Back to Journals » International Journal of General Medicine » Volume 14
The Identification of Alternative Polyadenylation in Stomach Adenocarcinomas Using the Genotype-Tissue Expression Project and the Cancer Genome Atlas– Stomach Adenocarcinoma Profiles
Authors Li J, Chen W, Cao Y, Li ZR
Received 12 July 2021
Accepted for publication 27 August 2021
Published 23 September 2021 Volume 2021:14 Pages 6035—6045
DOI https://doi.org/10.2147/IJGM.S329064
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Jian Li,1 Wen Chen,2 Yi Cao,3 Zheng-Rong Li3
1Department of Gastrointestinal Surgery, Jiangxi Provincial People’s Hospital Affiliated to Nanchang University, Nanchang, 330000, People’s Republic of China; 2Key Laboratory of Bioprocess Engineering of Jiangxi Province, College of Life Sciences, Jiangxi Science and Technology Normal University, Nanchang, People’s Republic of China; 3Department of Gastrointestinal Surgery, The First Affiliated Hospital of Nanchang University, Nanchang, 330000, People’s Republic of China
Correspondence: Zheng-Rong Li
Department of Gastrointestinal Surgery, The First Affiliated Hospital of Nanchang University, 1519 Dongyue Avenue, Nanchang County, Nanchang, 330000, People’s Republic of China
Tel +86 0791-86319546
Email [email protected]
Objective: Alternative polyadenylation (APA) is a common mechanism that is present in most human genes and determines the length of the messenger ribonucleic acid (mRNA) three prime untranslated region (3ʹ-UTR), which can give rise to changes in mRNA stability and localization. However, little is known about the specific changes related to APA in stomach adenocarcinomas (STADs).
Methods: We integrated RNA sequencing data from The Cancer Genome Atlas and Genotype-Tissue Expression project to comprehensively analyze APA events in 289 cases of STAD.
Results: Our results showed that APA events were widespread in patients with STAD and were rich in genes related to known STAD pathways. The APA events result in the loss of tumor-suppressing micro-ribonucleic acid (miRNA) binding sites and increased heterogeneity in the length of the 3ʹ-UTR altered genes. Survival analysis revealed that specific subsets of 3ʹ-UTR-altered genes independently characterized a poor prognostic cohort among patients with STAD, thereby indicating the potential of APA as a new prognostic biomarker.
Conclusion: Our single-cancer analysis showed that by losing miRNA regulation, APA can become a driving factor for regulating the expression of oncogenic genes in STAD and promote its development. Our research revealed that APA events regulated STAD genes that were functionally related, thereby providing a new approach for gaining a better understanding of the progress of STADs and a means for identifying new drug targets as avenues of treatment.
Keywords: alternative polyadenylation, stomach adenocarcinoma, heterogeneity, 3′-UTR alerted genes
Introduction
As the primary subtype of stomach cancer, stomach adenocarcinoma (STAD) is the fifth most frequently diagnosed malignant tumor and the third leading cause of cancer-related deaths in the world.1 Approximately 90% ~ 95% of all stomach cancer cases are STADs.2 The incidence of STAD in China is the highest in the world, accounting for 49.9% of global cases.3 Although the survival rate of patients with STADs has greatly improved in the past 20 years due to the development of treatment methods such as surgery, chemotherapy, and targeted therapy, its prognosis remains unsatisfactory.4 The five-year survival rate of patients with a STAD in China is 30.2% ~ 35.9%; in European countries, the five-year survival rate is only 10% ~ 30%.5
Genome-wide gene expression profiles have been established including The Cancer Genome Atlas (TCGA) to better understand the impact of clinical outcomes on genetic changes in tumors. Extensive sequencing data analyses have identified mutant genes (eg, TP53, ARID1A, FAT4, KMT2D, and PIK3CA) and dysregulated functional pathways (such as neuroactive ligand–receptor interaction, pancreatic secretion, and the calcium signaling pathway) related to STAD cases. However, the mechanism of gene expression dysregulation in patients with a STAD is still not fully understood.
Alternative polyadenylation (APA) is one of the key regulatory mechanisms of gene expression in tumors. As a post-transcriptional processing mechanism, APA can produce a different three prime untranslated region (3ʹ-UTR) on messenger ribonucleic acid (mRNA) transcripts via RNA polymerase II.6,7 The variability of the 3ʹ-UTR can change the micro-ribonucleic acid (miRNA) or RNA-binding protein regulatory sites of mRNA transcripts, thereby regulating the stability, function, expression, or subcellular localization of mRNA.8,9 In humans, 51% ~ 79% of genes express alternative 3ʹ-UTR, indicating that the transcripts corresponding to most genes have multiple polyadenylation sites (PAS).10 Alternative polyadenylation participates in many cells’ physiological processes, such as cell replication, proliferation, migration and invasion, chromatin signaling, pluripotent cell fate, and tissue aging.11–15 In addition, APA disorder is considered a driver of tumorigenesis.16,17 A large number of APA dysregulation events have been reported in many types of cancer, and 3ʹ-UTR-shortened events (61% ~ 98% of total APA events) are the principal consideration in this regard.18,19 These 3ʹ-UTR-shortened APA events regulate the expression of cancer-related genes by losing miRNA regulatory sites.15,20 The shortened APA events also regulate the expression of tumor suppressors by reducing competitive endogenous RNA interference.17,21 Due to a lack of control samples, existing studies on APA events in cases of STAD have primarily focused on pan-cancer APA analysis; the single-cancer APA study of STADs still requires further characterization.
We analyzed 289 STAD tumor data from the STAD study in TCGA (TCGA-STAD)22 to identify the distribution of APA in STAD. Since there was less data on normal tissue compared with cancer in TCGA related to STAD, we used data for 207 normal stomach tissue cases in the Genotype-Tissue Expression (GTEx) project as controls.23 We found 271 genes with highly recurrent dynamic APA events associated with STAD, demonstrating the additional prognostic capabilities of APA beyond common demographic and clinical variables, thereby expanding our knowledge of the mechanisms and consequences of APA regulation during STAD tumorigenesis.
Methods
Data Collection
The RNA-Seq data for 289 patients with STAD were downloaded from the TCGA-STAD database (https://portal.gdc.cancer.gov). The RNA-Seq data for 207 normal stomach tissue cases were downloaded from the GTEx database (dbGaP Study Accession: phs000424.v7.p2. https://gtexportal.org). These RNA-Seq data were both paired-end, non-strand-specific sequencing data. The data titles and the depth and length of the reads are listed in Table S1. Clinical data, such as age, TNM stage, gender, survival time, and status were also downloaded from the TCGA database.
Bioinformatics Analysis and Statistical Methods
After Sequence Read Archive data from GTEx was converted to FASTQ data, the same Pipeline process as in TCGA (https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/Expression_mRNA_Pipeline/) was used to generate BAM files to reduce the difference between the two cohorts.
Dynamic Analysis of Alternative Polyadenylation from RNA Sequencing Analysis
The aligned BAM files (converted to bedGraph files) were generated using BEDTools (v.2.29.0). The bedGraph files were provided as input to the dynamic analysis of alternative polyadenylation from RNA sequencing (DaPars) algorithm.18 The DaPars process (developed by Xia et al) is a regression-based algorithm that performs the de novo identification of dynamic APAs from standard RNA sequencing data.18 In this study, DaPars (Python 2.7.17) was used to process the bedGraph coverage files. The dynamic poly (A) sites in the RNA-Seq data were used to de novo identify and analyze differences in APA between the 3ʹ-UTR of tumor and normal samples. The output of DaPars is the PDUI score matrix for any given gene (row) in each sample (column).
The Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis
For the specific target gene, the R package clusterProfiler24 was used for the Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, as well as dot plot drawing using R-3.6.1.
The Micro-Ribonucleic Acid Binding Site
The highly conserved miRNA binding sites and their genomic location was downloaded from TargetScanHuman 7.2 (http://www.targetscan.org). Combining this list and DaPars’ prediction of the genetic coordinates of the lost 3ʹUTR, the missing highly conserved miRNA binding sites were counted.
Heterogeneity Analysis
We calculated the variance of each unique gene’s PDUI score in the tumor samples (Var [Tumor]) and normal samples (Var [Normal]) to measure their heterogeneity in the use of PAS. We plotted the variance difference between the two datasets (Var [Normal]–Var [Tumor]) using R-3.6.1. Homogeneity of variance testing was used to assess the heterogeneity of each gene.
Survival Analysis
We selected 21 genes with observably altered 3ʹ-UTRs and univariate prognostic value based on the P-value order and combined them with clinical covariates (gender, race, age, tumor stage, and surgical outcome) to conduct the study’s survival analysis. First, according to the PDUI score corresponding to the 21 genes, we selected the k-means grouping scheme with K = 3 to group patients with STADs. We used the standard Cox proportional hazard model implemented by the “survival” package in R to perform survival analysis, as well as Kaplan–Meier plots for each group of patients.25 We used the Log rank test to calculate the P-value.
Results
The mRNA-Seq data corresponding to 289 tumor samples was randomly selected from the TCGA-STAD study to analyze differences in the APA spectrum for comparing tumor and normal samples. Concurrently, considering the lack of data sources for normal comparable tissue in TCGA-STAD, 207 cases of normal gastric tissue data from the GTEx project were used as controls. The corresponding sample list is given in Table S1.
Since the data obtained from the TCGA-STAD project had been aligned to the hg38 genome, the data obtained from the GTEx project were processed using the TCGA data processing scheme, and the aligned BAM file was converted into a genomic coverage file for identifying 3ʹ-UTR differences. Finally, the dynamic analysis of an alternative DaPars algorithm using RNA-Seq data was employed to perform APA analysis on the coverage data to generate a percentage of distal PDUI scores for all of the given genes in each sample.18 The PDUI scores were obtained by quantifying the lengthened and shortened 3ʹ-UTR coverage levels. The gene corresponding to the lengthened 3ʹ-UTR (distal PAS) corresponded to a PDUI score close to 1, and the gene corresponding to the shortened 3ʹ-UTR (proximal PAS) corresponded to a PDUI score close to 0. The change in APA between the tumor and normal samples was characterized by ΔPDUI, which indicated the difference between the average PDUI scores of tumors and normal samples (ΔPDUI = MeanPDUI Tumor/MeanPDUI Normal). The ΔPDUI value was also used as a measure for correlating between the 3ʹ-UTR lengthening and shortening of the tumor samples. The final output was a PDUI matrix that included 1638 non-redundant unique genes, and each column included a tumor/normal sample (289 tumors + 207 normal = 496 columns; see Table S2).
Integrative Analysis of the Cancer Genome Atlas and the Cancer Genome Atlas–Stomach Adenocarcinoma Ribonucleic Acid Sequencing Data to Identify Dynamic Polyadenylation Events in Stomach Adenocarcinomas
We used a strict ΔPDUI threshold of = ±0.2 to further screen for significant APA events between the tumor and the normal samples (Figure 1A and B) to determine the extent of APA-mediated shortened and lengthened 3′-UTR in STADs. Although the ΔPDUI that corresponded to most genes did not exceed the threshold, the number of significantly shortened APA events was higher (the red dots, N = 149) compared with the significantly lengthened APA events (the blue dots, N = 122); all of the adjusted P-values corresponding to the significant APA event were less than 0.05. According to statistics, 55% of significant APA events were shortened 3ʹ-UTR events, which was consistent with the patterns observed in multiple pan-cancer and single-cancer analyses.17,18,26 The alternative lengths of STAD-related APA events were mainly distributed in 200–300 bp (Figure 1C). In addition, the number of shortened APA events with an alternative length greater than 500 bp was significantly larger compared with lengthened APA events (Figure 1C). We found that two genes in the Rho GTPases family of genes, CDC42 and RAC1, were significantly shortened APA events. The Rho GTPase family of enzymes is essential for multiple biological processes (including cell morphology, polarity, and migration) in many cancer types.27–31 In particular, the Rho GTPases family is also involved in the mechanism of tumorigenesis caused by the infection of gastric epithelial cells by Helicobacter pylori32 bacteria, microorganisms that are also the primary risk factor for stomach cancer.33 We visualized a 3ʹ-UTR coverage map of CDC42 and RAC1 between STAD and normal samples to confirm the shortening of the 3ʹ-UTR (Figure 1D). However, APA was not found in the most frequently mutated STAD genes (ie, TP53, ARID1A, FAT4, KMT2D, and PIK3CA; see Figure S1).
The KEGG pathway analysis of 271 significant 3ʹ-UTR-altered genes showed that these genes were mainly enriched in mRNA splicing and processing, as well as in oxidative phosphorylation pathways. Pan-cancer APA analysis identified similar pathways that were consistent with the presence of recurrent APA events in multiple cancer types.17,18 However, we observed that STAD-related pathways were also enriched, including non-alcoholic fatty liver disease, and epithelial cell signaling in Helicobacter pylori and salmonella infections (Figure 1E). Therefore, the alternative 3ʹ-UTR may regulate the activity of the STAD development pathway.
Alternative Polyadenylation Drives Gene Expression Disorders in Stomach Adenocarcinomas
The shortening of the 3ʹ-UTR during tumorigenesis can escape miRNA inhibition and up-regulate the expression of its parental gene.18 We calculated the number of highly conserved miRNA binding sites in the 3ʹ-UTR-shortened region (Figure 2A) to assess a pattern for the APA-mediated loss of miRNA binding sites in patients with STADs. Accordingly, we determined that 50% of the 3′-UTR-shortened genes lost at least one highly conserved miRNA binding site, suggesting that changes in the miRNA binding site pool were common patterns of APA-mediated regulation. In addition, by deriving the number of lost highly conserved miRNA binding sites, the miRNA family with the most missing miRNA binding sites was found to be miR-133a-3p, miR-133b, and miR-30-5p. (Figure 2B); miR-133a-3p is a tumor suppressor in many cancers and can regulate cell proliferation and differentiation.34–37 It has been reported that the overexpression of miR-133a-3p can prevent the activation of autophagy, destroy abnormal glutamine decomposition, and further inhibit the growth and metastasis of gastric cancer cells.38 In addition, miR-133 is reported to induce a negative adjustment of the CDC42-PAK pathway in gastric cancer.39 The analysis results of miRNA indicated that APA influenced the progress of STAD by changing the binding site-library of 3ʹ-UTR. To examine the consequences of the loss of miRNA binding sites in the 3ʹ-UTR, we calculated the differential gene expression between tumor and normal tissue types. The results indicated that the expression levels of genes with a shortened 3ʹ-UTR in tumors tended to be overexpressed (Figure 2C and D; P < 0.01, chi-square test).
The Heterogeneity of Proximal Polyadenylation Sites’ Use in Patients with Stomach Adenocarcinomas
The genetic and epigenetic mutations in tumors can promote the migration and invasion of cancer cells and, subsequently, stimulate the development of the tumor.40 Patients with pancreatic ductal adenocarcinoma showed significant heterogeneity in the use of proximal PAS.17 To identify whether patients with a STAD will also generate alternative 3ʹ-UTR lengths through APA heterogeneity and thus promote the progress of STADs, we compared the differences in the use of proximal PAS for all APA genes between STAD and normal samples. Taking CDC42 and ARF1 as examples, the PDUI scores in normal samples were closely distributed, while the distribution of PDUI scores in tumor samples was loose (Figure 3A and B). The distribution of PDUI in tumor samples had broader coverage, indicating that patients with a STAD showed greater heterogeneity in the use of proximal PAS. Although most genes did not show significant changes between normal and tumor samples (with a threshold of 0.015), the analysis of the use of proximal PAS in the case of all genes confirmed that 273 genes showed greater heterogeneity in tumor samples (the orange color), and only three genes showed significant heterogeneity in normal samples (the blue color, Figure 3C). There was no significant difference in the number of important heterogeneity genes between the 3′-UTR-shortened/lengthened genes (Figure S2). Using KEGG enrichment analysis, a subset of 273 genes was found to have primarily been enriched in pathways such as endocytosis, lysosome, and spliceosome pathways (Figure 3D).
Widespread Three Prime Untranslated Region Changes in Patients with Stomach Adenocarcinoma
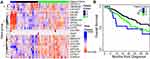
To further analyze the distribution of 3ʹ-UTR-altered genes in patients with STAD, we selected 235 genes from the 271 genes reviewed for changes in PDUI scores; the selected genes had corresponding PDUI values but did not have NA value (ΔPDUI)-based cluster analysis. It included 135 3′-UTR-shortened genes and 100 3′-UTR-lengthened genes (Figure 4). We found that a subset of genes (n = 34, top heatmap, Figure 4) reflected 3′-UTR-shortened genes (the color red, ΔPDUI ≤ –0.2) in more than 80% of patients. Among them, the ARF1 gene showed 3′-UTR-shortened events in 99.7% of patients. A small subset of genes (n = 18, bottom heatmap, Figure 4) was found in 3′-UTR-lengthened genes to be recurrently lengthened (the blue color, ΔPDUI ≥ 0.2) in most patients with a STAD. This showed that APA events were broadly distributed among patients with STADs. Hierarchical clustering was performed and, accordingly, patients with a STAD were divided into five categories (subgroups 1–5), which corresponded to different APA gene ΔPDUI distribution patterns. Subgroup 5 (the color purple) included 116 patients with more 3′-UTR-shortened genes and fewer 3′-UTR-lengthened genes. In contrast, subgroup 1 (the color blue) included 42 patients with more 3′-UTR-lengthened and fewer 3′-UTR-shortened genes. These subgroups were not related to the most frequently mutated STAD genes (ie, TP53, ARID1A, FAT4, KMT2D, and PIK3CA) or patient stage.
The Alternative Polyadenylation Events Contributed to the Assessment of the Prognosis of Patients with Stomach Adenocarcinomas
To further verify whether the APA event added prognostic information to patients with a STAD beyond the normal demographic and clinical factors (gender, race, age, tumor stage, and surgical outcome), we screened 21 genes that showed obvious 3ʹ-UTR changes and univariate prognostic value, as well as clustered patients with STADs, based on the PDUI scores of these 21 genes. Cluster analysis revealed three patient cohorts. Patients in cohort C showed an extensive degree of shortened 3ʹ-UTRs in all 21 genes; patients in cohort A showed shortening in the second group of genes, and patients in cohort B reflected primarily distal PAS (Figure 5A). There was a significant difference in the overall survival rate between cohorts A, B, and C (P = 0.04). The survival period of patients in cohort B was significantly longer than those in cohorts A (P = 0.05) and C (P = 0.03) (Figure 5B). In addition, the FPKM values of the most frequently mutated STAD genes (ie, TP53, ARID1A, FAT4, KMT2D, and PIK3CA) were compared. The results showed that there were differences in TP53, ARID1A, KMT2D, and PIK3CA among the three cohorts (Figure S3). The KMT2D and PIK3CA genes reflected relatively low expression in cohort B. Downregulation of KMT2D could potentially inhibit the proliferation and induce the apoptosis of gastric cancer,41 while the overexpression of PIK3CA may lead to a poor gastric cancer prognosis.42 Therefore, the APA distribution pattern can be used as an independent prognostic indicator in STAD cases, based on its potential for being a new prognostic biomarker.
Discussion
The disturbance of gene expression is the primary feature of cancer but our current understanding of its mechanism is limited.43 Recently, APA was identified as an important regulatory mechanism for the dysregulation of gene expression.17,44 Alternative polyadenylation is widely distributed in tumorigenesis. Some APA events are widely shared in different cancer types but many events are specific to tumor types.18,44,45 At present, APA analysis and research on STADs are limited. This work comprehensively characterized APA in patients with a STAD including its distribution, the loss of miRNA loci, changes in parental gene expression, heterogeneity, patient distribution, and prognostic information.
Tumor heterogeneity is a characteristic of malignant tumors. This means that during the growth of a tumor, its daughter cells show changes in morphological and phenotypic profiles following multiple divisions and proliferation, making the tumor different in terms of characteristics such as growth rate, invasive ability, sensitivity to drugs, and prognosis.46–48 Tumor heterogeneity can reflect different genetic backgrounds, such as differences in chromosome number and quality, different cell-case types, variable degrees of differentiation, and the diversity of cell evolution in different clinical stages.47,49 An in-depth study of tumor heterogeneity will help to analyze the mechanism and evolution of tumor formation and explain the reasons for the different reactivity in tumor treatments.46,48 Our data found that some of the genes of patients with STADs showed obvious heterogeneity to the extent of using proximal or distal PAS. The heterogeneity of these genes may be one of the key mechanisms causing tumor heterogeneity. Further study of the heterogeneity of the proximal PAS will support the screening of tumor biomarkers and therapeutic targets.
Endocytosis is the method by which large molecules or other cells enter a cell. The substance that enters the cell through the endocytosis pathway will participate in a series of life activities inside the cell, which can cause malignant transformations and contribute to the formation of cancer.50,51 The endocytic circulation of cells can promote the plasticity, infiltration, and metastasis of cancer. However, there is little research evidence that genetic changes in endoproteins cause high incidences of cancer in humans. Our data confirmed that in the case of STAD, both significant APA genes and heterogeneous APA genes were enriched in the endocytosis pathway. The 3ʹ-UTR changes to these genes may also participate in the endocytosis cycle of tumor cells and promote the progression of STADs.
We observed that the distribution of APA events varied among patients with a STAD but most of these patients had more 3′-UTR-shortened genes. Studies have shown that in proliferating cells, the upregulation of genes related to cell growth was consistent with shortening of the 3ʹ-UTR,52,53 which was consistent with the phenomenon we observed. We found that genes with a shortened 3ʹ-UTR were more likely to overexpress, possibly due to the regulation of missing miRNA binding sites. The miRNA can suppress gene expression by combining the complementary sequence in the 3ʹ-UTR of target mRNAs to degrade them, thereby preventing their translation.54 Therefore, APA can affect the interaction of miRNA–mRNA, which is an important aspect of post-transcriptional regulation and is considered a fundamental mediator of gene expression involved in many types of cancers.8 In addition, our data showed that the APA gene in patients with a STAD could be used as an independent prognostic indicator of STAD results. However, since this article is mainly based on the analysis of second-generation sequencing data, more confirmatory experiments are urgently needed; the description of results in this paper may have been overestimated due to a lack of experimental data.
Overall, our research revealed that APA events regulated STAD genes that were functionally related, thereby providing a new way for better understanding the progress of STADs and identifying new drug targets.
Data Sharing Statement
The data set is within the paper and its Supplementary material, with additional supporting material (RNA-seq data) at controlled-access portal providing data on TCGA (https://portal.gdc.cancer.gov/) and GTEx (dbGaP Study Accession: phs000424.v7.p2. https://gtexportal.org) according to NIH Genomic Data Sharing (GDS) Policy. These restrictions are imposed by NIH Data Access Committee. Access to these RNA-seq data will be granted by an NIH Data Access Committee and can be requested here (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login) for researchers who meet the criteria for access to controlled-access data. We confirm that we had no special access privileges to the data utilized that others would not have.
Acknowledgments
We appreciate Qin Su for assistance with the manuscript.
Funding
Funding information is not applicable.
Disclosure
The authors declare that they have no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
2. Kono T, Imai Y, Ichihara T, et al. Adenocarcinoma arising in gastric inverted hyperplastic polyp: a case report and review of the literature. Pathol Res Pract. 2007;203:53–56. doi:10.1016/j.prp.2006.08.010
3. Zhu Y-H, Jeong S, Wu M, et al. Dietary intake of fatty acids, total cholesterol, and stomach cancer in a Chinese population. Nutrients. 2019;11:1730. doi:10.3390/nu11081730
4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi:10.3322/caac.21551
5. Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet (London, England). 2018;391:1023–1075.
6. Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18:18–30. doi:10.1038/nrm.2016.116
7. Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi:10.1016/j.molcel.2011.08.017
8. Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14:496–506. doi:10.1038/nrg3482
9. Mayr C. Evolution and biological roles of alternative 3ʹUTRs. Trends Cell Biol. 2016;26:227–237. doi:10.1016/j.tcb.2015.10.012
10. Mayr C. What are 3ʹ UTRs doing? Cold Spring Harb Perspect Biol. 2019;11:10.
11. Wang L, Chen M, Fu H, Ni T, Wei G. Tempo-spatial alternative polyadenylation analysis reveals that 3′ UTR lengthening of Mdm2 regulates p53 expression and cellular senescence in aged rat testis. Biochem Biophys Res Commun. 2020;523:1046–1052. doi:10.1016/j.bbrc.2020.01.061
12. Lackford B, Yao C, Charles GM, et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 2014;33:878–889. doi:10.1002/embj.201386537
13. Brumbaugh J, Di Stefano B, Wang X, et al. Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell. 2018;172:106–120.e121. doi:10.1016/j.cell.2017.11.023
14. Chen M, Lyu G, Han M, et al. 3ʹ UTR lengthening as a novel mechanism in regulating cellular senescence. Genome Res. 2018;28:285–294. doi:10.1101/gr.224451.117
15. Chen X, Zhang JX, Luo JH, et al. CSTF2-induced shortening of the RAC1 3ʹUTR promotes the pathogenesis of urothelial carcinoma of the bladder. Cancer Res. 2018;78:5848–5862.
16. Miles WO, Lembo A, Volorio A, et al. Alternative Polyadenylation in Triple-Negative Breast Tumors Allows NRAS and c-JUN to Bypass PUMILIO Posttranscriptional Regulation. Cancer Res. 2016;76:7231–7241. doi:10.1158/0008-5472.CAN-16-0844
17. Venkat S, Tisdale AA, Schwarz JR, et al. Alternative polyadenylation drives oncogenic gene expression in pancreatic ductal adenocarcinoma. Genome Res. 2020;30:347–360. doi:10.1101/gr.257550.119
18. Xia Z, Donehower LA, Cooper TA, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3ʹ-UTR landscape across seven tumour types. Nat Commun. 2014;5:5274. doi:10.1038/ncomms6274
19. Feng X, Li L, Wagner EJ, Li W. TC3A: the Cancer 3′ UTR Atlas. Nucleic Acids Res. 2017;46:D1027–D1030. doi:10.1093/nar/gkx892
20. Li L, Wang D, Xue M, Mi X, Liang Y, Wang P. 3′UTR shortening identifies high-risk cancers with targeted dysregulation of the ceRNA network. Sci Rep. 2014;4:5406. doi:10.1038/srep05406
21. Masamha CP, Xia Z, Yang J, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–416. doi:10.1038/nature13261
22. Chang K, Creighton CJ, Davis C, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi:10.1038/ng.2764
23. GTEx. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi:10.1126/science.1262110
24. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi:10.1089/omi.2011.0118
25. Andersen PK, Gill RD. Cox’s regression model for counting processes: a large sample study. Annals Stat. 1982;1100–1120. doi:10.1214/aos/1176345976
26. Xiang Y, Ye Y, Lou Y, et al. Comprehensive Characterization of Alternative Polyadenylation in Human Cancer. J Natl Cancer Inst. 2018;110:379–389. doi:10.1093/jnci/djx223
27. Haga RB, Ridley AJ. Rho GTPases: regulation and roles in cancer cell biology. Small GTPases. 2016;7:207–221. doi:10.1080/21541248.2016.1232583
28. Hanna S, El-Sibai M. Signaling networks of Rho GTPases in cell motility. Cell Signal. 2013;25:1955–1961. doi:10.1016/j.cellsig.2013.04.009
29. Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi:10.1038/nature01148
30. Lin Y, Zheng Y. Approaches of targeting Rho GTPases in cancer drug discovery. Expert Opin Drug Discov. 2015;10:991–1010. doi:10.1517/17460441.2015.1058775
31. Nam S, Kim JH, Lee DH. RHOA in Gastric Cancer: functional Roles and Therapeutic Potential. Front Genet. 2019;10:438. doi:10.3389/fgene.2019.00438
32. Wessler S, Gimona M, Rieder G. Regulation of the actin cytoskeleton in Helicobacter pylori-induced migration and invasive growth of gastric epithelial cells. Cell Commun Signaling. 2011;9:27. doi:10.1186/1478-811X-9-27
33. Chiou CC, Chan CC, Sheu DL, Chen KT. Helicobacter pylori infection induced alteration of gene expression in human gastric cells. Gut. 2001;48:598–604. doi:10.1136/gut.48.5.598
34. Tao J, Wu D, Xu B, et al. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol Rep. 2012;27:1967–1975.
35. Zhang X-T, Zhang Z, Xin Y-N, Ma X-Z, Xuan S-Y. Impairment of growth of gastric carcinoma by miR-133-mediated Her-2 inhibition. Tumor Biol. 2015;36:8925–8930. doi:10.1007/s13277-015-3637-2
36. Zhou Y, Wu D, Tao J, Qu P, Zhou Z, Hou J. MicroRNA-133 inhibits cell proliferation, migration and invasion by targeting epidermal growth factor receptor and its downstream effector proteins in bladder cancer. Scand J Urol. 2013;47:423–432. doi:10.3109/00365599.2012.748821
37. Guo L, Huang W, Chen B, et al. gga-mir-133a-3p Regulates Myoblasts Proliferation and Differentiation by Targeting PRRX1. Front Genet. 2018;9:577. doi:10.3389/fgene.2018.00577
38. Zhang X, Li Z, Xuan Z, et al. Novel role of miR-133a-3p in repressing gastric cancer growth and metastasis via blocking autophagy-mediated glutaminolysis. J Exp Clin Cancer Res. 2018;37:320. doi:10.1186/s13046-018-0993-y
39. Cheng Z, Liu F, Wang G, Li Y, Zhang H, Li F. miR-133 is a key negative regulator of CDC42-PAK pathway in gastric cancer. Cell Signal. 2014;26:2667–2673. doi:10.1016/j.cellsig.2014.08.012
40. Hinohara K, Polyak K. Intratumoral Heterogeneity: more Than Just Mutations. Trends Cell Biol. 2019;29:569–579. doi:10.1016/j.tcb.2019.03.003
41. Xiong W, Deng Z, Tang Y, Deng Z, Li M. Downregulation of KMT2D suppresses proliferation and induces apoptosis of gastric cancer. Biochem Biophys Res Commun. 2018;504:129–136. doi:10.1016/j.bbrc.2018.08.143
42. Liang M, Shi B, Liu J, et al. Downregulation of miR203 induces overexpression of PIK3CA and predicts poor prognosis of gastric cancer patients. Drug Des Devel Ther. 2015;9:3607–3616.
43. Yaffe MB. Why geneticists stole cancer research even though cancer is primarily a signaling disease. Sci Signal. 2019;12:eaaw3483. doi:10.1126/scisignal.aaw3483
44. Yuan F, Hankey W, Wagner EJ. Alternative polyadenylation of mRNA and its role in cancer. Genes Dis. 2019;1–12. doi:10.1016/j.gendis.2019.10.011
45. Xue Z, Warren RL, Gibb EA, et al. Recurrent tumor-specific regulation of alternative polyadenylation of cancer-related genes. BMC Genom. 2018;19:536. doi:10.1186/s12864-018-4903-7
46. Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94. doi:10.1038/nrclinonc.2017.166
47. Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi:10.1038/nature12624
48. Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108:479–485. doi:10.1038/bjc.2012.581
49. Cassidy JW, Bruna A. Chapter 4 - Tumor Heterogeneity. In: Uthamanthil R, Tinkey P, editors. Patient Derived Tumor Xenograft Models. Academic Press; 2017:37–55.
50. Mellman I, Yarden Y. Endocytosis and cancer. Cold Spring Harb Perspect Biol. 2013;5:a016949. doi:10.1101/cshperspect.a016949
51. Lanzetti L, Di Fiore PP. Behind the Scenes: endo/Exocytosis in the Acquisition of Metastatic Traits. Cancer Res. 2017;77:1813–1817. doi:10.1158/0008-5472.CAN-16-3403
52. Lee S-H, Singh I, Tisdale S, Abdel-Wahab O. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature. 2018;561:127–131. doi:10.1038/s41586-018-0465-8
53. Singh P, Alley TL, Wright SM, et al. Global Changes in Processing of mRNA 3′ Untranslated Regions Characterize Clinically Distinct Cancer Subtypes. Cancer Res. 2009;69:9422. doi:10.1158/0008-5472.CAN-09-2236
54. Mao Z, Zhao H, Qin Y, et al. Post-Transcriptional Dysregulation of microRNA and Alternative Polyadenylation in Colorectal Cancer. Front Genet. 2020;11:64. doi:10.3389/fgene.2020.00064
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.