hengshui People’s Hospital), the 1964 Helsinki declaration and its later amendments, and with comparable ethical standards.
Consent for Publication
Informed consent was obtained from all participants included in the study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Natural Science Foundation of China project [grant nos. 82270019 and 82070011]; the Key Project of Jiangsu Commission of Health [grant no. K2019004]; Changsha Municipal Natural Science Foundation [grant no.Kq2014193]; Renshu Fund of Hunan Provincial People's Hospital [RS202105].
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Gona P, Van Dyke RB, Williams PL, et al. Incidence of opportunistic and other infections in HIV-infected children in the HAART era. JAMA. 2006;296(3):292–300. doi:10.1001/jama.296.3.292
2. Iroezindu MO. Disparities in the magnitude of human immunodeficiency virus-related opportunistic infections between high and low/middle-income countries: Is highly active antiretroviral therapy changing the trend? Ann Med Health Sci Res. 2016;6(1):4–18. doi:10.4103/2141-9248.180234
3. WHO Guidelines Approved by the Guidelines Review Committee. Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV. Geneva: World Health Organization; 2015.
4. Ramesh K, Gandhi S, Rao V. Clinical profile of human immunodeficiency virus patients with opportunistic infections: a descriptive case series study. Int J Appl Basic Med Res. 2015;5(2):119–123. doi:10.4103/2229-516X.157166
5. Scott L, da Silva P, Boehme CC, Stevens W, Gilpin CM. Diagnosis of opportunistic infections: HIV co-infections - tuberculosis. Curr Opin HIV AIDS. 2017;12(2):129–138. doi:10.1097/COH.0000000000000345
6. Kaplan JE, Benson C, Holmes KK, Brooks JT, Pau A, Masur H; Centers for Disease Control and Prevention (CDC); National Institutes of Health; HIV Medicine Association of the Infectious Diseases Society of America. Guidelines for prevention and treatment of opportunistic infections in HIV-infected adults and adolescents: recommendations from CDC, the National Institutes of Health, and the HIV Medicine Association of the Infectious Diseases Society of America. MMWR Recomm Rep. 2009;58(RR–4):1–207.
7. Solomon FB, Angore BN, Koyra HC, Tufa EG, Berheto TM, Admasu M. Spectrum of opportunistic infections and associated factors among people living with HIV/AIDS in the era of highly active anti-retroviral treatment in Dawro Zone hospital: a retrospective study. BMC Res Notes. 2018;11(1):604. doi:10.1186/s13104-018-3707-9
8. Pang W, Shang P, Li Q, et al. Prevalence of opportunistic infections and causes of death among hospitalized HIV-infected patients in Sichuan, China. Tohoku J Exp Med. 2018;244(3):231–242. doi:10.1620/tjem.244.231
9. Luo B, Sun J, Cai R, et al. Spectrum of opportunistic infections and risk factors for in-hospital mortality of admitted AIDS patients in Shanghai. Medicine. 2016;95(21):e3802. doi:10.1097/MD.0000000000003802
10. Mazuski JE, Sawyer RG, Nathens AB, et al. The Surgical Infection Society guidelines on antimicrobial therapy for intra-abdominal infections: evidence for the recommendations. Surg Infect (Larchmt). 2002;3(3):175–233. doi:10.1089/109629602761624180
11. Thorndike J, Kollef MH. Culture-negative sepsis. Curr Opin Crit Care. 2020;26(5):473–477. doi:10.1097/MCC.0000000000000751
12. Paul M, Kariv G, Goldberg E, et al. Importance of appropriate empirical antibiotic therapy for methicillin-resistant Staphylococcus aureus bacteraemia. J Antimicrob Chemother. 2010;65(12):2658–2665. doi:10.1093/jac/dkq373
13. Campion M, Scully G. Antibiotic use in the intensive care unit: optimization and de-escalation. J Intensive Care Med. 2018;33(12):647–655. doi:10.1177/0885066618762747
14. Yang L, Haidar G, Zia H, et al. Metagenomic identification of severe pneumonia pathogens in mechanically-ventilated patients: a feasibility and clinical validity study. Respir Res. 2019;20(1):265. doi:10.1186/s12931-019-1218-4
15. Chen X, Cao K, Wei Y, et al. Metagenomic next-generation sequencing in the diagnosis of severe pneumonias caused by Chlamydia psittaci. Infection. 2020;48(4):535–542. doi:10.1007/s15010-020-01429-0
16. Chen J, He T, Li X, Wang X, Peng L, Ma L. Metagenomic next-generation sequencing in diagnosis of a case of pneumocystis jirovecii pneumonia in a kidney transplant recipient and literature review. Infect Drug Resist. 2020;13:2829–2836. doi:10.2147/IDR.S257587
17. Carpenter ML, Tan SK, Watson T, et al. Metagenomic next-generation sequencing for identification and quantitation of transplant-related DNA viruses. J Clin Microbiol. 2019;57(12):12. doi:10.1128/JCM.01113-19
18. Jerome H, Taylor C, Sreenu VB, et al. Metagenomic next-generation sequencing aids the diagnosis of viral infections in febrile returning travellers. J Infect. 2019;79(4):383–388. doi:10.1016/j.jinf.2019.08.003
19. Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(suppl_2):S231–s240. doi:10.1093/cid/ciy693
20. Zhang Y, Cui P, Zhang HC, et al. Clinical application and evaluation of metagenomic next-generation sequencing in suspected adult central nervous system infection. J Transl Med. 2020;18(1):199. doi:10.1186/s12967-020-02360-6
21. Simner PJ, Buckwalter SP, Uhl JR, Wengenack NL. Identification of Mycobacterium species and Mycobacterium tuberculosis complex resistance determinants by use of PCR-electrospray ionization mass spectrometry. J Clin Microbiol. 2013;51(11):3492–3498. doi:10.1128/JCM.01408-13
22. Hodowanec AC, Pikis A, Komatsu TE, et al. Treatment and prevention of CMV disease in transplant recipients: current knowledge and future perspectives. J Clin Pharmacol. 2019;59(6):784–798. doi:10.1002/jcph.1363
23. Yu S, Yang Q, Wu J, et al. Clinical application of Epstein-Barr virus DNA loads in Epstein-Barr virus-associated diseases: a cohort study. J Infect. 2021;82(1):105–111. doi:10.1016/j.jinf.2020.11.027
24. Fang X, Mei Q, Fan X, et al. Diagnostic value of metagenomic next-generation sequencing for the detection of pathogens in bronchoalveolar lavage fluid in ventilator-associated pneumonia patients. Front Microbiol. 2020;11:599756. doi:10.3389/fmicb.2020.599756
25. Jiang J, Bai L, Yang W, et al. Metagenomic next-generation sequencing for the diagnosis of pneumocystis Jirovecii pneumonia in non-HIV-infected patients: a retrospective study. Infect Dis Ther. 2021;10(3):1733–1745. doi:10.1007/s40121-021-00482-y
26. Cao Q, Chen B, Chen L, et al. Expert consensus on the clinical application of metagenomics next-generation sequencing technology for detection of infectious pathogens in China. Chin J Infect Dis. 2020;38(11):681–689. doi:10.3760/cma.j.cn311365-20200731-00732
27. Peng JM, Du B, Qin HY, Wang Q, Shi Y. Metagenomic next-generation sequencing for the diagnosis of suspected pneumonia in immunocompromised patients. J Infect. 2021;82(4):22–27. doi:10.1016/j.jinf.2021.01.029
28. Liu L, Yuan M, Shi Y, Su X. Clinical performance of BAL metagenomic next-generation sequence and serum (1,3)-β-D-glucan for differential diagnosis of Pneumocystis jirovecii Pneumonia and Pneumocystis jirovecii colonisation. Front Cell Infect Microbiol. 2021;11:784236. doi:10.3389/fcimb.2021.784236
29. Han D, Li Z, Li R, Tan P, Zhang R, Li J. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. 2019;45(5–6):668–685. doi:10.1080/1040841X.2019.1681933
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
