Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 15
The Impact of Intestinal Microorganisms and Their Metabolites on Type 1 Diabetes Mellitus
Authors Zheng S , Luo Y , Xiao J
Received 24 December 2021
Accepted for publication 24 March 2022
Published 11 April 2022 Volume 2022:15 Pages 1123—1139
DOI https://doi.org/10.2147/DMSO.S355749
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ming-Hui Zou
Shu-Juan Zheng,1 Yi Luo,1,2 Jian-Hui Xiao1,2
1Zunyi Municipal Key Laboratory of Medicinal Biotechnology, Affiliated Hospital of Zunyi Medical University, Zunyi, 563003, People’s Republic of China; 2Guizhou Provincial Research Center for Translational Medicine, Affiliated Hospital of Zunyi Medical University, Zunyi, 563003, People’s Republic of China
Correspondence: Jian-Hui Xiao, Guizhou Provincial Research Center for Translational Medicine, Affiliated Hospital of Zunyi Medical University, 149 Dalian Road, HuiChuan District, Zunyi, 563003, People’s Republic of China, Email [email protected]
Background: Type 1 diabetes mellitus (T1DM) is an autoimmune disease with a complex etiology comprising numerous genetic and environmental factors; however, many of the mechanisms underlying disease development remain unclear. Nevertheless, a critical role has recently been assigned to intestinal microorganisms in T1DM disease pathogenesis. In particular, a decrease in intestinal microbial diversity, increase in intestinal permeability, and the translocation of intestinal bacteria to the pancreas have been reported in patients and animal models with T1DM. Moreover, intestinal microbial metabolites differ between healthy individuals and patients with T1DM. Specifically, short-chain fatty acid (SCFA) production, which contributes to intestinal barrier integrity and immune response regulation, is significantly reduced in patients with T1DM. Considering this correlation between intestinal microorganisms and T1DM, many studies have investigated the potential of intestinal microbiota in preventive and therapeutic strategies for T1DM.
Objective: The aim of this review is to provide further support for the notion that intestinal microbiota contributes to the regulation of T1DM occurrence and development. In particular, this article reviews the involvement of the intestinal microbiota and the associated metabolites in T1DM pathogenesis, as well as recent studies on the involvement of the intestinal microbiota in T1DM prevention and treatment.
Conclusion: Intestinal microbes and their metabolites contribute to T1DM occurrence and development and may become a potential target for novel therapeutics.
Keywords: type 1 diabetes mellitus, intestinal microorganisms, intestinal microbiota, dysbiosis, metabolites, prevention and treatment
Background
Type 1 diabetes mellitus (T1DM) is an autoimmune disease caused by islet atopic T cells that damage islet β cells, resulting in insufficient insulin secretion. According to recent estimates by the International Diabetes Federation, as of 2019, 463 million people, including 1.1 million children and adolescents under the age of 20 years, worldwide have been diagnosed with diabetes. Meanwhile, T1DM often has acute (diabetic ketoacidosis) or short-term (diabetic eye, kidney, and other diseases) complications caused by extreme blood sugar levels, which can lead to permanent disease and even death.
Environmental factors synergize with genetic susceptibility to promote TIDM development. In particular, environmental factors, including intestinal microorganisms, have central roles in T1DM pathogenesis.1 Typically, in healthy individuals, intestinal microorganisms are in a state of dynamic equilibrium; however, when this balance is disrupted, various chronic diseases can develop. In the context of T1DM, unique microbiome structures with poor stability and low diversity have been defined for patients with T1DM compared to healthy individuals.2,3 Meanwhile, patients with T1DM who have undergone treatment develop microbiota that resembles that of healthy individuals, thus supporting a role for the intestinal microbiota in T1DM development and/or pathogenesis. Accordingly, it has been postulated that intestinal microbiota may serve as a target for the prevention and treatment of T1DM. That is, restricting the growth of disease-associated microorganisms, or enhancing, within an effective range, the growth of microorganisms that favorably impact disease outcome, may prove to be successful strategies for treating or preventing T1DM. Current methods for disease prevention or treatment that influence gut microbiota include dietary fiber supplementation, probiotic intake, and breastfeeding, further proving that the intestinal microbiota is involved in the regulation of the disease.
The primary aim of the current article is to further investigate and collate the current literature related to the regulatory role of the intestinal microbiome in T1DM occurrence and development, while also providing new insights into the involvement of intestinal microbiota, and their metabolites, in T1DM pathogenesis.
Intestinal Microorganisms in T1DM
The intestinal tract, which contains a myriad of microbes, is the largest immune organ in the human body.4 The intestinal microbiota of healthy individuals primarily comprises Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, Deferribacteres, Tenericutes, and Verrucomicrobia.5 However, with age, as well as following changes in diet and environment, the composition of the intestinal microbiota becomes altered. Newborns are exposed to vaginal or epithelial microflora during natural delivery or cesarean section, respectively.6 This initial exposure influences subsequent microbial colonization of the infant’s intestinal tract, which will stimulate the development, and maturation, of the immune system.
Considering that T1DM is an autoimmune disease, and that the intestinal microbiome may significantly impact T1DM pathogenesis, studies have been conducted in various countries and regions to analyze the intestinal microflora of patients with T1DM. The findings of these studies reveal that in countries with a high incidence of autoimmune diseases, Bacteroides spp. are abundant in children with autoimmune diseases, while the lipopolysaccharides (LPS) secreted by Bacteroides are capable of inhibiting innate immune signals and endotoxin tolerance,7 leading to abnormal immune function. Moreover, in the T1DM model, intestinal microbial composition differs under distinct genetic backgrounds and ages.8–10 Meanwhile, in individuals with genetic susceptibility to T1DM, the metabolic pathways and predominant microbial strains within the intestinal microbiota are stable throughout infancy, whereas microbial diversity decreases following serum transformation, with an apparent increase in inflammatory tissues and activation of inflammatory pathways prior to T1DM development.11 In addition, significant differences have been reported in intestinal microbiota structure between patients with T1DM and healthy individuals. In particular, microorganism diversity and stability are low in patients with T1DM.2,3 For example, pregnant women with T1DM exhibited a relative shift toward a proinflammatory gut microbiota, with increased LPS-producing bacteria and decreased short-chain fatty acids (SCFA)-producing bacteria, resulting in increased levels of intestinal inflammatory markers within their stools and serum markers of intestinal epithelial damage.12
The intestinal barrier can prevent microorganisms from overstimulating the immune system and release bacteriostatic substances, such as antimicrobial peptides.13 Intriguingly, intestinal microbiota and its metabolites can also promote the expression of antimicrobial peptides.14 For instance, Liang et al reported that low cathelicidin-associated antimicrobial peptide (CRAMP) expression in the colon of neonatal non-obese diabetes mellitus (NOD, a spontaneous T1DM model) mice resulted in intestinal microbiota disorder and an increase in the production of intestinal type I interferon (I–IFN), which can promote the development of diabetes in adult mice.15 Meanwhile, supplementation with CRAMP caused reconstruction of the gut microbiota, thereby preventing development of diabetes in NOD mice.15 Hence, antimicrobial peptides have a critical role in maintaining intestinal homeostasis, raising the question of whether the concentration of antimicrobial peptides decreases in early stages of T1DM and, if so, do they offer novel insights regarding the pathogenesis, prevention, and treatment of T1DM? Indeed, antimicrobial peptides are involved in the maintenance and regulation of intestinal microbiota homeostasis.16 From a developmental perspective, antimicrobial peptides can be viewed as a defense system that maintains the balance between intestinal homeostasis and inflammation by generating appropriate antimicrobial responses, while intestinal microbial disorder disrupts the balanced interaction between gut microbiota and host antimicrobial peptides.
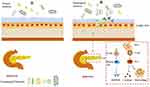
In addition, CD8+ T cells become activated and proliferate in NOD mice following the transfer of intestinal barrier-destroying bacteria, resulting in an increased incidence of diabetes.17 Thus, following intestinal barrier injury, intestinal bacteria interact with host immune components to activate immune cells, which then promote T1DM development. Indeed, impaired intestinal barrier function and increased permeability also occur before the onset of T1DM.18,19 Specifically, the intestinal permeability of germ-free (GF) NOD mice is increased following fecal transplantation from patients with T1DM,20 suggesting that alterations in the intestinal microbiota may cause intestinal barrier damage. However, the intestinal microbiota of patients appears to change following emergence of T1DM autoantibodies.21 That is, production of autoantibodies induces changes within the microbiota, which lead to impaired intestinal permeability. These events may facilitate contact between microorganisms and intestinal epithelial cells or bacterial translocation. The former triggers the intestinal mucosal immune response, while the latter may result in translocation of the microbes to other organs, including the pancreas, while also stimulating the immune response and cause pancreatic damage (Figure 1). Of note, bacterial translocation to the pancreas of diabetic mice is diminished following improved intestinal permeability.22 Moreover, translocated intestinal bacteria within the pancreatic lymph nodes (PLN) of diabetic mice, activate NOD2 receptors within the PLN, which in turn promote the production of proinflammatory Th1 and Th17 cells.23 Indeed, the initiation site of activation of diabetes-specific cells may be within the intestinal tract.24
Intestinal microbiota, as the main stimulator of the immune system, is involved in many diseases. However, the causal relationship between changes in gut microbes and disease occurrence, including of T1DM, as well as how intestinal microbes participate in the disease process, remain to be fully elucidated. In particular, the relationship between T1DM and intestinal microbiota continues to be investigated in the context of autoimmune diseases; however, it requires further investigation.
Metabolites of Intestinal Microorganisms in T1DM
Intestinal microorganisms can degrade carbohydrates to produce metabolites, such as SCFAs (including acetic acid, propionic acid, and butyric acid), secondary bile acids, and tryptophan derivatives.13 Recent studies have shown a potential causal association between gut microbes and blood metabolites. For instance, blood glutamate can reduce the abundance of Oxalobacter, while blood triglyceride concentration is affected by Oscillibacter and Alistipes, and members of Proteobacteria are affected by glutamate, alanine, 5-methyltetrahydrofolate, and selenium.25 Moreover, metabolites can provide an additional source of energy for intestinal cells and participate in host metabolism.26 For example, SCFAs can be absorbed by intestinal epithelial cells and, ultimately, enter the blood circulation, consequently affecting sugar storage of muscle, liver, and fat cells. Meanwhile, acetic acid can also reach the brain, and reduce appetite and food intake,27 thus, retarding the disease. SCFAs can also protect the integrity of the intestinal barrier by activating G-protein-coupled receptors, inhibiting histone deacetylase expression, increasing intestinal tight junction mRNA and protein expression, and reducing intestinal permeability. Similarly, bacterial tryptophan metabolites can regulate intestinal barrier function through aromatic hydrocarbon receptors.28
Although T1DM is an autoimmune disease, patients can also exhibit abnormal plasma metabolite levels. In fact, children with T1DM aged 6–18 years exhibit decreased fecal SCFA concentration compared to that of the control.20 Moreover, in a longitudinal study, early development of T1DM in infants (PT1D) was associated with decrease in the contents of sugar derivatives, amino acids, and fatty acids (including SCFAs) compared with those of controls (CTRs). Additionally, methionine lever is upregulated in PT1D, while islet autoantibody production is associated with decreased glutamate and aspartate levels in patients with only one serum autoantibody (P1Ab) compared with those of CTR.29 Another study examined PT1D, P1Ab, and CTRs and found abnormalities in plasma metabolites. Specifically, compared with that of the P1Ab and CTR groups, sphingomyelin lever was continuously downregulated in the PT1D group. Triacylglycerol and phosphatidylcholine levers were primarily downregulated in PT1D compared with those of P1Ab after three months.30 Moreover, plasma concentrations of free fatty acids differed between the healthy group and T1DM group,31 indicating that T1DM is accompanied by changes in plasma and feces metabolites. Alterations in metabolites are also associated with the development of T1DM-related complications, including higher baseline fumarate levels and lower amino acid levels (asparagine and glutamine) in patients with diabetic cardiovascular autonomic neuropathy,32 Moreover, plasma metabolites concentrations correlate with albuminuria lever in patients with T1DM in the early stages of diabetic nephropathy.33
Changes in metabolite levels have also been reported in various animal models of diabetes. A non-targeted longitudinal metabolomics study was carried out in female and male NOD mice and found different metabolic characteristics at different time points. That is, upon development of diabetes, mice had metabolite levels that differed significantly from those at other time points, among which the most influential metabolic pathways included lipid, branched-chain amino acid, carbohydrate, and oxidation pathways, accompanied by several biochemical alterations.34 Additionally, the key characteristics of T1DM were negatively correlated with the microbial metabolites, acetate and butyrate, in feces and blood,35 indicating that an increase in butyrate or acetate content may inhibit the occurrence, or development, of T1DM. This notion is further supported by a study reporting that butyrate synthase production is significantly reduced in T1DM animals,36 suggesting that supplementation with butyric acid, or other metabolites, at optimal time points, may delay T1DM occurrence.
With the advances in science and technology, new metabolites are being discovered, including microbial-derived SCFAs and tryptophan derivatives, which provide new testing methods for the treatments and prognoses of diseases. Furthermore, the difference in metabolite levels between healthy individuals and T1DM patients may offer insights into metabolic characteristics unique to T1DM that could serve as effective predictors of disease.
Intestinal Microbiota and Its Metabolites Affect the Occurrence and Development of T1DM by Regulating the Immune System
Intestinal microorganisms and their metabolites, as foreign antigens or pathogen-associated molecular patterns, can trigger an immune response and be recognized by pattern recognition receptors. Subsequently, via MYD88, or related signaling molecules, signal transduction is activated, resulting in recruitment of effector cells, and activation of APCs to produce various immune-related molecules.37 Thus, microorganisms can regulate the occurrence of autoimmune diabetes and contribute to immune regulation. Accordingly, studies have examined whether microbiota affects the occurrence of diseases by interacting with the immune system. Intestinal microorganisms reportedly accelerate the development of the disease by stimulating diabetic immunity cells. Specifically, the transporter polypeptide of Fusobacterium has strong homology with IGRP (autoantigen targeted by diabetic CD8+ T cells), which can be processed and presented by gut-associated APCs, stimulate IGRP-reactive CD8+ T cells, and accelerate diabetes onset with a strong correlation noted between the abundance of Fusobacterium and progression to diabetes.38 Additionally, the TCR-TG mice, in which most of the CD4+ T cells express TCR and pancreatic islet cells exhibit significant T cell infiltration, do not develop diabetes under normal circumstances. However, when dextran sulfate sodium (DSS) is used to induce colitis in the mice, the intestinal barrier integrity becomes compromised, resulting in activation and transfer of islet-reactive T cells from the intestinal mucosa to the pancreas, leading to diabetes. However, this phenomenon depends on the existence of intestinal microorganisms; otherwise, diabetes does not occur following DSS induction, indicating that the activation of islet reactive T cells requires the presence of the intestinal microbiome following intestinal barrier injury.39 That is, intestinal microorganisms can stimulate the activation and proliferation of immune cells associated with diabetes and participate in the pathogenesis of diabetes; however, it remains unclear which microbes lead to the activation of islet-reactive T cells. Nevertheless, Sun et al recently reported that microbiota-derived SCFA acted on the GPR43 receptor to activate the mTOR and STAT3 pathways in Th1 cells, thereby upregulating the expression of the transcription factor Blimp-1, which promote IL-10 production in Th1 cells and relieve intestinal inflammation.40 Meanwhile, another study showed that IL-10 knockout in TCR-Tg NOD mice increases CD4+ T cells activation and neutrophil infiltration in pancreatic islets to promote diabetes-related symptoms. The effect of IL-10 on neutrophil homeostasis was mediated by altering gut microbiota structure.41 However, the mechanism by which the gut microbiota regulates neutrophil homeostasis and their role in the development of T1DM remains to be elucidated. Collectively, this evidence suggests that the presence of microorganisms may stimulate the activation and proliferation of relevant immune cells, thereby promoting islet inflammation and exacerbating diabetes symptoms.
Interestingly, the incidence of T1DM in GF NOD mice does not differ from that of normal NOD mice. Meanwhile, following fecal transplantation from children with T1DM to GF NOD mice, the incidence of T1DM was lower in the fecal bacteria transplantation group than in the non-transplantation group,42 suggesting that gut microbial can affect the occurrence of T1DM in GF NOD. Meanwhile, the development of diabetes in NOD mice was observed reduced following colonization with specific Bacillus cereus, indicating that the restrictive microbiota may regulate disease development.43 For example, the transfer of a single strain of Akkermansia visiniopha to high incidence NOD mice can retard the onset of diabetes, by promoting mucus production and expression of the antimicrobial peptide Reg3 γ, while reducing the level of serum endotoxins and the expression of islet Toll-like receptor.44 Additionally, antibiotic exposure in early life causes rapid T1DM progression in NOD mice, whereas transplantation of maternal cecal microbiota significantly reduces the incidence of diabetes with concomitant restoration of microbiota, metabolites, and innate and adaptive immune effectors.45 These results indicate that the intestinal microbiota participates in the regulation of diabetes. Thus, regulation of intestinal microbiota may be an effective T1DM treatment strategy.
Intestinal microbiota can produce metabolites through the decomposition of carbohydrates, and these gut metabolites can affect disease progression by interacting with the immune system. Administration of metabolite-based dietary supplements (acetic acid and butyric acid modified high amylose corn resistant starch) to T1DM patients for six weeks results in remodeling of the gut microbiota structure, as well as altered immune profiles, immune tolerance, and improved glycemic control for the treatment of T1DM.46 Meanwhile, administration of a specially designed diet (fermented in the colon to produce large amounts of acetate or butyrate) to NOD mice, in which acetate influences the frequency of autoreactive T cells by interacting with GPR43 receptors, and butyrate modulates the number and function of Tregs by inhibiting HDAC expression or interacting with other butyrate receptors.35 Moreover, combined administration of acetate and butyrate enhances the integrity of the intestinal barrier and reduces the concentration of serum pro-inflammatory cytokines. Importantly, each diet of these diets has demonstrated protection against development of T1DM.35 Butyrate can also ameliorate the immune milieu of islets through enhancing the production of antimicrobial peptides in pancreatic endocrine cells in T1DM. In fact, following administration of diabetic mice with broad-spectrum antibiotics, the production of pancreatic antimicrobial peptides decreased, indicating that the intestinal microflora can regulate the pancreatic immune environment through SCFAs and antimicrobial peptides.47 Further evidence indicates that butyric acid can inhibit the inflammatory response aggravated by vancomycin, as well as the recruitment of IFN-γ+ T cells, by acting on pancreatic T cells and DCs, thus restraining the development of pancreas-guided autoimmunity in female offspring of NOD mice.48 Another study indicated that early administration of butyrate induces an increase in the number of intestinal Tregs that can migrate to the pancreas and PLN to restore immune tolerance during T1DM.49 In addition, long-term acetate supplementation treatment reduced the level of intestinal free IgA, prevented the binding of intestinal bacteria to IgA, inhibit the formation of germinal center B cell, and alleviate islet inflammation in T1DM mice.20
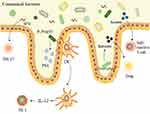
In the early stages of life, colonization by microorganisms is conducive to the development of the immune system, which not only participates in host immune regulation and promotes nutrient absorption but also functions in concert with the intestinal mucosal barrier to resist pathogen invasion. Simultaneously, the host provides an environment for microbiota to survive. Following colonization of the intestinal tract, microorganisms can affect the intestinal mucosal immune response through their own, or thallus components. For example, following colonization of GF mice with Bacteroides fragilis (B. fragilis), its bacterial capsular polysaccharide (PSA) was processed and presented by intestinal DCs, resulting in activation of CD4+ T cells and induction of cytokine production. PSA can also influence immune regulation by correcting T cell deficiency, Th1/Th2 imbalance, and guiding the development of lymphoid organs in GF mice.50 Meanwhile, colonization of GF mice with Clostridium causes the production of butyrate and other SCFAs. Butyrate can then inhibit the activity of HDAC and regulate the transcription factor SP1 to increase TGF-β expression.51 In addition, butyrate can metabolize to butyryl-CoA, which activates carnitine palmitoyltransferase 1A (CPT1A), enhance CPT1A-dependent fatty acid oxidation, and promote iTreg differentiation.52 Thus, providing an environment rich in TGF-β and Treg inducers, such as butyrate, affects the number and function of Foxp3+ Tregs in the colon.53 Segmented filamentous bacteria (SFB) colonization can induce the proliferation of Th17 cells in the lamina propria (Figure 2). That is, in the terminal ileum of mice containing Th17, SFB adheres to the surface of the intestinal epithelium.54 SFB and intestinal epithelial cells communicate by producing vesicles at the synaptic cleft, which enter intestinal epithelial cells and primarily contain SFB cell wall-related proteins, which can promote the proliferation of Th17 cells.55
In addition, metabolites produced by microorganisms exert immunomodulatory effects. Acetic acid has the ability to increase the number of Treg cells in mice, which promotes the differentiation of extrathymic Treg cells, depending on the intron enhancer CNS1. This promotes the production of peripheral spleen Treg cells and encourages dendritic cell (DC) stimulation of extrathymic Treg cell differentiation by inhibiting histone deacetylase activity.56 Butyrate also promotes IL-22 production by inhibiting histone deacetylase (HDAC) expression, activating GPR41 to promote the expression of aryl hydrocarbon receptor (AhR) and hypoxia-inducible factor 1α (HIF1α). Mechanistically, GPR41 promotes binding of HIF1α to the HRE region of the IL-22 promoter, inducing histone acetylation and enhancing IL-22 expression.57 Moreover, binding of secondary bile acid to its receptor TGR5 can reduce the activation of NLRP3 inflammatory bodies, induce CREB-mediated IL-10 production, and inhibit NF-κB signal transduction, while downregulating the production of proinflammatory cytokines by macrophages.13 In wild-type mice, oral administration of SCFAs activates the mTOR and STAT3 pathway in intestinal epithelial cells by interacting with GPR43, inducing the expression of antimicrobial peptides in intestinal epithelial cells.14
Taken together, these results suggest that intestinal microbiota and their metabolites influence the occurrence and development of T1DM by regulating the immune system, and may also serve as potential diagnostic indicators and therapeutic approaches for T1DM.
Intestinal Microbiota Influences T1DM Prevention and Treatment
Two primary treatments are administered for autoimmune diabetes, namely, insulin injection and islet organ transplant. However, both have their own limitations. Owing to the importance of intestinal microbiota in T1DM, many animal experiments and clinical studies have explored whether altering the intestinal microbiota can delay disease progression, or whether corresponding changes occur in intestinal microbial abundance following treatment administration. Hence, in this section, we review relevant literature related to whether intestinal microbiota impacts T1DM progression or treatment.
Dietary Supplements
Dietary interventions, as well as changes in the gut microbiome, and exposure to intestinal pathogens regulate the development of autoimmune diabetes.58,59 Gluten is the initiator of autoimmunity in celiac disease, and the autoimmune process stops when gluten is removed from the diet.60 Dietary interventions mainly refer to the use of dietary supplements such as probiotics, prebiotics, synbiotics, and dried yeast to improve T1DM symptoms and have been used in clinical trials (Table 1). Probiotics are living supplements that can maintain normal intestinal function. Prebiotics are food ingredients that promote the growth or activity of bacteria to improve human health, however, are not easily digested. Synbiotics are a combination of probiotics and prebiotics.61 Among them, probiotics are more widely used. Traditional probiotics, including Lactobacillus and Bifidobacterium, such as Lactobacillus in yogurt, promote digestion and absorption in the gastrointestinal tract and improve intestinal function.62
 |
Table 1 The Effect of Treatments on Intestinal Microbiota in T1DM Clinical Trial |
For people with diabetes, it is necessary to regulate diet. In particular, patients must reduce their intake of foods with high sugar content while increasing consumption of whole grains containing fibers. Indeed, administration of an anti-diabetic diet (infant formula) in young NOD mice retards disease onset.63 As aforementioned, colitis may be involved in T1DM occurrence, with mild colonic inflammation detected in young NOD mice. However, upon administration of an anti-diabetic diet to these mice, for two weeks after birth, the number of inflammatory DCs, as well as IL-17 and IL-23 abundance in the colon diminished, thereby reducing the occurrence of colonic inflammation, restoring colonic immune homeostasis, and decreasing the incidence of T1DM.63 Although these findings provide support for the notion that colitis is associated with T1DM, the associated relationship requires further clarification.
One way to alter gut microbes is to supplement dietary fiber, which can improve the microbiome diversity and benefit gut bacteria growth.64,65 Specifically, low-methoxyl pectin (LMP), a carbohydrate, has anti-inflammatory effects and reduces intestinal permeability.66,67 LMP supplementation also significantly reduces the incidence of diabetes in NOD mice, and this protective effect depends on the effect of LMP on the caecum microflora, by increasing the abundance of Firmicutes and TM7. The ratio of Firmicutes to Bacteroides not only affects carbohydrate metabolism but also increases production of SCFAs,68 compared with that of controls. Thus, LMP supplementation could improve the balance of the caecum environment. It can also regulate the pancreatic immune response by increasing the number of Foxp3+ Treg and decreasing the activation of NLRP3 inflammasome in both the pancreas and PLN of NOD mice.69 However, the precise mechanism by which immune cells in the gut-pancreas axis are induced to migrate from the caecum, or affect the immune environment of the pancreas, remains unclear.
Yeast β-glucan (YBG), a compound polysaccharide, can inhibit islet inflammation in pre-diabetic NOD mice by long-term oral administration and significantly retards the occurrence of T1DM. In animal experiments, immune regulation induced by oral YBG administration inhibits the infiltration of immune cells and destruction of β cells, resulting in delay of hyperglycemia in NOD mice. Meanwhile, YBG also impacts the intestinal microbiota. That is, YBG‐treated mice exhibit a significantly higher abundance of Verrucomicrobia and Bacteroidetes, as well as profoundly lower abundance of Proteobacteria and Firmicutes in fecal samples compared with those of controls. Verrucomicrobia and Bacteroidetes include many bacterial species that degrade polysaccharides. Hence, the protective effect of YBG is partially dependent on the intestinal microbiota, which was further demonstrated following antibiotic treatment of NOD mice.70 However, these findings must be verified in GF mice to ensure that the antibiotic treatment did not eliminate all bacteria.
As of March 5, 2022, 105 ongoing clinical studies have been conducted on T1DM treatment using dietary supplements (ClinicalTrials.gov), 14 of which were conducted using probiotics. Probiotics promote the diversity and stability of the host microbiota and regulate the host immune system.71 In a streptozotocin (STZ)-induced diabetic model, administration of probiotics effectively improved the values of various biochemical indicators of diabetes,72 reversed STZ-induced microbiota imbalance, restored intestinal barrier function, and reduced bacterial translocation and endotoxin levels.73 Moreover, probiotics can prevent T1DM development by regulating the intestinal environment at the microbiota and immune cell levels. For instance, after administration of VSL#3 probiotics to four-week-old NOD mice, the intestinal microbiota composition became altered as the relative abundance of Clostridia increased. These bacterial strains have strong pro-tolerance properties, which induce immune tolerance and reduce inflammation in the intestinal tissues.53,74 Specifically, the expression of IL-1β, an intestinal mucosal inflammatory component, is decreased, while that of indoleamine 2-3-dioxygenase (IDO) is increased. IDO inhibits the differentiation of Th1/Th17 and promotes the differentiation of Tregs, resulting in reduced incidence of diabetes in NOD mice.75
Although dietary supplements have shown clear success in animal studies, further clinical research is warranted. A recent clinical trial examined the effects of administering a dietary supplement based on SCFAs to patients with T1DM for 6 weeks; follow-up continued for 12 weeks. Concentrations of SCFAs in plasma and feces increased and were consistent with changes in microbiota composition and function. Although glycemic control and insulin requirements were not significantly impacted, patients with the highest concentrations of SCFAs exhibited superior glycemic control.46 Moreover, the TEDDY trial reported that early probiotic supplementation reduces the risk of islet autoimmunity in children with the highest genetic predisposition of T1DM.76 Further, in a single-center, double-blind, and randomized placebo-controlled trial, the intervention group was supplemented with probiotics for three months; the intervention group HbA1c and insulin bolus doses decreased significantly compared with those of the placebo group, with no adverse reactions reported.77 Meanwhile, in a 12-week randomized, placebo-controlled trial, the use of prebiotics (oligofructose-enriched insulin) significantly increased serum C-peptide concentrations, and permutational multivariate ANOVA showed that prebiotic treatment had a significant effect on microbial distribution between samples.78 In addition, in a double-blind clinical trial, continuous supplementation of synbiotics (Lactobacillus sporogenes GBI-30 (probiotic), maltodextrin and fructooligosaccharide (prebiotic)) for eight weeks significantly improved the levels for fasting blood glucose, hemoglobin A1c, insulin and other indicators.79
However, in a randomized double-blind placebo-controlled clinical trial, administration of probiotics in infancy did not affect pancreatic cellular autoimmunity nor diabetes development. This indicates that the use of probiotics does not impact β-cell autoimmunity nor the occurrence of T1DM.80 This phenomenon may be due to different patient responses and different probiotics. Moreover, in another double-blind, randomized controlled trial, oral Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12 were administered to children aged 8–17 years with newly diagnosed T1DM for 6 months and the follow-up was conducted for 12 months. However, no difference was observed in the area under the c-peptide level curve between placebo and treatment groups,81 Hence, the use of these two probiotics did not improve pancreatic β-cell function. Similarly, another TEDDY study analyzed the food records of 3358 children aged 9–48 months and found that intake of high soluble dietary fiber did not exert protective effects on T1DM occurrence.82
Collectively, we can preliminarily conclude that dietary supplements are effective treatment approaches in clinical trials. However, more large-scale clinical trials are required to verify their usefulness. In addition, the effects and underlying mechanism of action of dietary supplements on the intestinal microbiota in patients with T1DM also require further investigation.
Antibiotics
Antibiotics are the general class of antimicrobials that target bacteria, as well as certain fungi and parasites; however, owing to misuse of antibiotics, the intestinal microbiota can develop drug resistance.83 Moreover, the use of antibiotics is associated with a variety of diseases, including colorectal cancer.84 A dose-response relationship has even been reported between antibiotics use in infants and the incidence of childhood asthma.85 However, certain antibiotics, when administered responsibly, can have a positive effect on the intestinal microbiota by increasing the abundance of beneficial bacteria. In fact, this strategy has been adopted for the treatment of irritable bowel syndrome and hepatic encephalopathy.86
Although numerous studies have examined the role of antibiotics in T1DM, inconsistent results have been reported. For instance, in 2006, Brugman et al found that antibiotic treatment of bio-breeding diabetes-prone rats significantly delayed the incidence of diabetes, suggesting that antibiotics can destroy certain microbes, thus lowering the antigen load and reducing contact between bacterial and the immune system.59
Although T1DM has a complex etiology, the most significant contributing factor is production of autoantibodies against antigens of pancreatic cells. Under normal conditions, organisms do not produce antibodies that attack self-antigens. However, when certain foreign antigens have the same epitope as those of body cells, antibodies are produced against the foreign antigen, which can also attack self-cells. Alternatively, cells may express the foreign antigen receptors and become infected causing cytopathic effects.87 Indeed, microorganisms, as producers of foreign antigens, function as a contributing factor to T1DM autoimmunity. Supporting this view, a clear association has been reported between enterovirus and pancreatic autoimmunity or T1DM,88 as well as significant homology between microbial antigens and islet autoantigens.38 In the above article, the use of antibiotics reduced the abundance of microbiota, antigen load, and the incidence of T1DM,59 suggesting a correlation between the microflora and T1DM occurrence. Moreover, Kilham rat virus (KRV) can induce diabetes in a genetic susceptibility model, which is related to the innate immunity induced by KRV.89,90 It is also linked to transient changes in the intestinal microbiota as the abundance of Bifidobacterium and Clostridium increased after KRV infection.91 Similarly, the combination of trimethoprim and sulfamethoxazole exerts protective effects on KRV-induced diabetes models, particularly by reducing the abundance of Bifidobacterium and Clostridium and restoring intestinal microflora homeostasis.91
The use of antibiotics also destroys the stability of the intestinal microbiota and causes disease development. Increased incidence of diabetes in NOD mice treated with vancomycin or neomycin. Thus, increased incidence of diabetes and antibiotic-induced diabetes is related to microbial ecological imbalance, as well as a decrease in the abundance of protective bacteria and SCFA biosynthesis.92 Indeed, early use of antibiotics in NOD mice can alter the intestinal microbiota and its metabolic ability, as well as intestinal gene expression and T cell population, thus accelerating T1DM pathogenesis.93
In addition, the use of antibiotics during pregnancy impacts offspring. For instance, following administration of neomycin or vancomycin to pregnant NOD mice, the offspring of the neomycin-treated group were protected from T1DM development and exhibited immune tolerance via APCs, which had reduced specific self-antigen presentation function. In contrast, the incidence of diabetes in the offspring of the vancomycin-treated group was increased with more diabetes-causing T cells detected in the lymphoid organs.94 Moreover, treatment of NOD pregnant mice with vancomycin caused a decrease in the number of IL-17-producing cells in the lamina propria of the ileum in the offspring.95 These results indicate that antibiotics should be used with caution, not only in childhood but also during pregnancy.
Collectively, these results suggest that, owing to the varying impact elicited by different antibiotics, multiple factors must be taken into account when selecting an appropriate drug, including the risk to the specific patient for developing T1DM. This is particularly pertinent for pregnant persons or infants. However, further investigations, regarding which microorganisms negatively impact T1DM progression and development, as well as the impacts of each antibiotic on eliminating this harmful intestinal microbiota, are required.
Breast Milk
The early development of intestinal microbiota includes three stages: the first 3–14 months following delivery, is termed the development period; 15–30 months is the transition period; ≥31 months is the stable period, in which breastfeeding has the greatest influence on the microflora.96 Studies have shown that breast milk contains many active ingredients, including bacteria, antimicrobial peptides, antibodies, immune cells, and breast milk oligosaccharides.97 Among them, breast milk oligosaccharides can increase the abundance of beneficial Bifidobacterium and Lactobacillus in the intestine of an infant98 and can be used as bait receptors to bind to pathogenic bacteria, thus preventing pathogens from adhering to intestinal epithelial cells; breast milk oligosaccharides can regulate the expression of mucins and polysaccharides, exert immunomodulatory effects through Toll receptors,99 and reduce intestinal permeability.100 Notably, Bifidobacterium longum subsp. Infantis, which contain the human milk oligosaccharide-producing gene, only exist in breast-fed infants;101 Breast milk antibodies also influence the colonization and development of the intestinal microbiota and can help neonates develop immune tolerance and limit their immune response against the microbiota. SlgA can also maintain homeostasis within the intestinal microbiota.102 The one-carbon metabolite betaine in breast milk is also linked to the long-term metabolic health of the infant.103 Indeed, breastfeeding improves the survival and health of newborns while reducing the risk of infection and various developmental disorders.104 Given that breast milk can affect the composition of the gut microbiota, researchers have raised the question of whether breast milk positively impacts T1DM occurrence.
Epidemiological studies have shown that exclusive breastfeeding has a protective effect against T1DM onset; however, this association is weak.105 Nevertheless, the T1DM risk decrease with increased feeding time.106 Furthermore, breastfeeding prevents the occurrence of T1DM in NOD mice, helps avoid changes in the intestinal microbiota, reduces intestinal bacterial translocation in Peyer’s patch and pancreas through the microbial network, and protects DCs in the spleen. Specifically, it promotes the recruitment of DCs in the spleen by altering the abundance of Barnesiella, fecal B. fragilis, and B. vulgatus in Peyer’s patch.107 However, the precise active ingredients associated with these effects require further investigation. Breast milk oligosaccharide is currently the most studied active ingredient in breast milk. When NOD mice were given an oligosaccharide diet at the age of 4–10 weeks, inflammation of the pancreas was alleviated, and T1DM development was significantly delayed at 30 weeks of age, compared to those of the control group. This useful function may be achieved by beneficially altering the composition, and metabolism, of the intestinal microbiota, potentially by increasing the abundance of Akkermansia and Lachnospiraceae (a member of Clostridium cluster XIVa), which can degrade the mucin in the intestine and produce butyrate. Thus, intestinal immune abnormalities are regulated, and intestinal integrity is maintained.108 Furthermore, exclusive breastfeeding and prolonged breastfeeding (>6–12 months) can reduce the risk of T1DM in children,109 while also reducing the risk of developing diabetes in mothers.110
Stem Cells
In the last two decades, adult stem cells, including hematopoietic stem cells, amniotic epithelial cells and mesenchymal stem cells (MSCs), especially MSCs, have been widely used in the treatment of T1DM.111–113 MSCs-based therapy is considered as a promising new approach for the treatment of T1DM by reconstitution of immunotolerance and preservation of islet β-cell function. For example, intravenous infusion of MSCs primarily concentrates in the pancreas,114 which not only improves the pancreatic microenvironment and promotes the survival and regeneration of β cells but also regulates the pancreatic immune environment.115 MSCs can also differentiate into insulin-secreting cells, which are superior to MSCs in terms of lowering blood glucose and improving β-cell regeneration.116 Moreover, exosomes secreted by MSCs exert therapeutic effects.117–119
The MSCs mechanism of action is multifaceted. In an inflammatory state, MSCs can create a microenvironment with balanced inflammation and regeneration factors in damaged tissues.120 They are also involved in maintaining the balance of the intestinal microbiota.121,122 In fact, a recent study report that intestinal microbes are related to the efficacy of stem cell transplantation in T1DM treatment.22 This study found that the intestinal microflora of NOD mice can resist the therapeutic effect of adipose-derived stem cells, while the use of antibiotics can improve the curative effect of stem cells by altering the intestinal microbiota. Among them, the most influential microorganisms, after the use of antibiotics, is Bifidobacterium, the abundance of which increases. Bifidobacterium stimulates the production of mucin 2 and increases the thickness of colon mucus by enhancing the expression of TRPM7, thereby reducing gut bacterial translocation to the pancreas.22 Thus, high Bifidobacterium abundance is a protective factor against diabetes.123 The authors further demonstrated that intestinal microbiome imbalance in NOD mice impacts the efficacy of stem cell therapy. However, the mice in this study were killed one week after the last transplantation, which raises the question of whether stem cell treatment can alter the intestinal microbiota? Additionally, is a treatment time of one week sufficient for stem cells to exert therapeutic effects? In addition, fecal microbes only represent the microflora at the end of the colon;124 thus, it remains to be determined whether stem cells influence the full intestinal microbiota.
The success of animal experiments has allowed the use of stem cells in clinical trials. Most experiments choose autologous stem cells for transplantation, which largely avoids transplant rejection, while also offering superior therapeutic effects compared to those of allogeneic stem cell infusion.125 A clinical trial reported autologous bone marrow MSCs transplantation in five patients with T1DM. After three months, insulin usage and glycosylated hemoglobin content decreased, while leptin levers increased,126 indicating that MSC transplantation has high curative efficacy; however, this treatment regimen requires patients to continue insulin injections. Moreover, the transplantation is unlikely to be effective at all times.
Numerous studies have investigated intestinal microorganisms as predictive markers of disease occurrence and relapse. Thus, identification of intestinal microbial markers of disease recurrence or deterioration following stem cell transplantation may provide indications for when subsequent transplants are required.
Immunotherapy
Considering that T1DM is an autoimmune disease, many studies have examined the effects of various immunotherapies, all of which employ different mechanisms for controlling T1DM. For instance, antigen-specific therapy establishes immune tolerance by injecting insulin.127 Meanwhile, immunosuppressive agents can inhibit the proliferation of cells associated with proinflammatory immune response. Additionally, Tregs can be amplified either in vitro and subsequently injected back into the organism, or in vivo via specific stimulation.128
Many immunotherapy clinical trials have been undertaken; however, the results have not been significant.129 The intestinal microbiota composition significantly impacts immunotherapy efficacy. For example, when infliximab was used to treat children with Crohn’s disease, it enriched the SCFA production and bile salt hydrolase-producing bacteria, thereby inhibiting inflammation and restoring bile acid metabolism in children.130 The therapeutic effect of CD47 monoclonal antibody for the treatment of tumors is also affected by the composition of the intratumoral microbiota of mice. Specifically, enrichment of Bifidobacterium in the tumor can promote the response of tumor-bearing mice to CD47 monoclonal antibody.131 This indicates that the presence of specific microorganisms may optimize the effect of immunotherapy.
Tregs, a type of regulatory T cell, are functionally deficient in patients with T1DM and may be the cause of impaired immune tolerance;132 therefore, restoring the function of Tregs or stimulating their production in vivo is one of the primary strategies for the prevention and treatment of T1DM. Heligmosomoides polygyrus (HP) infection can prevent the occurrence of T1DM in STZ-induced diabetic mice and NOD mice. These observations are related to the effect of trehalose on the intestinal microbiota after HP infection, which increases the abundance of Ruminococcus that can induce suppressive CD8+ Treg cells in vivo.133 Hence, induction of Tregs expansion may affect the intestinal microbiota, and Tregs can be induced by altering the microbiota. The anti-CD3 monoclonal antibody can increase the number of intestinal Tr1 by changing the intestinal microbiota, while Tr1 can migrate to the periphery to inhibit the occurrence of T1DM.134 Moreover, the cecal microbiota of diabetic mice was unbalanced, in which the Bacteroidetes/Firmicutes ratio increases. Following IgM treatment, the cecal microbiota returned to normal state, hyperglycemia was improved, and insulin autoantibody content was reduced. Notably, transplantation of the cecal microbiota from IgM-treated mice showed similar therapeutic effects, suggesting that IgM treatment may exert therapeutic effects by altering the microbiota.135 Thus, immunotherapy may change the composition of the intestinal microbiota, and it is necessary to consider whether intestinal microbiota is the cause of the observed low success rate of immunotherapy clinical trials.
Conclusions
Millions of years of coevolution have enabled intestinal microbes to symbiotically exist in the human body, to the extent that alteration of the intestinal microbiota will initiate an immune response or cause the occurrence/aggravation of various diseases. Although numerous studies have demonstrated that intestinal microorganisms, and their metabolites, are involved in the pathogenesis of T1DM, and can be used as biomarkers, targets, and/or therapeutic agents for T1DM, the causal relationship between the gut microorganisms and T1DM requires further investigation. Moreover, research on the role of known or unknown intestinal microbial genera/species on T1DM development or progression remains in its infancy. In particular, the precise causes of changes in the intestinal microbiota composition, in the context of T1DM, as well as the corresponding function and mechanism of the differential intestinal microorganisms and their metabolites, remain unclear. Moreover, many of the reported clinical trials have small sample sizes, lack reproducibility, and are cross-sectional without longitudinal comparison. Moreover, most studies have used stool samples to detect and characterize the intestinal microbiota; however, fecal microorganisms only represent the microbial community of the terminal colon, whereas each intestinal segment contains unique microorganism communities. Nevertheless, the research to-date has demonstrated a clear relationship between intestinal microorganisms, and their metabolites, and the occurrence and development of T1DM. Thus, the intestinal microbiota almost certainly represents a promising target for T1DM preventatives and therapeutics; however, numerous unresolved concerns, as described herein, require further in-depth investigation.
Abbreviations
APC, antigen-presenting cell; AhR, aryl hydrocarbon receptor; ADSCs, adipose-derived stem cells; CRAMP, cathelicidin-associated antimicrobial peptide; CPT1A, carnitine palmitoyltransferase 1A; CTRs, controls; DC, dendritic cell; DSS, dextran sulfate sodium; GF, germ-free; HDAC, histone deacetylase; HIF1α, hypoxia-inducible factor 1α; HP, Heligmosomoides polygyrus; KRV, Kilham rat virus; I–IFN, type I interferon; LMP, low-methoxyl pectin; LPS, lipopolysaccharides; NOD, non-obese diabetes mellitus; NOD BDC2.5 TCR-Tg, non-obese diabetic (NOD) BDC2.5 TCR-transgenic (Tg); PLN, pancreatic lymph nodes; PSA, polysaccharide; P1Ab, individuals who tested positive for at least one antibody but did not progress to clinical T1DM; PT1D, progressors to T1DM; SCFAs, short-chain fatty acids; SFB, segmented filamentous bacteria; STZ, streptozotocin; T1DM, type 1 diabetes mellitus; YBG, yeast β-glucan.
Acknowledgments
The authors thank Editage for English editing and proofreading.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The authors are grateful to financial support from National Natural Science Foundation of China, PR China (No. 81460156), Science and Technology Innovation Leading Academics of National High-level Personnel of Special Support Program, PR China (No. GKFZ-2018-29), Guizhou High-Level Innovative Talent Support Program, PR China (No. QKH-RC-2022-0001), the Science and Technology Foundation of Guizhou, PR China (No. QKHQKHJC-ZK-2021-ZD-026), and S&T Foundation of Zunyi Science and Technology Bureau, PR China (No. ZSK-RC-2020-1).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zheng P, Li Z, Zhou Z. Gut microbiome in type 1 diabetes: a comprehensive review. Diabetes Metab Res Rev. 2018;34(7):e3043. doi:10.1002/dmrr.3043
2. Murri M, Leiva I, Gomez-Zumaquero JM, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11(1):46. doi:10.1186/1741-7015-11-46
3. de Goffau MC, Fuentes S, van den Bogert B, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57(8):1569–1577. doi:10.1007/s00125-014-3274-0
4. Desselberger U. The mammalian intestinal microbiome: composition, interaction with the immune system, significance for vaccine efficacy, and potential for disease therapy. Pathogens. 2018;7(3):57. doi:10.3390/pathogens7030057
5. Clavel T, Lagkouvardos I, Blaut M, Stecher B. The mouse gut microbiome revisited: from complex diversity to model ecosystems. Int J Med Microbiol. 2016;306(5):316–327. doi:10.1016/j.ijmm.2016.03.002
6. Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975. doi:10.1073/pnas.1002601107
7. Vatanen T, Kostic AD, d’Hennezel E, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(4):842–853. doi:10.1016/j.cell.2016.04.007
8. Mullaney JA, Stephens JE, Costello ME, et al. Type 1 diabetes susceptibility alleles are associated with distinct alterations in the gut microbiota. Microbiome. 2018;6(1):35. doi:10.1186/s40168-018-0417-4
9. Silverman M, Kua L, Tanca A, et al. Protective major histocompatibility complex allele prevents type 1 diabetes by shaping the intestinal microbiota early in ontogeny. Proc Natl Acad Sci U S A. 2017;114(36):9671–9676. doi:10.1073/pnas.1712280114
10. Hu Y, Peng J, Li F, Wong FS, Wen L. Evaluation of different mucosal microbiota leads to gut microbiota-based prediction of type 1 diabetes in NOD mice. Sci Rep. 2018;8(1):15451. doi:10.1038/s41598-018-33571-z
11. Kostic AD, Gevers D, Siljander H, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17(2):260–273. doi:10.1016/j.chom.2015.01.001
12. Roth-Schulze AJ, Penno M, Ngui KM, et al. Type 1 diabetes in pregnancy is associated with distinct changes in the composition and function of the gut microbiome. Microbiome. 2021;9(1):167. doi:10.1186/s40168-021-01104-y
13. Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol. 2020;38(1):23–48. doi:10.1146/annurev-immunol-070119-115104
14. Zhao Y, Chen F, Wu W, et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018;11(3):752–762. doi:10.1038/mi.2017.118
15. Liang W, Enée E, Andre-Vallee C, Falcone M, Sun J, Diana J. Intestinal cathelicidin antimicrobial peptide shapes a protective neonatal gut microbiota against pancreatic autoimmunity. Gastroenterology. 2021;162(4):1288–1302.e16. doi:10.1053/j.gastro.2021.12.272
16. Chung LK, Raffatellu M. G.I. pros: antimicrobial defense in the gastrointestinal tract. Semin Cell Dev Biol. 2019;88:129–137. doi:10.1016/j.semcdb.2018.02.001
17. Lee AS, Gibson DL, Zhang Y, Sham HP, Vallance BA, Dutz JP. Gut barrier disruption by an enteric bacterial pathogen accelerates insulitis in NOD mice. Diabetologia. 2010;53(4):741–748. doi:10.1007/s00125-009-1626-y
18. Neu J, Reverte CM, Mackey AD, et al. Changes in intestinal morphology and permeability in the biobreeding rat before the onset of type 1 diabetes. J Pediatr Gastroenterol Nutr. 2005;40(5):589–595. doi:10.1097/01.MPG.0000159636.19346.C1
19. Miranda M, Oliveira RP, Torres L, et al. Frontline science: abnormalities in the gut mucosa of non-obese diabetic mice precede the onset of type 1 diabetes. J Leukoc Biol. 2019;106(3):513–529. doi:10.1002/JLB.3HI0119-024RR
20. Huang J, Pearson JA, Peng J, et al. Gut microbial metabolites alter IgA immunity in type 1 diabetes. JCI Insight. 2020;5(10). doi:10.1172/jci.insight.135718.
21. Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12(3):154–167. doi:10.1038/nrendo.2015.218
22. Lv W, Graves DT, He L, et al. Depletion of the diabetic gut microbiota resistance enhances stem cells therapy in type 1 diabetes mellitus. Theranostics. 2020;10(14):6500–6516. doi:10.7150/thno.44113
23. Costa FR, Françozo MC, de Oliveira GG, et al. Gut microbiota translocation to the pancreatic lymph nodes triggers NOD2 activation and contributes to T1D onset. J Exp Med. 2016;213(7):1223–1239. doi:10.1084/jem.20150744
24. Jaakkola I, Jalkanen S, Hänninen A. Diabetogenic T cells are primed both in pancreatic and gut-associated lymph nodes in NOD mice. Eur J Immunol. 2003;33(12):3255–3264. doi:10.1002/eji.200324405
25. Liu X, Tong X, Zou Y, et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat Genet. 2022;54(1):52–61. doi:10.1038/s41588-021-00968-y
26. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19(1):55–71. doi:10.1038/s41579-020-0433-9
27. Kim CH. Microbiota or short-chain fatty acids: which regulates diabetes. Cell Mol Immunol. 2018;15(2):88–91. doi:10.1038/cmi.2017.57
28. Scott SA, Fu J, Chang PV. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc Natl Acad Sci USA. 2020;117(32):19376–19387. doi:10.1073/pnas.2000047117
29. Lamichhane S, Kemppainen E, Trošt K, et al. Circulating metabolites in progression to islet autoimmunity and type 1 diabetes. Diabetologia. 2019;62(12):2287–2297. doi:10.1007/s00125-019-04980-0
30. Lamichhane S, Ahonen L, Dyrlund TS, et al. A longitudinal plasma lipidomics dataset from children who developed islet autoimmunity and type 1 diabetes. Sci Data. 2018;5(1):180250. doi:10.1038/sdata.2018.250
31. Sobczak A, Pitt SJ, Smith TK, Ajjan RA, Stewart AJ. Lipidomic profiling of plasma free fatty acids in type-1 diabetes highlights specific changes in lipid metabolism. Biochim Biophys Acta Mol Cell Biol Lipids. 2021;1866(1):158823. doi:10.1016/j.bbalip.2020.158823
32. Mathew AV, Jaiswal M, Ang L, Michailidis G, Pennathur S, Pop-Busui R. Impaired amino acid and TCA metabolism and cardiovascular autonomic neuropathy progression in type 1 diabetes. Diabetes. 2019;68(10):2035–2044. doi:10.2337/db19-0145
33. Winther SA, Henriksen P, Vogt JK, et al. Gut microbiota profile and selected plasma metabolites in type 1 diabetes without and with stratification by albuminuria. Diabetologia. 2020;63(12):2713–2724. doi:10.1007/s00125-020-05260-y
34. Buchwald P, Tamayo-Garcia A, Ramamoorthy S, Garcia-Contreras M, Mendez AJ, Ricordi C. Comprehensive metabolomics study to assess longitudinal biochemical changes and potential early biomarkers in nonobese diabetic mice that progress to diabetes. J Proteome Res. 2017;16(10):3873–3890. doi:10.1021/acs.jproteome.7b00512
35. Mariño E, Richards JL, McLeod KH, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18(5):552–562. doi:10.1038/ni.3713
36. Tanca A, Palomba A, Fraumene C, Manghina V, Silverman M, Uzzau S. Clostridial butyrate biosynthesis enzymes are significantly depleted in the gut microbiota of nonobese diabetic mice. mSphere. 2018;3(5). doi:10.1128/mSphere.00492-18
37. Tabibian JH, Kenderian SS. The microbiome and immune regulation after transplantation. Transplantation. 2017;101(1):56–62. doi:10.1097/TP.0000000000001444
38. Tai N, Peng J, Liu F, et al. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J Exp Med. 2016;213(10):2129–2146. doi:10.1084/jem.20160526
39. Sorini C, Cosorich I, Lo Conte M, et al. Loss of gut barrier integrity triggers activation of islet-reactive T cells and autoimmune diabetes. Proc Natl Acad Sci USA. 2019;116(30):15140–15149. doi:10.1073/pnas.1814558116
40. Sun M, Wu W, Chen L, et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun. 2018;9(1):3555. doi:10.1038/s41467-018-05901-2
41. Huang J, Tan Q, Tai N, et al. IL-10 deficiency accelerates type 1 diabetes development via modulation of innate and adaptive immune cells and gut microbiota in BDC2.5 NOD mice. Front Immunol. 2021;12:702955. doi:10.3389/fimmu.2021.702955
42. Neuman V, Cinek O, Funda DP, et al. Human gut microbiota transferred to germ-free NOD mice modulate the progression towards type 1 diabetes regardless of the pace of beta cell function loss in the donor. Diabetologia. 2019;62(7):1291–1296. doi:10.1007/s00125-019-4869-2
43. King C, Sarvetnick N, von Herrath M. The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS One. 2011;6(2):e17049. doi:10.1371/journal.pone.0017049
44. Hänninen A, Toivonen R, Pöysti S, et al. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2018;67(8):1445–1453. doi:10.1136/gutjnl-2017-314508
45. Zhang XS, Yin YS, Wang J, et al. Maternal cecal microbiota transfer rescues early-life antibiotic-induced enhancement of type 1 diabetes in mice. Cell Host Microbe. 2021;29(8):1249–1265.e9. doi:10.1016/j.chom.2021.06.014
46. Bell KJ, Saad S, Tillett BJ, et al. Metabolite-based dietary supplementation in human type 1 diabetes is associated with microbiota and immune modulation. Microbiome. 2022;10(1):9. doi:10.1186/s40168-021-01193-9
47. Sun J, Furio L, Mecheri R, et al. Pancreatic β-cells limit autoimmune diabetes via an immunoregulatory antimicrobial peptide expressed under the influence of the gut microbiota. Immunity. 2015;43(2):304–317. doi:10.1016/j.immuni.2015.07.013
48. Jia L, Cao M, Chen H, et al. Butyrate ameliorates antibiotic-driven type 1 diabetes in the female offspring of nonobese diabetic mice. J Agric Food Chem. 2020;68(10):3112–3120. doi:10.1021/acs.jafc.9b07701
49. Jacob N, Jaiswal S, Maheshwari D, et al. Butyrate induced Tregs are capable of migration from the GALT to the pancreas to restore immunological tolerance during type-1 diabetes. Sci Rep. 2020;10(1):19120. doi:10.1038/s41598-020-76109-y
50. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi:10.1016/j.cell.2005.05.007
51. Martin-Gallausiaux C, Béguet-Crespel F, Marinelli L, et al. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci Rep. 2018;8(1):9742. doi:10.1038/s41598-018-28048-y
52. Hao F, Tian M, Zhang X, et al. Butyrate enhances CPT1A activity to promote fatty acid oxidation and iTreg differentiation. Proc Natl Acad Sci U S A. 2021;118(22):22. doi:10.1073/pnas.2014681118
53. Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi:10.1126/science.1198469
54. Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi:10.1016/j.cell.2009.09.033
55. Ladinsky MS, Araujo LP, Zhang X, et al. Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science. 2019;363(6431):6431. doi:10.1126/science.aat4042
56. Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi:10.1038/nature12726
57. Yang W, Yu T, Huang X, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. 2020;11(1):4457. doi:10.1038/s41467-020-18262-6
58. Vaarala O. Is the origin of type 1 diabetes in the gut. Immunol Cell Biol. 2012;90(3):271–276. doi:10.1038/icb.2011.115
59. Brugman S, Klatter FA, Visser JT, et al. Antibiotic treatment partially protects against type 1 diabetes in the bio-breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes. Diabetologia. 2006;49(9):2105–2108. doi:10.1007/s00125-006-0334-0
60. Serena G, Camhi S, Sturgeon C, Yan S, Fasano A. The role of gluten in celiac disease and type 1 diabetes. Nutrients. 2015;7(9):7143–7162. doi:10.3390/nu7095329
61. Raman M, Ambalam P, Kondepudi KK, et al. Potential of probiotics, prebiotics and synbiotics for management of colorectal cancer. Gut Microbes. 2013;4(3):181–192. doi:10.4161/gmic.23919
62. Mazlyn MM, Nagarajah LH, Fatimah A, Norimah AK, Goh KL. Effects of a probiotic fermented milk on functional constipation: a randomized, double-blind, placebo-controlled study. J Gastroenterol Hepatol. 2013;28(7):1141–1147. doi:10.1111/jgh.12168
63. Alam C, Valkonen S, Palagani V, Jalava J, Eerola E, Hänninen A. Inflammatory tendencies and overproduction of IL-17 in the colon of young NOD mice are counteracted with diet change. Diabetes. 2010;59(9):2237–2246. doi:10.2337/db10-0147
64. Belizário JE, Napolitano M. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front Microbiol. 2015;6:1050. doi:10.3389/fmicb.2015.01050
65. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi:10.1016/j.cell.2016.05.041
66. Sun Y, He Y, Wang F, Zhang H, de Vos P, Sun J. Low-methoxyl lemon pectin attenuates inflammatory responses and improves intestinal barrier integrity in caerulein-induced experimental acute pancreatitis. Mol Nutr Food Res. 2017;61(4):1600885. doi:10.1002/mnfr.201600885
67. Sahasrabudhe NM, Beukema M, Tian L, et al. Dietary fiber pectin directly blocks toll-like receptor 2-1 and prevents doxorubicin-induced ileitis. Front Immunol. 2018;9:383. doi:10.3389/fimmu.2018.00383
68. Komaroff AL. The microbiome and risk for obesity and diabetes. JAMA. 2017;317(4):355–356. doi:10.1001/jama.2016.20099
69. Wu C, Pan LL, Niu W, et al. Modulation of gut microbiota by low methoxyl pectin attenuates type 1 diabetes in non-obese diabetic mice. Front Immunol. 2019;10:1733. doi:10.3389/fimmu.2019.01733
70. Gudi R, Perez N, Johnson BM, et al. Complex dietary polysaccharide modulates gut immune function and microbiota, and promotes protection from autoimmune diabetes. Immunology. 2019;157(1):70–85. doi:10.1111/imm.13048
71. Shi CW, Cheng MY, Yang X, et al. Probiotic lactobacillus rhamnosus gg promotes mouse gut microbiota diversity and T cell differentiation. Front Microbiol. 2020;11:607735. doi:10.3389/fmicb.2020.607735
72. Yadav R, Dey DK, Vij R, Meena S, Kapila R, Kapila S. Evaluation of anti-diabetic attributes of Lactobacillus rhamnosus MTCC: 5957, Lactobacillus rhamnosus MTCC: 5897 and Lactobacillus fermentum MTCC: 5898 in streptozotocin induced diabetic rats. Microb Pathog. 2018;125:454–462. doi:10.1016/j.micpath.2018.10.015
73. Chung PH, Wu YY, Chen PH, Fung CP, Hsu CM, Chen LW. Lactobacillus salivarius reverse diabetes-induced intestinal defense impairment in mice through non-defensin protein. J Nutr Biochem. 2016;35:48–57. doi:10.1016/j.jnutbio.2016.05.013
74. Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi:10.1038/nature12331
75. Dolpady J, Sorini C, Di Pietro C, et al. Oral probiotic VSL#3 prevents autoimmune diabetes by modulating microbiota and promoting indoleamine 2,3-dioxygenase-enriched tolerogenic intestinal environment. J Diabetes Res. 2016;2016:7569431. doi:10.1155/2016/7569431
76. Uusitalo U, Liu X, Yang J, et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr. 2016;170(1):20–28. doi:10.1001/jamapediatrics.2015.2757
77. Kumar S, Kumar R, Rohilla L, Jacob N, Yadav J, Sachdeva N. A high potency multi-strain probiotic improves glycemic control in children with new-onset type 1 diabetes mellitus: a randomized, double-blind, and placebo-controlled pilot study. Pediatr Diabetes. 2021;22(7):1014–1022. doi:10.1111/pedi.13244
78. Ho J, Nicolucci AC, Virtanen H, et al. Effect of prebiotic on microbiota, intestinal permeability, and glycemic control in children with type 1 diabetes. J Clin Endocrinol Metab. 2019;104(10):4427–4440. doi:10.1210/jc.2019-00481
79. Zare Javid A, Aminzadeh M, Haghighi-Zadeh MH, Jamalvandi M. The effects of synbiotic supplementation on glycemic status, lipid profile, and biomarkers of oxidative stress in type 1 diabetic patients. a placebo-controlled, double-blind, randomized clinical trial. Diabetes Metab Syndr Obes. 2020;13:607–617. doi:10.2147/DMSO.S238867
80. Savilahti E, Härkönen T, Savilahti EM, Kukkonen K, Kuitunen M, Knip M. Probiotic intervention in infancy is not associated with development of beta cell autoimmunity and type 1 diabetes. Diabetologia. 2018;61(12):2668–2670. doi:10.1007/s00125-018-4738-4
81. Groele L, Szajewska H, Szalecki M, et al. Lack of effect of Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12 on beta-cell function in children with newly diagnosed type 1 diabetes: a randomised controlled trial. BMJ Open Diabetes Res Care. 2021;9(1):e001523. doi:10.1136/bmjdrc-2020-001523
82. Beyerlein A, Liu X, Uusitalo UM, et al. Dietary intake of soluble fiber and risk of islet autoimmunity by 5 y of age: results from the TEDDY study. Am J Clin Nutr. 2015;102(2):345–352. doi:10.3945/ajcn.115.108159
83. Tamma PD, Miller MA, Cosgrove SE. Rethinking how antibiotics are prescribed: incorporating the 4 moments of antibiotic decision making into clinical practice. JAMA. 2019;321(2):139–140. doi:10.1001/jama.2018.19509
84. Wan QY, Zhao R, Wang Y, Wu Y, Wu XT. Antibiotic use and risk of colorectal cancer: a meta-analysis of 412 450 participants. Gut. 2020;69(11):2059–2060. doi:10.1136/gutjnl-2020-320826
85. Donovan BM, Abreo A, Ding T, et al. Dose, timing, and type of infant antibiotic use and the risk of childhood asthma. Clin Infect Dis. 2020;70(8):1658–1665. doi:10.1093/cid/ciz448
86. Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65(11):1906–1915. doi:10.1136/gutjnl-2016-312297
87. Ylipaasto P, Klingel K, Lindberg AM, et al. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia. 2004;47(2):225–239. doi:10.1007/s00125-003-1297-z
88. Blanter M, Sork H, Tuomela S, Flodström-Tullberg M. Genetic and environmental interaction in type 1 diabetes: a relationship between genetic risk alleles and molecular traits of enterovirus infection. Curr Diab Rep. 2019;19(9):82. doi:10.1007/s11892-019-1192-8
89. Guberski DL, Thomas VA, Shek WR, et al. Induction of type I diabetes by Kilham’s rat virus in diabetes-resistant BB/Wor rats. Science. 1991;254(5034):1010–1013. doi:10.1126/science.1658938
90. Needell JC, Brown MN, Zipris D. Involvement of adipose tissue inflammation and dysfunction in virus-induced type 1 diabetes. J Endocrinol. 2018;238(1):61–75. doi:10.1530/JOE-18-0131
91. Hara N, Alkanani AK, Ir D, et al. Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol. 2012;189(8):3805–3814. doi:10.4049/jimmunol.1201257
92. Brown K, Godovannyi A, Ma C, et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. 2016;10(2):321–332. doi:10.1038/ismej.2015.114
93. Livanos AE, Greiner TU, Vangay P, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol. 2016;1(11):16140. doi:10.1038/nmicrobiol.2016.140
94. Hu Y, Jin P, Peng J, Zhang X, Wong FS, Wen L. Different immunological responses to early-life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J Autoimmun. 2016;72:47–56. doi:10.1016/j.jaut.2016.05.001
95. Candon S, Perez-Arroyo A, Marquet C, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One. 2015;10(5):e0125448. doi:10.1371/journal.pone.0125448
96. Stewart CJ, Ajami NJ, O’Brien JL, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–588. doi:10.1038/s41586-018-0617-x
97. Nadimpalli ML, Bourke CD, Robertson RC, Delarocque-Astagneau E, Manges AR, Pickering AJ. Can breastfeeding protect against antimicrobial resistance. BMC Med. 2020;18(1):392. doi:10.1186/s12916-020-01862-w
98. Karav S, Le Parc A, Leite Nobrega de Moura Bell JM, et al. Oligosaccharides released from milk glycoproteins are selective growth substrates for infant-associated bifidobacteria. Appl Environ Microbiol. 2016;82(12):3622–3630. doi:10.1128/AEM.00547-16
99. Kong C, Faas MM, de Vos P, Akkerman R. Impact of dietary fibers in infant formulas on gut microbiota and the intestinal immune barrier. Food Funct. 2020;11(11):9445–9467. doi:10.1039/D0FO01700K
100. Chleilat F, Klancic T, Ma K, Schick A, Nettleton JE, Reimer RA. Human milk oligosaccharide supplementation affects intestinal barrier function and microbial composition in the gastrointestinal tract of young Sprague Dawley rats. Nutrients. 2020;12(5):1532. doi:10.3390/nu12051532
101. Vatanen T, Franzosa EA, Schwager R, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562(7728):589–594. doi:10.1038/s41586-018-0620-2
102. Atyeo C, Alter G. The multifaceted roles of breast milk antibodies. Cell. 2021;184(6):1486–1499. doi:10.1016/j.cell.2021.02.031
103. Ribo S, Sánchez-Infantes D, Martinez-Guino L, et al. Increasing breast milk betaine modulates Akkermansia abundance in mammalian neonates and improves long-term metabolic health. Sci Transl Med. 2021;13:587. doi:10.1126/scitranslmed.abb0322
104. Bode L, Raman AS, Murch SH, Rollins NC, Gordon JI. Understanding the mother-breastmilk-infant “triad”. Science. 2020;367(6482):1070–1072. doi:10.1126/science.aaw6147
105. Cardwell CR, Stene LC, Ludvigsson J, et al. Breast-feeding and childhood-onset type 1 diabetes: a pooled analysis of individual participant data from 43 observational studies. Diabetes Care. 2012;35(11):2215–2225. doi:10.2337/dc12-0438
106. Hall K, Frederiksen B, Rewers M, Norris JM. Daycare attendance, breastfeeding, and the development of type 1 diabetes: the diabetes autoimmunity study in the young. Biomed Res Int. 2015;2015:203947. doi:10.1155/2015/203947
107. Sane F, Scuotto A, Pierrat V, Kacet N, Hober D, Romond MB. Diabetes progression and alterations in gut bacterial translocation: prevention by diet supplementation with human milk in NOD mice. J Nutr Biochem. 2018;62:108–122. doi:10.1016/j.jnutbio.2018.08.017
108. Xiao L, Van’t Land B, Engen PA, et al. Human milk oligosaccharides protect against the development of autoimmune diabetes in NOD-mice. Sci Rep. 2018;8(1):3829. doi:10.1038/s41598-018-22052-y
109. Lampousi AM, Carlsson S, Löfvenborg JE. Dietary factors and risk of islet autoimmunity and type 1 diabetes: a systematic review and meta-analysis. EBioMedicine. 2021;72:103633. doi:10.1016/j.ebiom.2021.103633
110. Moon JH, Kim H, Kim H, et al. Lactation improves pancreatic β cell mass and function through serotonin production. Sci Transl Med. 2020;12(541):541. doi:10.1126/scitranslmed.aay0455
111. Ye L, Li L, Wan B, et al. Immune response after autologous hematopoietic stem cell transplantation in type 1 diabetes mellitus. Stem Cell Res Ther. 2017;8(1):90. doi:10.1186/s13287-017-0542-1
112. Luo Y, Cheng YW, Yu CY, et al. Effects of hyaluronic acid on differentiation of human amniotic epithelial cells and cell-replacement therapy in type 1 diabetic mice. Exp Cell Res. 2019;384(2):111642. doi:10.1016/j.yexcr.2019.111642
113. Pixley JS. Mesenchymal stem cells to treat type 1 diabetes. Biochim Biophys Acta Mol Basis Dis. 2020;1866(4):165315. doi:10.1016/j.bbadis.2018.10.033
114. Li L, Hui H, Jia X, et al. Infusion with human bone marrow-derived mesenchymal stem cells improves β-cell function in patients and non-obese mice with severe diabetes. Sci Rep. 2016;6(1):37894. doi:10.1038/srep37894
115. Sohni A, Verfaillie CM. Mesenchymal stem cells migration homing and tracking. Stem Cells Int. 2013;2013:130763. doi:10.1155/2013/130763
116. Domouky AM, Hegab AS, Al-Shahat A, Raafat N. Mesenchymal stem cells and differentiated insulin producing cells are new horizons for pancreatic regeneration in type I diabetes mellitus. Int J Biochem Cell Biol. 2017;87:77–85. doi:10.1016/j.biocel.2017.03.018
117. Nojehdehi S, Soudi S, Hesampour A, Rasouli S, Soleimani M, Hashemi SM. Immunomodulatory effects of mesenchymal stem cell-derived exosomes on experimental type-1 autoimmune diabetes. J Cell Biochem. 2018;119(11):9433–9443. doi:10.1002/jcb.27260
118. Mahdipour E, Salmasi Z, Sabeti N. Potential of stem cell-derived exosomes to regenerate β islets through Pdx-1 dependent mechanism in a rat model of type 1 diabetes. J Cell Physiol. 2019;234(11):20310–20321. doi:10.1002/jcp.28631
119. Sabry D, Marzouk S, Zakaria R, Ibrahim HA, Samir M. The effect of exosomes derived from mesenchymal stem cells in the treatment of induced type 1 diabetes mellitus in rats. Biotechnol Lett. 2020;42(8):1597–1610. doi:10.1007/s10529-020-02908-y
120. Shi Y, Wang Y, Li Q, et al. Immunoregulatory mechanisms of mesenchymal stem and stromal cells in inflammatory diseases. Nat Rev Nephrol. 2018;14(8):493–507. doi:10.1038/s41581-018-0023-5
121. Ikarashi S, Tsuchiya A, Kawata Y, et al. Effects of human adipose tissue-derived and umbilical cord tissue-derived mesenchymal stem cells in a dextran sulfate sodium-induced mouse model. Biores Open Access. 2019;8(1):185–199. doi:10.1089/biores.2019.0022
122. Sun J, Ding X, Liu S, Duan X, Liang H, Sun T. Adipose-derived mesenchymal stem cells attenuate acute lung injury and improve the gut microbiota in septic rats. Stem Cell Res Ther. 2020;11(1):384. doi:10.1186/s13287-020-01902-5
123. Traversi D, Rabbone I, Scaioli G, et al. Risk factors for type 1 diabetes, including environmental, behavioural and gut microbial factors: a case-control study. Sci Rep. 2020;10(1):17566. doi:10.1038/s41598-020-74678-6
124. Tang Q, Jin G, Wang G, et al. Current sampling methods for gut microbiota: a call for more precise devices. Front Cell Infect Microbiol. 2020;10:151. doi:10.3389/fcimb.2020.00151
125. Thakkar UG, Trivedi HL, Vanikar AV, Dave SD. Insulin-secreting adipose-derived mesenchymal stromal cells with bone marrow-derived hematopoietic stem cells from autologous and allogenic sources for type 1 diabetes mellitus. Cytotherapy. 2015;17(7):940–947. doi:10.1016/j.jcyt.2015.03.608
126. Ulyanova O, Askarov M, Kozina L, et al. Autologous mesenchymal stem cell transplant in patients with type 1 diabetes mellitus. Exp Clin Transplant. 2019;17(Suppl 1):236–238. doi:10.6002/ect.MESOT2018.P100
127. Ludvigsson J. Autoantigen treatment in type 1 diabetes: unsolved questions on how to select autoantigen and administration route. Int J Mol Sci. 2020;21(5):1598. doi:10.3390/ijms21051598
128. Yu H, Paiva R, Flavell RA. Harnessing the power of regulatory T-cells to control autoimmune diabetes: overview and perspective. Immunology. 2018;153(2):161–170. doi:10.1111/imm.12867
129. Rapini N, Schiaffini R, Fierabracci A. Immunotherapy strategies for the prevention and treatment of distinct stages of type 1 diabetes: an overview. Int J Mol Sci. 2020;21(6):2103. doi:10.3390/ijms21062103
130. Wang Y, Gao X, Zhang X, et al. Microbial and metabolic features associated with outcome of infliximab therapy in pediatric Crohn’s disease. Gut Microbes. 2021;13(1):1–18. doi:10.1080/19490976.2021.1900996
131. Shi Y, Zheng W, Yang K, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med. 2020;217(5). doi:10.1084/jem.20192282.
132. Lindley S, Dayan CM, Bishop A, Roep BO, Peakman M, Tree TI. Defective suppressor function in CD4(+)CD25(+) T-cells from patients with type 1 diabetes. Diabetes. 2005;54(1):92–99. doi:10.2337/diabetes.54.1.92
133. Shimokawa C, Kato T, Takeuchi T, et al. CD8(+) regulatory T cells are critical in prevention of autoimmune-mediated diabetes. Nat Commun. 2020;11(1):1922. doi:10.1038/s41467-020-15857-x
134. Yu H, Gagliani N, Ishigame H, et al. Intestinal type 1 regulatory T cells migrate to periphery to suppress diabetogenic T cells and prevent diabetes development. Proc Natl Acad Sci USA. 2017;114(39):10443–10448. doi:10.1073/pnas.1705599114
135. Chhabra P, Spano AJ, Bowers D, et al. Evidence for the role of the cecal microbiome in maintenance of immune regulation and homeostasis. Ann Surg. 2018;268(3):541–549. doi:10.1097/SLA.0000000000002930
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.