Back to Journals » OncoTargets and Therapy » Volume 8
The histone demethylase PHF8 promotes prostate cancer cell growth by activating the oncomiR miR-125b
Authors Ma Q, Chen Z, Jia G, Lu X, Xie X, Jin W
Received 25 March 2015
Accepted for publication 19 May 2015
Published 10 August 2015 Volume 2015:8 Pages 1979—1988
DOI https://doi.org/10.2147/OTT.S85443
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Daniele Santini
Qiang Ma,* Zhuo Chen,* Guojin Jia, Xueqiang Lu, Xuefeng Xie, Wei Jin
Department of Urology, Jinshan Hospital, Fudan University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Aims: Prostate cancer (PCa) is the most frequently diagnosed malignancy in men. However, the underlying mechanism is not fully understood. In this study, we aim to research the molecular mechanisms underlying the initiation and progression of PCa.
Results: Plant homeodomain finger protein 8 (PHF8) is upregulated in human PCa tissues and cell lines. PHF8 knockdown attenuates growth and cellular transformation of PCa cells. PHF8 depletion induces PCa cell apoptosis by activating proapoptotic proteins and inactivating antiapoptotic proteins. Furthermore, miR-125b is a target of PHF8, and miR-125b seems to be essential for the hyper proliferation of PCa cells in the presence of PHF8.
Conclusion: In conclusion, we identify the histone demethylase PHF8 as an oncogenic protein in human PCa. These findings indicate PHF8 as a potential candidate for clinical intervention.
Keywords: PHF8, prostate cancer, apoptosis, miR-125b
Introduction
Prostate cancer (PCa) is the most frequently diagnosed malignancy in men and the second leading cause of cancer deaths among men in Western countries.1 There is still an urgent need for appropriate diagnostic and prognostic markers for PCa, in addition to the established serum protease prostate-specific antigen. Although the involvement of certain genes and chromosomal aberrations in prostate carcinogenesis has been suggested,2,3 the molecular mechanisms underlying the initiation and progression of PCa are poorly understood.
Plant homeodomain finger protein 8 (PHF8) is a JmjC domain-containing protein and erases repressive histone marks, including H4K20me1 and H3K9me1/2.4–6 PHF8 binds to H3K4me3, an active histone mark usually located at transcription start sites, through its plant homeodomain, and is thus recruited and enriched at promoters of target genes.7,8 Chromatin immunoprecipitation-sequencing data from HeLa cells show that approximately 72% of PHF8-binding sites are at promoters.5 Also, PHF8 regulates the cell cycle biological processes via removing H4K20me1 from the promoters of certain E2F1-regulated genes.4 Interestingly, altering PHF8 levels in HeLa cells affects H4K20me1 methylation only in late G2/M and early G1 stages of the cell cycle, not globally.4 PHF8 also binds to rRNA gene promoters and demethylates H3K9me2/1 to activate rRNA synthesis,6,9 functioning as an activator.10 However, the role of PHF8 in human diseases, especially carcinogenesis, is largely unknown.
MicroRNAs have been widely considered in the initiation and development of cancer. Both local and circulating microRNAs have been reported to participate in human PCa.11–14 For instance, the oncomiR miR-125b has been demonstrated to promote the development of PCa, and the underlying mechanism has been widely investigated.13,15–19 Nevertheless, how miR-125b is regulated in PCa remains elusive.
In the present work, we find that PHF8 is overexpressed in human PCa tissues and cell lines. The results show that PHF8 regulates PCa cell growth, cellular transformation, and survival by regulating the oncomiR miR-125b.
Materials and methods
Patient identification and pathology evaluation
Radical prostatectomy specimens were available from 89 patients treated at the Jinshan Hospital, Fudan University (Shanghai) between 2001 and 2009. Tumors were diagnosed and classified according to the Gleason system. The 19 adjacent normal tissues were obtained from PCa patients undergoing surgery. A written form of informed consent was obtained from all the patients, and the study was approved by the Clinical Research Ethics Committee of Fudan University.
Immunohistochemical assay
Tissues were fixed with 4% neutral formalin. Cancer or adjacent tissue sections were subjected to immunohistochemical staining with anti-PHF8 antibody. Paraffin sections were subjected to high-temperature antigen retrieval, 3 minutes in a pressure cooker in 0.01 M citrate buffer pH 6.0. Slides were treated with 3% H2O2 for 15 minutes, blocked in 5% normal horse serum in phosphate-buffered saline for 20 minutes, and stained with anti-PHF8 antibody in 5% normal goat serum overnight at 4°C. Secondary antibody was used according to VECTASTAIN ABC® kits, followed by DAB staining. The areas of total prostate tumors and PHF8-positive areas were quantified using ImageJ. The average percentage of PHF8-positive area is 31%. This median value was used to cut off the subgroups of all immunohistochemical variables in our data. The patients were then divided into two groups: PHF8 high expression group (≥31% PHF8-positive/total tissue cores, n=41) and PHF8 low expression group (<31% PHF8-positive/total tissue cores, n=48).
Western blot
PCa tissues and cells were lysed with cell lysis buffer (Beyotime Institute of Technology, Shanghai, People’s Republic of China). Total proteins were applied to SDS–polyacrylamide gel. After electrophoresis, the proteins were transferred to poly(vinylidene fluoride) membranes, followed by blocking in the buffer containing 5% fat-free dry milk. The membranes were then probed with indicated antibodies overnight, and then washed and incubated with HRP-conjugated secondary antibodies (Zhongsan Jinqiao Biotechnology, Beijing, People’s Republic of China) for 1.5 hours, and finally, visualized using Chemiluminescent ECL reagent (Vigorous Biotechnology, Beijing, People’s Republic of China). The following antibodies were used: anti-GAPDH (Cell Signaling Technology, Beverly, MA, USA), anti-PHF8 (Cell Signaling Technology), anti-poly(ADP-ribose) polymerase (Abcam, Cambridge, UK), anti-caspase 3 (Abcam), anti-Bax (Abcam), anti-Bcl-2 (Santa Cruz Biotechnology Inc., Dallas, TX, USA), and anti-p21 (Santa Cruz).
Quantitative real-time PCR
Total RNA was extracted from cells or tissues with TRIzol®, and cDNA was synthesized from 1 μg of RNA with one-Step RT-PCR Kit. q-PCR was performed with the SYBR Green detection method on an ABI-7500 RT-PCR system (Thermo Fisher Scientific, Waltham, MA, USA). GAPDH was used as housekeeping gene. The primers used are listed in Table S1.
Cell lines and cell culture
The human prostate carcinoma cell lines LNCaP, DU145, and PC3 were obtained from the American Type Culture Collection ([ATCC], Manassas, VA, USA), PTN2 from Sigma-Aldrich Co. (St Louis, MO, USA), and BPH-1 from YRGenge (Changsha, Hunan, the People’s Republic of China), and were grown in RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific).
Package of retrovirus and transduction
Control sh-RNA and specific sh-RNAs targeting PHF8 were purchased from Thermo Fisher Scientific, and the corresponding sequences were cloned into the pSIREN-RetroQ plasmid (Addgene, Cambridge, MA, UK) for retrovirus production with 293T cells. The targeting sequences are listed in Table S2. For transduction, 293T cells were incubated with virus-containing supernatant in the presence of 8 mg/mL polybrene. After 48 hours, infected cells were selected with puromycin (2 mg/mL). Then, the clones were picked and cultured for further experiment.
For transfection, the locked nucleic acid (LNA)-antimiR-125b or LNA-control (Exiqon A/S, Skelstedet, Vedbaek, Denmark) was delivered at a final concentration of 50 nM using Lipofectamine® 2000 reagent (Thermo Fisher Scientific).
Cell proliferation assay
Cell proliferation rate was tested with MTT assay. The MTT assay was performed using MTT Cell Proliferation and Cytotoxicity Assay Kit (R&D Systems, Inc., Minneapolis, MN, USA) according to the manufacturer’s protocol.
Soft sugar colony formation assay
PCa cells were suspended in 1.5 mL complete medium supplemented with 0.45% low melting point agarose (Thermo Fisher Scientific). The cells were placed in 35 mm tissue culture plates containing 1.5 mL complete medium and agarose (0.75%) on the bottom layer. The plates were incubated at 37°C with 5% CO2 for 2 weeks. Cell colonies were stained with 0.005% crystal violet and analyzed using a microscope.
In vivo tumor growth experiment
Xenograft mice experiments were performed as described previously.20 In brief, equal numbers of PHF8 cells expressing either control or PHF8 knockdown vectors were injected subcutaneously, within 30 minutes of harvesting, over the right and left flanks in male nu/nu mice between 4 weeks and 6 weeks of age. Tumor growth was monitored using calipers. The mice were killed, and the tumors were weighted 3 weeks after inoculation. N=10 in each group. The experimental protocol used in this study has been reviewed and approved by the Jinshan Hospital affiliated to Fudan University -Institutional Animal Care and Use Committee on their ethical procedures and scientific care.
Apoptotic assays
Cells were washed twice with cold PBS, and then resuspended in 1× binding buffer (BD Biosciences, San Jose, CA, USA) at a concentration of 1×106 cells/mL. Then, 100 mL of the solution (1×105 cells) was transferred to a 5 mL culture tube and stained with 5 mL each of allophycocyanin–annexin V (BD Biosciences) and 50 mg/mL propidium iodide (Thermo Fisher Scientific). The cells were gently mixed and incubated at room temperature for 15 minutes. For assessment of apoptosis, 400 mL of 1× binding buffer was then added to each tube and the samples were analyzed by flow cytometry using a LSRII instrument (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
The induction of apoptosis was also monitored by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) method. The TUNEL assay was performed according to the guidelines recommended by the TUNEL Apoptosis Kit (R&D Systems, Inc.).
Luciferase assay
To generate the luciferase reporter vectors, an internal ribosome entry site was amplified from pMSCV-PIG and cloned into the BglII site of pGL3-Basic (Promega Corporation, Fitchburg, WI, USA). miR-125b promoter fragments21 were amplified from human genomic DNA and cloned into the XhoI site. Twenty-four hours before transfection, 7×104 cells were plated per well in a 24-well plate. pGL3 constructs (100 ng) plus 1 ng of the Renilla luciferase plasmid phRL-SV40 (Promega) were transfected using FuGENE 6 (Hoffman-La Roche Ltd., Basel, Switzerland). Twenty-four hours after transfection, luciferase assays were performed using the dual luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla PHF8-mediated transactivation of miR-125b luciferase activity for each transfected well. Full-length human PHF8 was cloned into the pcDNA4 vector for expression in mammalian cells. For each experimental trial, wells were transfected in triplicate and each well was assayed in triplicate. For each condition tested, the luciferase activity was normalized to the activity produced from empty vector.
Statistical analysis
All the values are expressed as the mean ± standard error of mean (SEM). Statistical differences between the two groups were determined using Student’s t-test. The correlation of PHF8 immunoreactivity with patients’ clinicopathological variables was analyzed by the χ2 test or Fisher’s exact test. The Kaplan–Meier method was used to estimate overall survival. Survival differences according to PHF8 or miR-125b expression were analyzed by the log-rank test. Regression analysis was performed using GraphPad Prism®. P-values of <0.05 were considered statistically significant.
Results
PHF8 is upregulated in human PCa tissues and cell lines
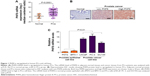
To investigate the potential role of the histone demethylase PHF8 in human PCa, we first profiled the expression pattern of PHF8 in human normal prostate tissues and cancer tissues. We analyzed 19 normal tissues and 89 PCa tissues and found that PHF8 was upregulated in human PCa tissues at both mRNA and protein levels (Figure 1A and B). Furthermore, we analyzed the expression of PHF8 in three human PCa cell lines (LNCaP, PC-3, and DU145) and normal prostate epithelial cell lines (PNT2 and BPH-1). The results showed that PHF8 mRNA levels were high in PCa cells compared to normal epithelial cells (Figure 1C). Taken together, these findings demonstrate that PHF8 is upregulated in human PCa and indicate the potential role of PHF8 in human PCa.
PHF8 regulates PCa growth and cellular transformation
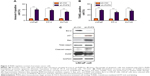
Since our results indicated that PHF8 is involved in human PCa, we explored whether PHF8 regulates the cellular behaviors of PCa cells. Therefore, we knocked down PHF8 in PCa cell lines, LNCaP, PC-3, and DU145. We found that PHF8 knockdown reduced cellular proliferation rate of PCa cells in vitro (Figure 2A–C). We next probed the potential contribution of PHF8 to the transformative properties of PCa cells. We observed that PHF8-depleted cells possessed reduced colony-forming activity in LNCaP, PC-3, and DU145 cells (Figure 2D–F). Furthermore, we generated prostate cells with stably PHF8 knockdown and performed xenograft experiment. The results showed that PHF8 knockdown significantly inhibited the growth of PCa cells in vivo (Figure 2G–I). These evidences demonstrate that PHF8 promotes PCa cell growth and cellular transformation.
PHF8 regulates apoptosis of PCa cells
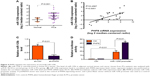
To investigate whether PHF8 regulates PCa cell proliferation and colony formation by orchestrating survival, we performed fluorescence-activated cell sorting to detect apoptotic cells. We found that PHF8 knockdown significantly increased the number of apoptotic cells in LNCaP, PC-3, and DU145 cells (Figure 3A). We next performed TUNEL assay to investigate whether DNA damage is involved in apoptosis induced by PHF8 knockdown. Markedly, PHF8 knockdown increased the number of TUNEL-positive cells in PCa cells, indicating that PHF8 knockdown induced DNA damage (Figure 3B). Finally, we performed Western blot to analyze the apoptotic signaling pathways. The results indicated that PHF8 knockdown activated apoptotic pathway and inactivated antiapoptotic pathway. In detail, PHF8 knockdown increased the protein level of Bax, p21, cleaved caspase 3, and cleaved PARP, while the antiapoptotic protein Bcl-2 was downregulated when PHF8 was knocked down (Figures 3C and S1). Taken together, PHF8 regulates survival of human PCa cells.
PHF8 promotes miR-125b expression in human PCa
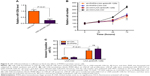
miR-125b is widely accepted as an oncogenic microRNA, and its role has been deeply investigated in human PCa. miR-125b was reported to be an important antiapoptotic factor and regulated drug resistance in human PCa. Our q-PCR results also showed that miR-125b was upregulated in human PCa tissues (Figure 4A), which was consistent with previous reports.13 Furthermore, we performed linear regression analysis to figure out whether miR-125b level is correlated with PHF8 expression in human PCa. The results indicated that miR-125b level was significantly and positively correlated with PHF8 level in human PCa tissues (Figure 4B), indicating that PHF8 may regulate miR-125b. We next knocked down PHF8 in LNCaP cells and found that PHF8 knockdown significantly reduced the level of miR-125b (Figure 4C). Finally, our luciferase assay demonstrated that PHF8 overexpression enhanced the promoter activity of miR-125b (Figure 4D). Taken together, these results demonstrate that PHF8 regulates miR-125b expression in human PCa.
MiR-125b is critically for PHF8 function in PCa
As we have demonstrated that PHF8 regressed the expression of miR-125b, we wanted to know whether miR-125b is essential for the function of PHF8 in human PCa. We knocked down miR-125b in LNCaP cells using specific LNA-antimiR-125b (Figure 5A). We found that miR-125b knockdown reduced LNCaP cell proliferation and induced apoptosis (Figure 5B and C). However, PHF8 knockdown was unable to affect LNCaP cell proliferation and apoptosis when miR-125b was knocked down (Figure 5B and C). These results demonstrate that miR-125b is critically essential for the function of PHF8 in human PCa.
Discussion
PHF8 functions essentially in many developmental and disease processes. Mutations in PHF8 are associated with X-linked mental retardation and cleft lip/cleft palate.10,22–24 Histone demethylation modulated by PHF8 is suggested to play a critical role in neuronal differentiation, brain, and craniofacial development.4,25 Recently, PHF8 was reported to participate in several types of cancers, including leukemia, esophageal squamous cell carcinoma (ESCC), non-small-cell lung cancer, and PCa. In acute promyelocytic leukemia, PHF8 governs retinoic acid response. All-trans retinoic acid (ATRA) sensitivity depends on the enzymatic activity and phosphorylation status of PHF8, which in turn can be pharmacologically manipulated to resurrect ATRA sensitivity to resistant cells.26 Oncogenic features of PHF8 in ESCC were also reported.27 Currently, PHF8 was shown to regulate lung cancer cells growth and survival by targeting miR-21.28
Björkman et al29 carried out a systematic, genome-wide analysis of the functional significance of 615 epigenetic proteins in PCa cells. They identified that PHF8 was overexpressed in PCa with an impact on cell proliferation, migration, and invasion.29 Inconsistent with the finding of Björkman et al29 we found that PHF8 was overexpressed in human PCa tissues and cell lines (Figure 1). In the present study, we also found that PHF8 knockdown inhibits cellular proliferation of PCa cells (LNCaP, PC-3, and DU145) in vitro and in vivo (Figure 2). Furthermore, cellular transformation of PCa cells was also reduced by PHF8 knockdown (Figure 2). In addition, we showed that PHF8 knockdown induces apoptosis in PCa cells by upregulating apoptotic protein (Bax, p-21, cleaved caspase 3, and cleaved PARP) and downregulating the antiapoptotic protein Bcl-2. Noticeably, DNA damage is involved in this effect (Figure 3). Together with the previous reports, our evidence indicates that PHF8 regulates cellular proliferative, transformative, and survival behaviors of cancer cells.
The oncomiR miR-125b has been reported to participate in human PCa.19 miR-125b was found to have the ability of rendering LNCaP cells resistant to androgen withdrawal. It was found to be androgen regulated and one of its targets, BAK1, was identified as being involved in how these PCa cells undergo apoptosis functionally.30 miR-125b promotes growth of PCa xenograft tumor through targeting proapoptotic genes.18 Both p53-dependent and p53-independent pathways are involved in miR-125b-mediated apoptosis in PCa.16 For a long time, the underlying mechanism by which miR-125b is regulated remained unknown. We found that PHF8 could activate the promoter of miR-125b and promote its expression in human PCa cells (Figure 4). PHF8 knockdown induced apoptosis in PCa cells, and p53 may be involved in this process, because p21, a downstream target of p53 was upregulated when PHF8 was knocked down (Figure 3), which may be attributed to the downregulation of miR-125b. Finally, we demonstrated that miR-125b was critically essential for PHF8 effects on proliferation and apoptosis of PCa cells (Figure 5).
Conclusion
In conclusion, we identify the histone demethylase PHF8 as an oncogenic protein in human PCa. These findings indicate PHF8 as a potential candidate for clinical intervention.
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics, 2003. CA Cancer J Clin. 2003;53:5–26. | ||
Karan D, Lin MF, Johansson SL, Batra SK. Current status of the molecular genetics of human prostatic adenocarcinomas. Int J Cancer. 2003;103:285–293. | ||
Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–2434. | ||
Qi HH, Sarkissian M, Hu GQ, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. | ||
Liu W, Tanasa B, Tyurina OV, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. | ||
Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17:445–450. | ||
Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–887. | ||
Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and polycomb-repressive complex 2. Genes Dev. 2008;22:1345–1355. | ||
Zhu Z, Wang Y, Li X, et al. PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res. 2010;20:794–801. | ||
Laumonnier F, Holbert S, Ronce N, et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet. 2005;42:780–786. | ||
Lin HM, Castillo L, Mahon KL, et al. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. Br J Cancer. 2014;110:2462–2471. | ||
Alshalalfa M, Bader GD, Goldenberg A, Morris Q, Alhajj R. Detecting microRNAs of high influence on protein functional interaction networks: a prostate cancer case study. BMC Syst Biol. 2012;6:112. | ||
Sun D, Layer R, Mueller AC, et al. Regulation of several androgen-induced genes through the repression of the miR-99a/let-7c/miR-125b-2 miRNA cluster in prostate cancer cells. Oncogene. 2014;33:1448–1457. | ||
Nguyen HC, Xie W, Yang M, et al. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate. 2013;73:346–354. | ||
Feng N, Xu B, Tao J, et al. A miR-125b binding site polymorphism in bone morphogenetic protein membrane receptor type IB gene and prostate cancer risk in China. Mol Biol Rep. 2012;39:369–373. | ||
Amir S, Ma AH, Shi XB, Xue L, Kung HJ, Devere White RW. Oncomir miR-125b suppresses p14(ARF) to modulate p53-dependent and p53-independent apoptosis in prostate cancer. PLoS One. 2013;8:e61064. | ||
Giangreco AA, Vaishnav A, Wagner D, et al. Tumor suppressor microRNAs, miR-100 and -125b, are regulated by 1,25-dihydroxyvitamin D in primary prostate cells and in patient tissue. Cancer Prev Res (Phila). 2013;6:483–494. | ||
Shi XB, Xue L, Ma AH, Tepper CG, Kung HJ, White RW. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate. 2011;71:538–549. | ||
Lin KY, Zhang XJ, Feng DD, et al. miR-125b, a target of CDX2, regulates cell differentiation through repression of the core binding factor in hematopoietic malignancies. J Biol Chem. 2011;286:38253–38263. | ||
Han W, Xin Z, Zhao Z, et al. RNA-binding protein PCBP2 modulates glioma growth by regulating FHL3. J Clin Invest. 2013;123:2103–2118. | ||
Kim JK, Noh JH, Jung KH, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. | ||
Koivisto AM, Ala-Mello S, Lemmelä S, Komu HA, Rautio J, Järvelä I. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin Genet. 2007;72:145–149. | ||
Loenarz C, Ge W, Coleman ML, et al. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum Mol Genet. 2010;19:217–222. | ||
Kleine-Kohlbrecher D, Christensen J, Vandamme J, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38:165–178. | ||
Qiu J, Shi G, Jia Y, et al. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res. 2010;20:908–918. | ||
Arteaga MF, Mikesch JH, Qiu J, et al. The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell. 2013;23:376–389. | ||
Sun X, Qiu JJ, Zhu S, et al. Oncogenic features of PHF8 histone demethylase in esophageal squamous cell carcinoma. PLoS One. 2013;8:e77353. | ||
Shen Y, Pan X, Zhao H. The histone demethylase PHF8 is an oncogenic protein in human non-small cell lung cancer. Biochem Biophys Res Commun. 2014;451:119–125. | ||
Björkman M, Östling P, Härmä V, et al. Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion. Oncogene. 2012;31:3444–3456. | ||
DeVere White RW, Vinall RL, Tepper CG, Shi XB. MicroRNAs and their potential for translation in prostate cancer. Urol Oncol. 2009;27:307–311. |
Supplementary materials
  | Table S1 Primers for qPCR |
  | Table S2 sh-RNA used to targeting PHF8 |
  | Figure S1 Relative level of cleaved PARP related to Figure 3C. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.