Back to Journals » OncoTargets and Therapy » Volume 13
The Long Non-Coding RNA-RoR Promotes the Tumorigenesis of Human Colorectal Cancer by Targeting miR-6833-3p Through SMC4
Authors Li X, Chen W, Jia J, You Z, Hu C, Zhuang Y, Lin Z, Liu Y, Yang C, Xu R
Received 17 November 2019
Accepted for publication 3 February 2020
Published 27 March 2020 Volume 2020:13 Pages 2573—2581
DOI https://doi.org/10.2147/OTT.S238947
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Xinyu Li,1,* Wen Chen,2,* Jing Jia,1,* Zhicheng You,3 Changjin Hu,4 Yihuang Zhuang,1 Zhibin Lin,1 Yan Liu,1 Chunkang Yang,5 Rongyu Xu1
1Quanzhou First Hospital, Quanzhou, Fujian Province, People’s Republic of China; 2The Second Affiliated Hospital of Fujian Medical University, Quanzhou, Fujian Province, People’s Republic of China; 3Yongchun County Hospital, Quanzhou, Fujian Province, People’s Republic of China; 4Jinjiang Hospital of Traditional Chinese Medicine Affiliated to Fujian University of Traditional Chinese Medicine, Jinjiang, Fujian Province, People’s Republic of China; 5Fujian Tumor Hospital, Fuzhou, Fujian Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chunkang Yang
Department of Gastroenterology, Fujian Tumor Hospital, Fuzhou, Fujian Province, People’s Republic of China
Email [email protected]
Rongyu Xu
Department of Surgical Oncology, Quanzhou First Hospital, Quanzhou, Fujian Province, People’s Republic of China
Email [email protected]
Background: Long non-coding RNA regulator of reprogramming (LINC-RoR) has shown different expressions in a variety of tumors as a stem cell inducer through reprogramming regulation. However, its role and regulation mechanisms in colorectal cancer (CRC) are still unclear.
Materials and Methods: Quantitative real-time PCR and Western blot were performed to examine LINC-RoR expression in paired CRC samples and cell lines. The relationship of LINC-RoR expression with clinicopathological characteristics and clinical outcomes was analyzed. The biological functions of LINC-RoR were studied by MTS and colony formation in vitro. Cell apoptosis was analysed by the flow cytometry. The Dual-luciferase reporter assays and RIP assays were performed to explore the regulatory relationship of LINC-RoR.
Results: In this study, we found that LINC-RoR was upregulated in CRC cell lines and tissues. High expression of LINC-RoR was associated with poorer survival time and multivariate analysis results showed that LINC-RoR was an independent risk factor of tumor malignancy progression. Overexpression of LINC-RoR promoted the cell proliferation and knocked down it can reverse the effect in vitro. The regulatory network of LINC-ROR/miR-6833-3p/SMC4 was predicted with bioinformatics analysis tools and validated via dual-luciferase reporter assays and RIP. Further study revealed that in overexpression LINC-RoR cell lines the expression of miR-6833-3p was downregulated and miR-6833-3p can inhibit its target gene SMC4, the apoptosis-related protein.
Conclusion: We concluded that LINC-RoR functions as an oncogene in CRC through the miR-6833-3p/SMC4 pathway.
Keywords: LINC-RoR, colorectal cancer, miR-6833-3p, SMC4, non-coding RNA, apoptosis
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumor of digestive system and it is a major cause of cancer-related mortality worldwide.1 In United States there were about 145,600 newly diagnosed CRC patients in 2018, leading to about 51,020 deaths.2 The 5-year cancer-free survival rate of CRC was only 52.8%, and it has high rates of recurrence and metastasis. Despite the standardized surgical treatment have been widespread use, the treatment of CRC remains a great challenge.3,4 Therefore, there is an urgent need to elucidate the mechanism for tumorigenesis and to develop potential therapeutic targets for CRC patients.
Non-coding RNAs such as microRNA (miRNA) and long non-coding RNA (lncRNA) have been put forward as epigenetic regulator for many years. Long non-coding RNAs (lncRNAs) are one type of non-coding RNAs with a length of >200 nucleotides and no protein-coding ability.5,6 They are aberrantly expressed in a variety of cancers, and they play key roles in promoting and maintaining tumorigenesis, demonstrating their clinical potential therapeutic targets.7–10 Although not usually translated into protein, they almost involve in every stage of gene expression.11,12 In addition, more and more evidences reveal that the lncRNAs may function through their intergenic trait‐associated regions.13 In recent years, many lncRNAs, such as the lnc-GNAT1-1, PVT1, SNHG12, DILC. XIST and LAMC2-1 have shown abnormal expression and been demonstrated to be involved in multiple development, differentiation and tumorigenesis of CRC.14–19 More recently, studies have suggested that some lncRNAs may competitively bind with miRNAs, therefore leading to aberrant expression of its target messenger RNAs.20,21 In this study, we discussed the role of LINC-RoR in the regulation of miR‐6833‐3p/SMC4 pathway in detail.
Materials and Methods
Patients and Tissues
Twenty-four CRC patients’ tumor tissues and paired normal tissue adjacent to tumor were obtained from Fujian Tumor Hospital during 2010–2012. All the patients were diagnosed as colon cancer or rectal cancer without radiotherapy or chemotherapy before surgery. The study was undertaken with the approval of the Fujian tumor hospital institutional ethics committee board. All patients signed the written consent form.
Cell Culture
NCM460, SW480, HT-29, HCT-116 cell lines were obtained from the Sixth affiliated Hospital of Sun Yat-sen University (Authenticated by STR profiling, CAMS/PUMC). Cells were cultured in RPMI-1640 (Gibco, Carlsbad, CA) mixed with 10% fetal bovine serum (Gibco, Carlsbad, CA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Solarbio, Beijing, China). All the cells were cultured in an incubator at 37°C with 5%CO2. The LINC-RoR stable overexpression cell lines were constructed. Full-length cDNA LINC-RoR and negative control (NC) were synthesized by GenePharma (Shanghai, China) and cloned into SW 480 cell line with the recombinant lentiviral ZsGreen Puro vector (Inovogen Tech. Co).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Assay
The total RNAs of cells were isolated with TRIzol reagent (ComWin Biotech, Beijing, China), and the cDNA was generated using TransScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). The quantitative real-time PCR was performed using a RT-PCR system (TransGen Biotech, Beijing, China) on the Applied Biosystems 7500 Detection System (Applied Biosystems, Foster City, CA, USA) according to the manufacturers’ instructions. with the following specific primers:. LINC-ROR forward, 5ʹ‐CTTGATGGC ATTGTCGCTAA‐3ʹ and reverse, 5ʹ‐TCCAGTGGCTGTGCTAGATG‐3ʹ; β‐actin forward, 5ʹ‐CCACTGGCATCGTGATGGA‐3ʹ and reverse, 5ʹ‐CGCT CGGTGAGGATCTTCAT‐3ʹ; miR-6833-3p: stem-loop primer: GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACctgaggaa, forward: TTTCTCTCTCCACTTCCTCAGGTCGT, reverse: TGAGGTGCTGTGCGTGAC. U6: forward: CTCGCTTCGGCAGCACA, reverse: AACGCTTCACGAATTTGCGT
SMC4 forward: CGCCTCCAGCAATGACCAAT, reverse: CCCCAGCATAGGATTTGAAGTT; siRNA against LINC-RoR (coding region sense was 5′-GGAGAGGAAGCCUGAGAGUdTdT-3′, antisense was 5′-ACUCUCAGGCUUCCUCUCCdTdT-3′)
Cell Proliferation Assay
Cell viability was measured by CellTiter 96® AQueous One Solution Cell Proliferation Assay Kit (Promega, USA). Cells were divided into the LINC-RoR overexpression group and NC group. According to the protocol, all cells (2×103) were seeded in 96-well plates and cultured for 24, 48, 72, and 96 hrs. Then 20 μL of the reagent was added into each well and incubated at 37°C for another 2 hrs. Absorbance was detected at 490 nm with a microplate reader (BioTek Instruments, USA).
Flow Cytometry Analysis of Cell Apoptosis
For cell apoptosis assays, flow cytometer (FACScan; BD Biosciences) was used to analyse with the Annexin V‐APC/7‐aminoactinomycin D (7‐AAD) Apoptosis Detection Kit (KeyGen, China). After seeded, digested, washed and precooling, we added the Annexin V‐APC and 7‐AAD into the sample. The samples were incubated at room temperature in dark place and quantified using flow cytometry analysis (BD Accuri™ C6, USA).
Colony Formation Assay
SW480 cells, overexpression LINC-RoR SW480 cells and NC SW480 cells were seeded into 6-well plates and cultured 2 weeks. After fixation with methanol, the colonies were stained with crystal violet in methanol and the area percentage of colonies was calculated.
Western Blot Analysis
Total protein of cells were extracted using RIPA buffer with protease inhibitors (Solarbio, Beijing, China). The concentrations of extracted proteins were measured with BCA Protein Assay Kit (Thermo Fisher Scientific) using multimode microplate reader (Biotek, USA). Then 50μg total proteins were diluted and were denatured. Proteins were separated by 10% SDS gel electrophoresis and transferred onto polyvinylidene difluoride membranes. After blocked with tris buffered saline-tween with 5% nonfat milk (BD Bioscience, USA), the membrane was incubated with primary antibodies against SMC4 (1:1000; Abcam, Cambridge, MA, USA), or β-actin (1:3000; ZSGBBIO, Beijing, China). Then the membranes were incubated with anti-rabbit or anti-mouse secondary antibodies (1:5000; ZSGB-BIO, Beijing, China), and detected by SuperSignal West Femto Trial Kit (Thermo Fisher Scientific, USA).
Dual-Luciferase Reporter Assay
The SW480 cells were seeded in a 48‐well plate, after incubating for 24 hrs and co-transfected with a mixture of miRNA mimics and conditioned vector pmirGLO (Genewiz, Suzhou, China). Then we detected the luciferase activity with the dual‐luciferase reporter assay system (Promega Corporation, Madison, WI) and the multimode microplate reader (Biotek, USA). Then we calculated the fluorescence intensity with the ratio of firefly luciferase (hLuc) activity/Renilla luciferase (hRluc) activity.
RNA Immunoprecipitation
The RNA immunoprecipitation (RIP) assay was performed using a Magna RIP™ RNABinding Protein Immunoprecipitation Kit (Millipore, USA) according to the manufacturer’s protocols. Cells were cross-linked with 1% formaldehyde, incubated overnight with antibodies, including rabbit polyclonal IgG (Millipore, USA) as NC and anti-SMC4 (Abcam, USA), and recovered with protein G Dynabeads. RNA obtained after stringent washing and extraction with TRIzol was analyzed by qRT-PCR. To detect the expression level of miR‐6833‐3p.
Statistical Analysis
All statistical analyses were performed in the R software environment (version 3.4.0; http://r-project.org/), and p < 0.05 was considered significant in all statistical analyses. Graphs were presented by using GraphPad Prism 7 Software (GraphPad, San Diego, CA) and R. Differences between groups were assessed using the χ 2 Student’s t-test or one‐way analysis of variance (ANOVA). Three independent experiments were repeated at least.
Results
Upregulation of LINC-RoR Level in CRC Tissues and Cell Lines
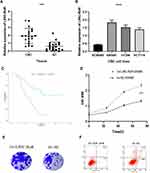
The LINC-RoR expression level in clinical tissues was assessed by qRT-PCR. Firstly we compared the expression of LINC-RoR in 24 CRC tissues and their paired adjacent tissues. We found that the expression level of LINC-RoR in CRC tissues was significantly upregulated compared with their adjacent tissues (p < 0.0001) (Figure 1A). Then we compared the LINC-RoR expression level in normal and CRC cell lines. The expression of LINC-RoR in NCM460 was lower than SW480, HT296 and HCT 116 (Figure 1B).
The Prognostic Value of LINC-RoR in CRC Patients
To investigate the prognostic value of LINC-RoR in CRC, we conducted a Kaplan-Meier (K-M) survival analysis with the data of 47 CRC patients (information listed in Table 1). The association of LINC-RoR expression level with OS was shown in a Kaplan-Meier survival analysis (Figure 1C). We found that CRC patients with high LINC-RoR expression level had shorter OS compared with those who had low LINC-RoR expression level. The p value=0.00937. After the Cox regression analysis, we found that T stage and the level of LINC-RoR were independent risk factors for CRC (Table 2). Thus, we speculated that LINC-RoR may act as an oncogene in CRC.
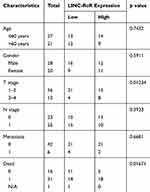 |
Table 1 Association of LINC-RoR Expression with Clinicopathological Parameters in Patients with Colorectal Cancer (n = 48) |
 |
Table 2 Cox Regression Analysis Results |
Overexpression LINC-RoR Promotes Cell Viability, Proliferation and Inhibit Apoptosis in CRC Cell Lines
To explore the biological function of LINC-RoR upregulation in CRC, we constructed LINC-RoR stable overexpression and NC cell line with SW480 cell line using recombinant lentiviruses. Then we compared cell viability between overexpression SW480 cell line and NC SW480 cell line using the MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay. Results showed that the optical density (OD) values of overexpression LINC-RoR group were higher than NC group and normal group (p = 0.0434) (Figure 1D). The phenomenon demonstrated that overexpression LINC-RoR can enhance the CRC cell viability. The colony formation assay results (Figure 1E) also showed that the number of SW480 cell colonies increased significantly in LINC-RoR overexpression group compared with the NC group, suggested that upregulated LINC-RoR expression promoted CRC cell proliferation.
Furthermore, the flow cytometric analysis was also conducted to detect the cell apoptosis between these two groups. The apoptotic cells percent of LINC-RoR overexpression group was reduced by 9.74%±2.13%. indicated that LINC-RoR overexpression can inhibit apoptosis in SW480 cell line (Figure 1F).
LINC-RoR Can Bind to miR-6833-3p: QRT-PCR, Luciferase Reporter Assay and RIP
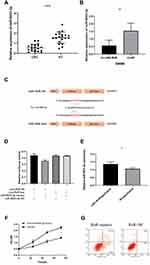
As lncRNAs can bind to miRNA as competitive miRNA sponge, to investigate whether LINC-RoR plays such function in CRC, we supposed that some miRNAs can directly bind LINC-RoR, too. To find the miRNAs that target LINC-RoR, DIANA tools (http://carolina.imis.athena-innovation.gr/), an online bioinformatics websites were used to analysis. We identified 13 targets miRNAs with Bind Score more than 0.9. Then we tested the miRNAs expression level in overexpression LINC-RoR SW480 cell lines. Among these miRNAs, only miR-6833-3p was downregulated significantly (p=0.0033) (Figure 2A). Then we compared the expression of miR-6833-3p in CRC tissues and nor tissue. The results showed that miR-6833-3p was downregulated in CRC tissue (Figure 2B). To validate the bioinformatics prediction, a wild‐type (WT) and mutant pmirGLO‐LINC-RoR vector were inserted with the binding sites of miR‐6833‐3p (Figure 2C). The hLuc2 and hRluc was set as a primary reporter and the control reporter, respectively. The ratio of hLuc/hRluc was tested as fluorescence intensity. We found that the fluorescence intensity of WT LINC-RoR reporter vector was downregulated by miR‐6833‐3p mimics, compared with the other two groups (Figure 2D). This finding demonstrated that miR‐6833‐3p could bind to LINC‐RoR. Then to further validate the regulatory relationship of LINC-RoR and miR-6833-3p, we conducted an anti‐AGO2 RIP assay. Every procedure was followed as the manufacturer’s protocol. After pulling down, the RNA immunoprecipitates were detected by qRT‐PCR. The results showed that the expression of miR‐6833‐3p in the LINC-RoR overexpression group was significantly higher than the NC group (p=0.0331; Figure 2E).
miR-6833‐3p Can Reverse the Effect Induced by LINC-RoR
As overexpression of LINC-RoR can promote proliferation and inhibit apoptosis in SW480, to investigate whether this effect could be reversed by miR‐6833‐3p, we performed a functional rescue assay. MiR‐6833‐3p mimics or miR‐NC were transfected into the LINC-RoR stable overexpression SW480 cell line. MTS assay and flow cytometry were conducted. Results showed that the group transfected with miR-6833‐3p mimics could inhibit proliferation and promote apoptosis, which is opposite of the function of LINC-RoR (Figure 2F and G).
LINC-RoR Regulates SMC4 Expression Through miR‐6833‐3p
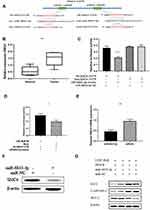
To explore the biological role of miR‐6833‐3p in CRC, we further predicted the mRNA binding sites in DIANA‐micro-CDS software and Targetscan Human7.1. Threshold was wet to 0.9 and predicted genes were screened by publication search. Then we selected 9 genes as potential targets and qRT-PCR was used to detect relative expression. Finally, Structural maintenance of chromosomes 4 (SMC4) was discovered as the binging sites (Figure 3A). Results showed that the SMC4 expression was upregulated in CRC tissues. (p = 0.0043; Figure 3B). Dual‐luciferase assay was performed to determine whether miR‐6833-3p could directly bind with SMC4. We selected two predictive binding sites of SMC4 and miR‐6833‐3p for mutation. Results suggested that only the fluorescence intensity of Wt-SMC4–3ʹ‐UTR reporter vector with miR‐6833‐3p mimics group was significantly reduced in all four groups (p <0.0001; Figure 3C). To further investigate the role of LINC-RoR, we conducted another dual‐luciferase analysis: Group 1 (LINC-RoR + Wt-SMC4 + miR‐6833‐3p mimics) and Group 2 (NC + Wt-SMC4 + miR‐6833‐3p mimics). The results revealed that the fluorescence intensity in Group 2 was reduced compared with Group 1 (Figure 3D), p =0.0088. These results demonstrated that SMC4 expression can be regulated by LINC-RoR/miR‐6833‐3p as ceRNA manner. Then we detected the expression levels of SMC4 by qRT‐PCR and Western blot in SW480 cell line. Results showed that the expression of SMC4 in Group 1 (miR‐6833‐3p mimics) was significantly lower than Group 2 (miR‐NC) (p = 0.0013; Figure 3E). Western blot results also revealed that SMC4 expression was downregulated in the miR‐6833‐3p mimics group (Figure 3F). Furthermore, we transfected si‐SMC4/si‐ NC into the two groups (LINC-RoR and NC) respectively. The transfection efficiency was detected by qRT‐PCR. Then we conducted Western blot to detect the expression level of SMC4 and apoptosis‐related proteins respectively. Results showed that after knockdown of LINC‐RoR, Bcl‐2 was downregulated, Bax and Cleaved Caspase‐3 were upregulated (Figure 3G). Combining with above results, we can conclude that LINC-RoR could upregulate SMC4 expression through targeting miR‐6833‐3p and inhibit apoptosis.
Discussion
Recently numbers of studies have demonstrated that lncRNAs could exhibit tumorigenicity through diverse modes among different cancers.22 As far as we know, LINC‐RoR has show ectopic expression levels in many malignant tumors. For example, Li et al21 elucidated that LINC-ROR acted as a sponge for miR-145 to derepress the expression of target gene ZEB2 in hepatocellular carcinoma, thereby inducing EMT and promoting HCC metastasis. Thiele et al have indicated that LINC-RoR was associated with the progression of CRC.23 Moreover, Li et al have outlined that LINC RoR can influence the cell proliferation via p53.24 Yang et al revealed that LINC-RoR promoted the resistance of radiotherapy for human CRC cells by targeting the p53/miR-145 pathway.25 In our research, we verified that LINC-RoR was upregulated in CRC tissues and cell lines compared with adjacent normal tissues and normal colorectal cell line. At the same time, Kaplan-Meier analysis showed that CRC patients with different expression level of LINC-RoR have different survival time. High level LINC-RoR was connected with shorter live. Then we conducted the in vitro examination to detect the function of LINC-RoR in CRC. We found that overexpression of LINC-RoR can increase cell viability, promote cell proliferation ability and inhibit cell apoptosis in SW480 cell line. These findings revealed that LINC-RoR may play a role in the tumorigenesis of CRC. Moreover, we established the lncRNA–miRNA–mRNA regulatory networks of LINC-RoR-miR-6833-3p-SMC4. As hundreds of publications have described the influence of ceRNA regulation in normal and disease cells. That means some sites of miRNAs’ 3ʹ‐UTR can bind to lncRNAs and then compose a so-called RNA‐induced silencing complex.26 lncRNAs can influence the expression of many genes and further regulate broad biological processed by this way. In our research, firstly we detected the different expression levels of LINC-RoR between tissues and cell lines and confirmed its prognostic value in a certain number of long follow-up clinical patients. Secondly, we explored the function of LINC-RoR in vitro. Thirdly we predicted the binding miRNAs with bioinformatics tools and validated with different methods. Lastly, we further predicted the target gene of miR-6833-3p and validated it. Based on the above evidences, we set a new interaction network between LINC-RoR and SMC4 mediated by miR‐6833‐3p, which is also can be named as LINC‐RoR/miR‐6833‐3p/SMC4 axis. Overexpression or low expression of LINC-RoR can promote or inhibit the expression of SMC4 and miR‐6833‐3p mimics can offset this effect. The regulatory relationships were validated by dual-luciferase reporter assay and RNA immunoprecipitation analysis. All these findings demonstrated our perspective that LINC-RoR can regulate SMC4 expression in a ceRNA model. Few studies have reported about miR-6833-3p, however, SMC4 was a core subunit of condensin I and II, which are large protein complexes, is involved in chromosome condensation, and has been associated with tumorigenesis.27
In conclusion, we highlighted a novel LINC-RoR/miR-6833-3p/SMC4 regulatory network of CRC and LINC-RoR was involved in cell apoptosis in CRC by targeting miR‐6833‐3p/SMC4 axis in vitro.
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
1. Sinicrope FA. Lynch syndrome–associated colorectal cancer. N Engl J Med. 2018;379(8):764–773.
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
3. Dienstmann R, Mason MJ, Sinicrope FA, et al. Prediction of overall survival in stage II and III colon cancer beyond TNM system: a retrospective, pooled biomarker study. Ann Oncol. 2017;28(5):1023–1031.
4. Suenaga M, Akiyoshi T, Shinozaki E, et al. A feasibility study of capecitabine and oxaliplatin for patients with stage II/III colon cancer ACTOR Study. Anticancer Res. 2018;38(3):1741–1747.
5. Mercer TR
6. Liu B, Ye B, Yang L, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18(5):499.
7. Tutino V, Defrancesco ML, Tolomeo M, et al. The expression of riboflavin transporters in human colorectal cancer. Anticancer Res. 2018;38(5):2659–2667.
8. Lea MA, Kim H. Effects of biguanides on growth and glycolysis of bladder and colon cancer cells. Anticancer Res. 2018;38(9):5003–5011.
9. Tei M, Otsuka M, Suzuki Y, Kishi K, Tanemura M, Akamatsu H. Safety and feasibility of single-port surgery for colon cancer in octogenarians. Anticancer Res. 2018;38(5):2967–72.
10. Dai W, Feng Y, Mo S, et al. Transcriptome profiling reveals an integrated mRNA–lncRNA signature with predictive value of early relapse in colon cancer. Carcinogenesis. 2018;39(10):1235–1244.
11. Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154(1):240–251
12. Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927.
13. Li X, Wu Z, Fu X, Han W. lncRNAs: insights into their function and mechanics in underlying disorders ScienceDirect. Mutat Res Rev Mutat. 2014;762:1–21.
14. Ye C, Shen Z, Wang B, et al. A novel long non-coding RNA lnc-GNAT1-1 is low expressed in colorectal cancer and acts as a tumor suppressor through regulating RKIP-NF-κB-Snail circuit. J Exp Clin Cancer Res. 2016;35(1):187.
15. Ye C, Shen Z, Wang B, et al. A functional polymorphism in lnc-LAMC2-1:1 confers risk of colorectal cancer by affecting miRNA binding. Carcinogenesis. 2016;35(1):187.
16. Shang AQ, Wang WW, Yang YB, et al. Knockdown of long noncoding RNA PVT1 suppresses cell proliferation and invasion of colorectal cancer via upregulation of microRNA-214-3p. Am J Physiol Gastrointest Liver Physiol. 2019;317(2):G222–G232.
17. Liu Y, Zhou J, Wang S, Song Y, Zhou J, Ren F. Long non-coding RNA SNHG12 promotes proliferation and invasion of colorectal cancer cells by acting as a molecular sponge of microRNA-16. Exp Ther Med. 2019;18(2):1212–1220.
18. Song H, He P, Shao T, Li Y, Li J, Zhang Y. Long non-coding RNA XIST functions as an oncogene in human colorectal cancer by targeting miR-132-3p. J buon. 2017;22(3):696–703.
19. Gu LQ, Xing XL, Cai H, et al. Long non-coding RNA DILC suppresses cell proliferation and metastasis in colorectal cancer. Gene. 2018;666:18–26.
20. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14
21. Li C, Lu L, Feng B, et al. The lincRNA-ROR/miR-145 axis promotes invasion and metastasis in hepatocellular carcinoma via induction of epithelial-mesenchymal transition by targeting ZEB2. Sci Rep. 2017;7(1):1–4.
22. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981.
23. Thiele JA, Hosek P, Kralovcova E, et al. lncRNAs in non-malignant tissue have prognostic value in colorectal cancer. Int J Mol Sci. 2018;19(9):2672.
24. Li H, Jiang X, Niu X. Long non-coding RNA reprogramming (ROR) promotes cell proliferation in colorectal cancer via affecting P53. J Exp Clin Res. 2017;23:919.
25. Yang P, Yang Y, An W, et al. The long noncoding RNA-ROR promotes the resistance of radiotherapy for human colorectal cancer cells by targeting the p53/miR-145 pathway. J Gastroenterol Hepatol. 2017;32(4):837–845.
26. Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75(3):467–484. doi:10.1007/s00018-017-2626-6
27. Zhou B
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.