Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 17
The LINC01176-miR-218-5p-IL-36G Network is Responsible for the Pathogenesis of Psoriasis by Promoting Inflammation
Authors Zhao Z, Cheng J, Sun W, Zhu J , Lu S, Feng Y, Song Z, Yang Y, Wu X
Received 30 October 2023
Accepted for publication 25 December 2023
Published 3 January 2024 Volume 2024:17 Pages 1—12
DOI https://doi.org/10.2147/CCID.S444265
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
Zongfeng Zhao,1,* Jie Cheng,2,* Wanqun Sun,1 Jian Zhu,3 Sheng Lu,3 Yanyan Feng,4 Zhendong Song,5 Yali Yang,6 Xiujuan Wu3
1Department of Scientific Research, Shanghai Xuhui Central Hospital, Zhongshan-Xuhui Hospital, Fudan University, Shanghai, People’s Republic of China; 2Department of Urology, Shanghai Xuhui Central Hospital, Zhongshan-Xuhui Hospital, Fudan University, Shanghai, People’s Republic of China; 3Department of Dermatology, Shanghai Xuhui Central Hospital, Zhongshan-Xuhui Hospital, Fudan University, Shanghai, People’s Republic of China; 4Chengdu Second People’s Hospital, Chengdu, Sichuan Province, People’s Republic of China; 5WLSA Shanghai Academy, Shanghai, People’s Republic of China; 6Department of Dermatology, Shanghai Ninth Hospital Affiliated to Shanghai Jiao Tong University, School of Medicine, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yali Yang, Department of Dermatology, Shanghai Ninth Hospital affiliated to Shanghai Jiao Tong University, School of Medicine, No. 639, Manufacturing Bureau Road, Huangpu District, Shanghai, 200011, People’s Republic of China, Email [email protected] Xiujuan Wu, Department of Dermatology, Shanghai Xuhui Central Hospital, Zhongshan-Xuhui Hospital, Fudan University, No. 966, Huaihai Middle Road, Xuhui District, Shanghai, 200031, People’s Republic of China, Email [email protected]
Purpose: Psoriasis is an incurable chronic inflammatory skin disease. The exact function and regulatory mechanism of non-coding RNA upregulation in psoriasis remains to be elucidated. The aim of this study was to analyse the role of the lncRNA-miRNA-mRNA network of psoriasis and LINC01176 in the pathogenesis of psoriasis.
Patients and Methods: We performed miRNA, lncRNA, and mRNA sequencing analysis in pretreatment and treatment psoriatic tissues and normal tissues, constructed an lncRNA-miRNA-mRNA coexpression network and screened mRNA-associated pathways using bioinformatics analysis. We further validated the regulatory role of LINC01176-miR-218-5p on the proliferation and inflammation of the psoriatic model by dual-luciferase reporter assay, cell transfection, CCK-8 method, TUNEL staining and animal model construction method. An lncRNA-miRNA-mRNA coexpression network was successfully constructed by RNA-seq data analysis.
Results: We obtained the relationship between LINC01176, miR-218-5p and IL36-G. Analysis of the apoptotic and proliferative capacity of the transfected cells showed that miR-218-5p up-regulation significantly inhibited cell proliferation and promoted apoptosis. A mouse model of psoriasis was successfully established. Phenotypic observations revealed that keratin-forming cells in mice coated with LINC01176-shRNA emulsifier were significantly lower than those in the model group and close to those in the normal group. HE and immunohistochemical experiments were performed, and the results showed the role and mechanism of action of LINC01176-shRNA. Suppression of LINC01176 significantly inhibited the expression of IL-36G in psoriatic tissues. LINC01176 showed a targeting and positive correlation with IL36-G expression.
Conclusion: Our study shows that LINC01176 promotes the proliferation and invasion of keratinocytes and inhibits apoptosis by targeting miR-218-5p, which acts as a repressor of the psoriasis-associated IL-36G. The shRNA-LINC01176 emulsion showed potential treatment capability in alleviating symptoms of psoriasis.
Keywords: psoriasis, LINC01176, IL-36G, inflammation
Introduction
Psoriasis is a common chronic, recurrent, immune-mediated disease of the skin and joints.1 The underlying pathological mechanisms include complex interactions between the innate and adaptive immune systems.2 The prevalence is approximately 3% worldwide but varies by region, with the highest prevalence in Caucasians (3.6%).3 In addition to affecting the skin, psoriasis also affects the quality of life of patients due to its embarrassing appearance.4 The risk of suicide for people with psoriasis is reported to be as high as 30%, equivalent to life-threatening diseases such as heart diseases, diabetes, and depression.5 Treatment options for psoriasis depend on the severity of the disease, comorbidities, and access to health care. Mild to moderate psoriasis can be treated with a combination of topical glucocorticoids, vitamin D analogues, and phototherapy, while moderate to severe psoriasis usually requires systemic treatment.6 In the past decade, biologics targeting tumour necrosis factor-α, interleukin (IL)-23, and IL-17 have been developed and approved for the treatment of psoriasis.7 These biologics dramatically improve skin and joint symptoms in patients with moderate-to-severe psoriasis and psoriatic arthritis.8 Despite extensive research into psoriasis treatment and pathogenesis, a complete cure remains elusive, largely due to the complex and multifaceted nature of the factors and mechanisms involved in its development.
Long non-coding RNAs (lncRNAs) are a type of non-coding RNA with a length of >200 nt.9 LncRNAs are hypothesized to be involved in many important biological processes such as intracellular homeostasis, genomic imprinting, immunity, and development.10 In addition, lncRNAs have a role in various aspects of gene expression regulation through different mechanisms. LncRNAs and microRNAs are usually interconnected and act upon each other to regulate gene expression.11 LncRNAs have recently been reported to play a key role in psoriasis. In 2005, Sonkoly et al12 identified and characterized a new long non-coding RNA, named the psoriasis susceptibility-related RNA gene induced by stress (PRINS), which is increased in uninvolved epidermis from psoriatic patients compared with epidermis from healthy individuals. Szegedi et al showed that PRINS regulates psoriasis susceptibility and the cellular stress response and is involved in the pathogenesis of psoriasis by directly reacting with NPM (nucleolar phosphoprotein) or regulating the expression of the G1P3 protein, leading to altered functions of keratinocytes.13,14 Zhou et al8 identified some key lncRNAs related to the pathogenesis of psoriasis through statistical analysis, but there were no further functional validations such as cell transfection analyses.
Building upon these insights, we conducted comprehensive sequencing of psoriatic and paired tissues to identify a coexpression network involving LINC01176-miR-218-5p-IL36G. Subsequently, we carried out rigorous validation of its biological functions in both cellular and animal models.
Materials and Methods
Collection of Clinical Samples
In this study, we collected tissue samples from six patients who had been pathologically diagnosed with psoriasis and had not received any other drug treatments or reported other skin complaints. All patients were sourced from Shanghai Xuhui Central Hospital, and all provided written informed consent. Ethical approval for all experimental procedures was obtained from the Ethics Committee of Shanghai Xuhui Central Hospital, Fudan University Zhongshan Xuhui Hospital (Approval document No. 2021–103). Clinical psoriasis tissue samples obtained before any treatments, after treatment with an IL-17A inhibitor, and adjacent healthy tissue samples (normal skin tissue located 5 cm away from the psoriatic tissue, referred to as the NC cohort) were collected and properly preserved.
RNA Isolation
The tissues were extracted and stored at −80°C, followed by the addition of TRIzol for thorough grinding and homogenization. The resulting mixture was centrifuged to isolate the supernatant containing RNA. To remove impurities, the RNA was washed with isopropyl alcohol and 75% ethanol, and then centrifuged again to precipitate the RNA. Subsequently, an appropriate quantity of DEPC water was added to fully dissolve the precipitate, completing the RNA sample preparation process.
LncRNA/mRNA and miRNA Library Preparation and Sequencing
The RNA samples underwent purification using the Ribo-Zero Golden kit (Epicentre, Illumina, USA), while the TruSeq total RNA sample preparation kit (Illumina, USA) was applied for the sequencing of both long non-coding RNA (lncRNA) and messenger RNA (mRNA). Following library preparation, the quality of the samples was validated using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, USA).
For microRNA (miRNA) analysis, the TruSeq Small RNA Library Preparation Kit (Illumina, USA) was employed to prepare the library. Subsequently, all libraries were diluted to 2nM and pooled at a 1:1 ratio for sequencing using the Novaseq 6000 platform (Illumina, USA). The sequencing was performed in a paired-end mode with a read length of 150bp.
Reverse Transcription-Quantitative (RT-q) PCR
Quantitative Polymerase Chain Reaction (qPCR) was conducted for IL36-G, LINC01176, and miR-218-5p following the instructions provided in the respective kits, after the reverse transcription of RNA. Data analysis was performed using the 2-ΔΔCq calculation method.15 The primers were designed as follows, for IL36-G Foward Primer 5’-AGGAAGGGCCGTCTATCAATC, Reverse Primer 5’-CACTGTCACTTCGTGGAACTG, for LINC01176, Forward Primer, 5’-CGGATAACAAGGAGCTAAAGG, Reverse Primer 5’-TCCACTTGAATCCTCTAGTCC, for miR-218-5p, Reverse Transcription Primer 5’-AGCCTTGGCTCAAGCGTGGCTATCTGCCTGCGGTACG, Forward Primer 5’-CGTACCGCAGGCAGATAGCCACCCTTGAGCCAAGGCT, universal-Reverse Primer 5’-TGTCGTGGAGTCGGCAATTC.
Coexpression Network and Pathway Analysis
The data from the NC cohort, pretreatment, and IL-17A inhibitor-treated skin samples underwent several preprocessing steps. These included filtering through trimmomatic to remove adapters and low-quality reads, Hisat2 alignments against the reference genome (GRCh38), and quantification of fragments using FeatureCounts with the GTF file (GRCh38.99) to obtain an expression matrix. Subsequently, singular value decomposition was performed using R’s “prompt” function for clustering analysis of differentially expressed lncRNAs and mRNAs based on count values. Differential gene analysis was carried out using “edgeR” with a false discovery rate (FDR) threshold of <0.05. For miRNA prediction, the ENCORI database was utilized to generate two tables: one for miRNAs predicted by differentially expressed lncRNAs and another for miRNAs predicted by differentially expressed mRNAs. The miRNAs predicted from both tables were intersected to construct the lncRNA-miRNA-mRNA network.A ceNetwork focusing on key miRNA genes related to the NLRP family and IL36 family was constructed. Finally, functional analysis was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology Biological Process (GOBP) to explore related biological functions.
Dual-Luciferase Report Assay
HaCaT cells were cultured and seeded into 24-well culture plates at a density of 1*105 cells per well, the cells were then incubated overnight in a cell culture incubator. For transfection experiments, Either the wild-type (WT) plasmid (containing the wild-type LINC01176 or IL36G sequence along with a luciferase reporter gene) or the mutant (MUT) plasmid (containing the mutant LINC01176 or IL36G sequence along with a luciferase reporter gene) was added to each experimental group. Additionally, 20 pmol of miR-218-5p mimics or mimics control were co-transfected into the cells, following the instructions provided with the transfection reagent. After 48 hours of transfection, the cells were harvested, and luciferase activity was measured using the Luciferase Reporter Gene Assay Kit.
LINC01176 Targets miR-218-5p to Regulate IL-36G Expression-qPCR Verification
HaCaT cells were cultured and seeded into 6-well culture plates at a density of 3*105 cells per well. For transfection, following the instructions provided with the transfection reagent, 4 µg of the LINC01176 over-expression plasmid or an empty plasmid, along with 80 pmol of mir-218-5p mimics or mimics control, were added to each experimental group. The cells were then incubated in a cell incubator for 6 hours. After the incubation period, the medium was replaced with fresh complete medium, and the cells were returned to the incubator for an additional 48 hours. To verify the effects of the transfection, qPCR was performed to assess the expression levels of miR-218-5p and IL-36G.
CCK8 Proliferation Assay
The transfected cells were seeded in a 96-well cell culture plate at a density of 1*104 cells per well. To assess changes in the proliferation capacity of the cells over time, measurements were taken at 0 hours, 24 hours, 48 hours, and 72 hours after the medium replacement using the CCK-8 assay method. Specifically, the old medium was discarded, and 95 µL of fresh medium was added to each well along with 5 µL of CCK-8 solution. The plate was then incubated at 37°C for 1–3 hours.Following incubation, the optical density (OD) of each well was measured at 450 nm using iMark(Bio-Rad).
Apoptosis Assay
The transfected cells were seeded at a density of 1*105 cells per well in a 24-well culture plate and incubated in a cell culture incubator for 48 hours.Following trypsinization, the cells were washed twice with PBS and once with binding buffer.To prepare the cells for flow cytometry analysis, the cell pellet was resuspended in binding buffer, and the cell concentration was adjusted to 1*106 cells/mL. Subsequently, 5μL of annexin V-FITC and 5 µL of propidium iodide were added to 100 µL of the cell suspension and mixed thoroughly. The cell suspension was then shielded from light and allowed to incubate at room temperature for 15–20 minutes.After the incubation period, binding buffer was added to the cell suspension to bring the total volume to 500 µL, and the mixture was thoroughly mixed. Finally, the cell suspension was analyzed using a flow cytometer.
shRNA Liposome Emulsifier Formulation
The shRNA-LINC01176 plasmid was diluted with phosphate buffer (PBS) to create a solution with a concentration of 50 µg/µL of shRNA, then six microliters of the shRNA solution (equivalent to 300 µg) were mixed with 100 µL of Vivo-jetPEI® reagent for solution A. Additionally, Solution B was prepared by diluting 50 µL of Vivo-jetPEI® buffer with 50 µL of sterile water. Solution B was gently added to Solution A, followed by gentle vortexing. The resulting mixture was incubated for 15 minutes at room temperature, resulting in the preparation of transfection mix referred to as the LINC01176- shRNA liposome emulsifier mix.
Psoriasis Animal Model
Eighteen C57 male mice (4 weeks old, SPF) were randomly selected and divided into 3 groups of 6 each (divided into the control group, psoriasis model group, and LINC01176-shRNA treatment group). A 2 cm*3 cm area on the back of the mice was selected for hair removal, and the corresponding treatment drugs were administered separately. The control group (NC) received a uniform application of petroleum jelly. The psoriasis model group (OC) received an application of 5% imiquimod cream. The LINC01176-shRNA treatment group first received 5% imiquimod cream followed by LINC01176-shRNA liposome emulsifier mixture 2 hours later. The mice were kept separately to avoid skin inflammation caused by fighting during mixing male mice. At the end of ten consecutive days of treatment, the mice were anaesthetized with ether and euthanized. Specimens of mouse skin lesions and normal skin tissue were taken for HE staining and immunohistochemistry.
Immunohistochemical Staining
The skin samples from the mice were embedded and sectioned for histological staining using hematoxylin and eosin (HE). The expression of IL-36G proteins (PA5-81,417, Thermo Fisher Scientific) was detected by immunohistochemistry. Images of the stained sections were captured and analyzed through scanning for further examination and analysis.
Results
RNA Sequencing Revealed the hsa_LINC_01176-miR-218-5p-IL-36G Axis
RNA sequencing analysis was performed on each group of skin lesions. RNAseq deconvolution can decode microenvironments. A heatmap of the relative expression of immune cells is shown in Figure 1a. The results of principal component analysis (PCA) showed significant differences between groups, with the greatest distance between the normal and psoriasis groups, while the treatment group was in between the two groups, indicating good sample reproducibility and a significant treatment effect (Figure 1b). The HCA clustering results also showed significant differences between normal and psoriasis group and good sample reproducibility (Figure 1c).
The results of the trend gene heatmap showed that IL-36G and LINC01176 were significantly increased in the psoriasis group compared with the normal group, and the gene expression of the treatment group decreased to a similar level as in the normal group. miR-218-5p was significantly decreased in the psoriasis group, and the gene expression of the treatment group increased to a similar level as that in the normal group (Figure 1d). qPCR showed that both IL-36G and LINC01176 were significantly increased in the psoriasis group compared to the normal group (P< 0.001, P<0.001) and that the expression levels of the genes IL-36G and LINC01176 were significantly decreased in the treated group compared to the psoriasis group (P<0.01, P<0.05), as shown in Figure 2a and b. miR-218-5p expression was significantly lower in the psoriasis group than in the normal group (p<0.01) and increased to a similar level in the treatment group (p<0.05), as shown in Figure 2c. The trend of gene expression levels was consistent with the sequencing results.
GOBP and KEGG analyses of the key lncRNAs revealed that IL-36G was involved in the HTLV-1 infection pathway, proteoglycans in the cancer pathway, thermogenesis pathway, cytokine-cytokine receptor interaction pathway, cornification pathway, inflammatory response to antigenic stimulus pathway, and positive regulation of the JNK cascade pathway (Figure 2d and e). Comparing the differentially expressed lncRNA and mRNA predicted miRNA tables and taking the intersection to perform a ceNetwork revealed that they were linked by miRNA (miR-218-5p) (Figure 3).
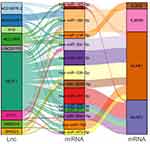 |
Figure 3 The network of LINCRNA-miRNA regulating IL-36G. |
LINC01176 Targets miR-218-5p and Affects Downstream IL-36G in Hacat Cell Line Model
A search of the lncTarD and starBase databases revealed complementary sequences between the 3’UTR of miR-218-5p and LINC01176 or IL-36G (Figure 4a and b). In the dual-luciferase reporter gene assay, LINC01176 or IL-36G (Wild or MUT) plasmids were transfected into HaCat cells, followed by transfection of miR-218-5p mimics or mimics control. The results showed that miR-218-5p mimics significantly inhibited the luciferase activity of the LINC01176-WT reporter molecule compared to the normal group (P<0.01) (Figure 4c), and miR-218-5p mimics significantly inhibited the luciferase activity of the IL-36G-WT reporter molecule (P<0.01) (Figure 4d). The qPCR results in Figure 4e and f show that the LINC01176 vector group significantly inhibited the expression of miR-218-5p (P<0.05) and significantly increased the expression of IL-36G (P<0.001) compared to the NC group. The LINC01176 vector+miR-218-5p mimics group significantly promoted the expression of miR-218-5p (P<0.01) and significantly inhibited the expression of IL-36G (P<0.001) compared to the LINC01176 vector group. The above results demonstrate that miR-218-5p is a downstream target of LINC01176 and that IL-36G is negatively regulated by miR-218-5p. That is, LINC01176 positively regulates IL-36G expression by inhibiting miR-218-5p.
LINC01176 and miR-218-5p Had Opposite Effects on HaCaT Cells
The CCK-8 assay (Figure 5a) showed that LINC01176 significantly promoted the proliferation of HaCaT cells (48 h, P<0.05; 72 h; P<0.001). However, miR-218-5p mimics inhibited cell proliferation (48 h, P<0.01; 72 h, P<0.01). Similarly, flow cytometry showed that LINC01176 significantly inhibited apoptosis in HaCaT cells (P<0.01), and miR-218-5p mimics significantly promoted apoptosis (P<0.01) (Figure 5b–e).
The Intervention of LINC01176 in a Mouse Model of Psoriasis
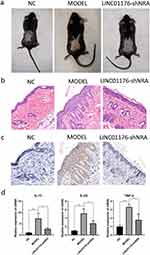
We successfully established the psoriasis mouse model group, and the degree of skin damage in the model group was increasingly serious. Ten days after treatment, erythema, thin scales, skin wrinkles, thickening, and scaly detachment appeared. Compared with the OC group, the LINC01176 shRNA treatment group had lighter and thinner skin erythema and fewer silvery-white scales (Figure 6a).
The results of HE staining showed that compared with the NC group, the epithelial layer of the OC group was thickened, the keratosis of the stratum corneum was incomplete, and the infiltration of dermatitis cells was obvious, which was similar to the histological manifestations of psoriasis. Compared with the OC group, the Stratum Spinosum of the LINC01176-shRNA treatment group became thinner, and the epidermis gradually returned to normal (Figure 6b). Compared with NC group, the expression of IL-36G in the OC group increased significantly; Compared with OC group, the expression of IL-36G in the LINC01176-shRNA treatment group was significantly decreased, similar to that of the NC group (Figure 6c). The qPCR results showed that the gene expression levels of inflammatory factors IL-17 (P<0.01), IL-23 (P<0.01), and TNF-α (P<0.001) were significantly increased in the OC group relative to the NC group; Comparing with the OC group, the gene expression levels of inflammatory factor IL-17 (P<0.05), IL-23 (P<0.05), and TNF-α (P<0.05) had significantly decreased gene expression levels in the LINC01176-shRNA treatment group (Figure 6d). This finding indicates that LINC01176 and IL-36G are positively correlated with the severity of psoriasis.
Discussion
Psoriasis is a chronic, recurrent, and immune-mediated inflammatory disease that affects 2–3% of the world population.16 The roles of various cytokines in psoriasis have been extensively studied over the last few decades.17 Therefore, cytokine-targeted drugs have been developed, including adalimumab (which targets tumour necrosis factor-alpha), secukinumab (which targets IL-17A), and ustekinumab (which targets IL-12 and IL-23 P40).18 However, the role of lncRNAs in psoriasis remains largely unclear.
lncRNAs are a class of genes that do not encode proteins and play a key role in cell metabolism, senescence, and apoptosis.19 lncRNAs have important roles in immune and inflammatory pathways by regulating gene expression through multiple mechanisms.20 Some lncRNAs (eg, MEG3, AL162231.4, and NONHSAT044111) were down-regulated during psoriasis pathogenesis, whereas other lncRNAs (eg, PRINS, MIR31HG, RP6-65G23.1, MSX2P1, SLC6A14-1:1, and NR_003062) were up-regulated.21 According to ceRNA theory, lncRNAs act as “sponges” for miRNAs, reducing the effect of miRNAs on their target genes and thus promoting their expression.22 For example, lncRNA-MSX2P1 activates S100A71 and facilitates the growth of IL-22-stimulated keratinocytes by inhibiting miR-6731-5p,23 and lncRNA H19 regulates the differentiation of keratinocytes by increasing desmoglein 1 expression through sponging miR-130b-3p.24 YW et al showed that LncRNA XIST was highly expressed in the serum of patients with psoriasis, and was positively correlated with disease severity and inflammation, XIST may regulate keratinocyte proliferation and inflammation via regulating miR-338-5p/IL-6 axis.25
In the present study, psoriasis-associated lncRNA-miRNA-mRNA networks were constructed based on ceRNA theory, and the DEGs were also subjected to GOBP enrichment analysis and KEGG pathway enrichment analysis. We have identified a molecular mechanism for the action of LINC01176. Dual-luciferase reporter gene analysis revealed that LINC01176 could target miR-218-5p and regulate the expression of IL-36G. GOBP analysis of the key lncRNAs revealed that IL-36G was involved in the cornification pathway, inflammatory response to antigenic stimulus pathway, and positive regulation of the JNK cascade pathway. These findings align with prior research. Upregulation of miR-218-5p inhibits the invasion and migration of PCa cells. The expression of miR-218-5p was decreased in bone metastatic prostate cancer tissues and serum samples, which was positively correlated with poor clinicopathological features and bone metastasis-free survival of prostate cancer patients.26 The level of miR-218-5p in GBC was significantly lower than that in adjacent noncancerous tissues, which was also related to the prognosis of patients.27 Serum levels of IL-36G are closely linked to psoriasis disease activity (as measured by PASI) and decrease under treatment with anti-TNFα drugs. IL-36G has a highly specific positive predictive diagnostic value for psoriasis.28
However, this study has some limitations to consider. First, a single biological signaling molecule may not provide a complete cure. Additionally, the prognosis and staging of LINC01176, miR-218-5, and IL-36G in psoriasis were not investigated, necessitating further research.
Conclusion
In conclusion, this study proposes a new and important psoriasis-related Cerna network based on ceRNA theory. These findings suggest that LINC01176 plays a key role in the development of psoriasis. LINC01176 can act as a ceRNA for miR-218-5p and promote keratinocyte proliferation and invasion and inhibit apoptosis by increasing the expression of IL-36G. The present study provides insight into the molecular mechanisms of psoriasis, and this may help to identify novel therapeutic targets for the treatment of psoriasis in the future.
Ethical Approval
The present study was approved by the Ethics Committee of The Shanghai Xuhui Central Hospital, Zhongshan-Xuhui Hospital, Fudan University (approval number 2021-103). All enrolled patients signed an informed consent form before surgery. This study complies with the Declaration of Helsinki.
Funding
This work was supported by the Major Project of Shanghai Xuhui District Medical Research Fund (grant number SHXH202002), the General Project of Shanghai Science and Technology Commission (grant number 20ZR1432100), Medical Education Collaborative Innovation Foundation of Jiangsu University (grant number JDYY2023093), the Jiangsu University Medical Education Collaborative Innovation Fund (grant number JDYY2023093).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Langley RG, Krueger GG, Griffiths C. Psoriasis: epidemiology, clinical features, and quality of life. Ann Rheumatic Dis. 2005;64(suppl 2):ii18–23. doi:10.1136/ard.2004.033217
2. Kamiya K, Kishimoto M, Sugai J, Komine M, Ohtsuki M. Risk factors for the development of psoriasis. Int J Mol Sci. 2019;20(18):4347. doi:10.3390/ijms20184347
3. Armstrong AW, Mehta MD, Schupp CW, Gondo GC, Bell SJ, Griffiths CEM. Psoriasis prevalence in adults in the United States. JAMA dermatol. 2021;157(8):940–946. doi:10.1001/jamadermatol.2021.2007
4. Krueger G, Koo J, Fau - Lebwohl M, et al. The impact of psoriasis on quality of life: results of a 1998 national psoriasis foundation patient-membership survey. Arch Dermatol. 2001;137(3):280–284.
5. Kurd SK, Robert PD, Daihung DO, Gelfand JM. The risk of depression, anxiety, and suicidality in patients with psoriasis: a population-based cohort study. Arch Dermatol. 2010;146(8):891–895. doi:10.1001/archdermatol.2010.186
6. Mrowietz U, Kragballe K, Reich K, et al. Definition of treatment goals for moderate to severe psoriasis: a European consensus. Arch Dermatol Res. 2011;303:1. doi:10.1007/s00403-010-1080-1
7. Blauvelt A, Chiricozzi A. The immunologic role of il-17 in psoriasis and psoriatic arthritis pathogenesis. Clin Rev Aller Immunol. 2018;55:379–390. doi:10.1007/s12016-018-8702-3
8. Zhou Q, Yu Q, Gong Y, et al. Construction of a lncrna-miRNA-mRNA network to determine the regulatory roles of lncrnas in psoriasis. Exp Ther Med. 2019;18(5):4011–4021. doi:10.3892/etm.2019.8035
9. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
10. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi:10.1016/j.cell.2013.02.012
11. Antonini D, Mollo MR, Missero C. Research techniques made simple: identification and characterization of long noncoding RNA in dermatological research. J Invest Dermatol. 2017;137(3):e21–6. doi:10.1016/j.jid.2017.01.006
12. Sonkoly E, Bata-Csorgo Z, Pivarcsi A, et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, prins. J Biol Chem. 2005;280(25):24159–24167. doi:10.1074/jbc.M501704200
13. Szegedi K, Göblös A, Bacsa S. Expression and functional studies on the noncoding RNA, prins. Int J Mol Sci. 2013;14(1):205–225. doi:10.3390/ijms14010205
14. Szegedi K, Sonkoly E, Nagy N. The anti-apoptotic protein g1p3 is overexpressed in psoriasis and regulated by the non-coding RNA, prins. Exper Dermatol. 2010;19(3):269–278. doi:10.1111/j.1600-0625.2010.01066.x
15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
16. Zhang PA-O, Wu MX. A clinical review of phototherapy for psoriasis. Lasers Med Sci. 2018;33:173–180. doi:10.1007/s10103-017-2360-1
17. de Alcantara CC, Reiche EMV, Simão ANC. Cytokines in psoriasis. Advan Clin Chem. 2021;100:171–204.
18. Nograles KE, Krueger JG. Anti-cytokine therapies for psoriasis. Exp Cell Res. 2011;317(9):1293–1300. doi:10.1016/j.yexcr.2011.01.024
19. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011
20. Wu GC, Pan HF, Leng RX, et al. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmunity Rev. 2015;14(9):798–805. doi:10.1016/j.autrev.2015.05.004
21. Ghafouri-Fard S, Eghtedarian R, Taheri M, Rakhshan A. The eminent roles of ncRNAs in the pathogenesis of psoriasis. Non-Coding RNA Res. 2020;5(3):99–108. doi:10.1016/j.ncrna.2020.06.002
22. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of cerna crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
23. Qiao M, Li R, Zhao X, Yan J, Sun Q. Up-regulated lncrna-msx2p1 promotes the growth of il-22-stimulated keratinocytes by inhibiting mir-6731-5p and activating s100a7. Exp Cell Res. 2018;363(2):243–254. doi:10.1016/j.yexcr.2018.01.014
24. Li CX, Li HG, Huang LT, et al. H19 lncrna regulates keratinocyte differentiation by targeting mir-130b-3p. Cell Death Dis. 2017;8(11):e3174–e3174. doi:10.1038/cddis.2017.516
25. Wang Y, Jiang F, Chen F, Zhang D, Wang J. LncRNA XIST Engages in psoriasis via sponging miR-338-5p to regulate keratinocyte proliferation and inflammation. Skin Pharmacol Physiol. 2022;35(4):196–205. doi:10.1159/000523781
26. Peng P, Chen T, Wang Q, et al. Decreased mir-218-5p levels as a serum biomarker in bone metastasis of prostate cancer. Oncol Res Treat. 2019;42(4):165–181. doi:10.1159/000495473
27. Wang H, Zhan M, Xu SW, et al. Mir-218-5p restores sensitivity to gemcitabine through prkce/mdr1 axis in gallbladder cancer. Cell Death Dis. 2017;8(5):e2770. doi:10.1038/cddis.2017.178
28. D’ErmeAM, Wilsmann-TheisD, WagenpfeilJ, et al.Il-36γ (il-1f9) is a biomarker for psoriasis skin lesions. J Invest Dermatol. 2015;135(4):1025–1032. doi:10.1038/jid.2014.532
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.