Back to Journals » International Journal of General Medicine » Volume 15
The Features of BRCA1 and BRCA2 Germline Mutations in Hakka Ovarian Cancer Patients: BRCA1 C.536 A>T Maybe a Founder Mutation in This Population
Authors Luo Y, Wu H , Huang Q, Rao H, Yu Z, Zhong Z
Received 31 December 2021
Accepted for publication 1 March 2022
Published 10 March 2022 Volume 2022:15 Pages 2773—2786
DOI https://doi.org/10.2147/IJGM.S355755
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Yu Luo,1,2,* Heming Wu,2,3,* Qingyan Huang,2,3 Hui Rao,2,3 Zhikang Yu,2,3 Zhixiong Zhong2,3
1Department of Gynaecology, Meizhou People’s Hospital (Huangtang Hospital), Meizhou Academy of Medical Sciences, Meizhou, People’s Republic of China; 2Guangdong Provincial Key Laboratory of Precision Medicine and Clinical Translational Research of Hakka Population, Meizhou People’s Hospital (Huangtang Hospital), Meizhou Academy of Medical Sciences, Meizhou, People’s Republic of China; 3Center for Precision Medicine, Meizhou People’s Hospital (Huangtang Hospital), Meizhou Academy of Medical Sciences, Meizhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhikang Yu; Zhixiong Zhong, Center for Precision Medicine, Guangdong Provincial Key Laboratory of Precision Medicine and Clinical Translational Research of Hakka Population, Meizhou People’s Hospital (Huangtang Hospital), Meizhou Academy of Medical Sciences, No. 63 Huangtang Road, Meijiang District, Meizhou, 514031, People’s Republic of China, Tel +753-2131-591, Email [email protected]; [email protected]
Objective: To investigate the frequencies of BRCA1 and BRCA2 mutations in Chinese Hakka patients with ovarian cancer.
Methods: The protein coding regions and exon intron boundary regions of the BRCA gene were sequenced using genomic DNA isolated from the lymphocytes of patients with next-generation sequencing. The patients’ family history and clinical records were collected.
Results: A total of 195 patients with ovarian cancer were included in the study, and 52 distinct variants of the BRCA gene were identified. It was found that 64 patients (64/195, 32.8%) had BRCA gene mutations, including 32 patients (50.0%) with BRCA1 mutation, 27 patients (42.2%) with BRCA2 mutation, and 5 patients (7.8%) with both mutations. Furthermore, 22 pathogenic mutations were detected in 26 patients, 2 likely pathogenic variants in 2 patients, 12 variants of uncertain significance in 20 patients, and 16 likely benign variants in 24 patients. The mutations were mainly found to occur in exons 8, 14, and 17 of BRCA1 and exons 10, 11, 14, and 15 of BRCA2. The results showed that the BRCA genes possess different mutation hotspots in different ethnic groups. In addition, recurrent mutations were noted in many patients. BRCA1 c.536 A>T, considered a founder mutation, was identified in 10 patients (15.63%, 10/64), followed by BRCA1 c.2635 G>T (6.25%, 4/64) and BRCA2 c.2566 T>C (6.25%, 4/64).
Conclusion: The BRCA1 c.536 A>T could be considered to be a founder mutation in this ovarian cancer population. This recurrent BRCA1 mutation has rarely been observed in other ethnic groups. Our findings are expected to provide valuable data for clinical consultation and for designing individualized treatment for ovarian cancer.
Keywords: BRCA gene, ovarian cancer, variants, Hakka population
Introduction
Ovarian cancer is one of the most common cancers in women and a leading cause of death in women. Increased risk factors for cancer have led to an upward trend in the incidence of the disease globally.1,2 There are several risk factors for ovarian cancer, such as genetic predisposition, ovulation, endometriosis, dietary factors, and ethnicity/race. Ovarian cancer can occur sporadically in any woman, including those without any notable risk factors.3
Ovarian cancer is thought to be divided into two main subtypes: type I and type II. Ovarian carcinoma type I includes patients with endometrioid carcinoma, low-grade serous carcinoma, low grade adenocarcinoma carcinoma, mucinous carcinoma, clear cell carcinoma, and transitional cell carcinoma, which are mostly confined to one ovary and are associated with a good prognosis. Ovarian carcinoma type II includes patients with high-grade serous carcinoma, undifferentiated carcinoma, carcinosarcoma, and high grade adenocarcinoma. Unlike ovarian cancer type I, it is highly aggressive and progresses rapidly. Ovarian carcinoma type II is poorly differentiated, and is generally absent in early ovarian lesions.4,5 The diagnosis of ovarian cancer involves pelvic examination, detection of serum tumor markers detection, blood cell analysis, ultrasonography, computed tomography (CT), magnetic resonance imaging (MRI), and other imaging techniques as well as the joint application of these detection methods. However, these tests used in the early diagnosis and condition monitoring of ovarian cancer have certain limitations; hence, the early diagnostic markers and disease characteristics of ovarian cancer need to be studied and clarified.
Studies have identified that family history and genetic factors are important risk factors for ovarian cancer. Ovarian cancer susceptibility genes include breast cancer susceptibility gene 1 (BRCA1), breast cancer susceptibility gene 2 (BRCA2), RAD51 recombinase (RAD51) gene family,6 BRCA1-associated RING Domain protein 1 (BARD1) gene,7 and murine double minute (MDM) gene family.8,9 The two major ovarian cancer susceptibility genes, BRCA1 (MIM #113705) and BRCA2 (MIM #600185), have been explored the most.10 BRCA1 is located on chromosome 17q21 (containing 22 exons) and encodes and expresses a multi-domain protein containing 1863 amino acids.11 BRCA2 is located on chromosome 13q12-q13 (containing 27 exons) and encodes and expresses a multidomain protein containing 3418 amino acids. BRCA1 is similar to BRCA2, but there is no significant homology in the exon region. BRCA1 plays a crucial role in important cell activities and in maintaining gene stability.12–14 BRCA2 is thought to play a key role in the repair of double-strand breaks and in the partial regulation of RAD51 response via chromosome recombination mechanism.15–17 Corresponding studies have also confirmed the correlation between the genetic marker on the chromosome 17q and the susceptibility to breast and ovarian cancer in some patients with a family history of these diseases.18 The results of a genome-wide association analysis involving 15 high-risk breast cancer families revealed that BRCA2, localized on the chromosome 13q12-13, is associated with breast cancer.19 Both BRCA1 and BRCA2 are tumor-suppressor genes, and mutations in these genes are seen in some patients with cancer. Mutations in the human BRCA gene may be race-specific in a given region and region-specific in a given ethnic group.20,21
The Hakka is a Han ethnic group with a unique genetic background and originate from the Hakka ancestors of the Han nationality in Central China. They migrated southward for many times and fused with the ancient Yue residents in Guangdong, Fujian and Jiangxi.22 Meizhou City is located in the northeast of Guangdong Province and has a large Hakka population. The BRCA1/2 mutations and the characteristics of BRCA1/2-associated ovarian cancer in the Hakka population remain unclear. This study retrospectively analyzed the results of the BRCA gene in patients with ovarian cancer among the Hakka population using next-generation sequencing.
Materials and Methods
Participants
Enrollment for the ovarian cancer BRCA gene mutation screening trial was conducted between January 2018 and May 2021 of subjects visiting the Meizhou People’s Hospital (Huangtang Hospital), Meizhou Academy of Medical Sciences. Signed informed consent forms were obtained from all participants. The baseline data, including the general information, related medical history, hematological parameters, and staging (according to the AJCC 8th edition staging) of the enrolled subjects were collected. The ovarian cancer patients were categorized into three groups according to the pathological type: (1) type 1, patients with endometrioid, mucinous, clear cell, low-grade serous, and low grade adenocarcinoma, benign serous cystadenoma or Mullerian cystadenoma; (2) type 2, high grade serous, undifferentiated, carcinosarcoma, and high grade adenocarcinoma; and (3) others, including granulosa cell tumors, borderline tumors, and tumors with unavailable grade or histology data. The study was conducted on the basis of the Declaration of Helsinki, and was supported by the Ethics Committee of the Meizhou People’s Hospital (Huangtang Hospital).
Detection of Serum Tumor Markers and Inflammatory Markers in the Sampled Blood
The subjects’ blood samples (3 mL) were collected at the time of admission and at 2–3 days before treatment and the serum was immediately separated. Serum tumor markers analysis was performed on the Luminex 200 system (Luminex Corporation, Austin, USA) to measure the concentration of serum carcinoembryonic antigen (CEA), carbohydrate antigen 199 (CA199), carbohydrate antigen 125 (CA125), and alpha-fetoprotein (AFP) using relevant quantitative detection kits (TELLGEN Life Science and Technology Co., Ltd., China). After adding the samples and the labeled antibodies into each well, the reaction was performed in the dark at 37°C for 5 min, after which the fluorescence-encoded microspheres cross-linked with antibodies were added. After the reaction, the plate was incubated for 60 min in a darkroom at 37°C, the termination solution was added to terminate the reaction, and the microsphere signals of each well were read on the Luminex 200 system.
The blood sample (2 mL) was collected via venipuncture of an antecubital vein from each subject and collected in a tube containing ethylenediaminetetraacetic acid (EDTA) as an anticoagulant. The erythrocyte correlative indices were detected by using the Sysmex XE-2100 Blood Analyzer (Sysmex Corporation, Japan) according to the standard operating procedures (SOP). The blood routine results were collected before treatment and the inflammation index was calculated according to the following formulas: SII = platelet × neutrophil/lymphocyte,
SIRI = monocyte × neutrophil/lymphocyte,
NLR = neutrophil/lymphocyte,
PLR = platelet/lymphocyte,
LMR = lymphocyte/monocyte.
BRCA1 and BRCA2 Mutations Were Detected by Next-Generation Sequencing (NGS)
The peripheral blood sample (2 mL) was collected from each participant and collected in a tube containing EDTA as an anticoagulant. Genomic DNA was extracted by using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. DNA concentration and purity were quantified using the Nanodrop 2000TM Spectrophotometer (ThermoFisher Scientific, Waltham, MA). The DNA samples were sequenced after library construction, template preparation and template enrichment according to standard operating procedures of the Life Technology Company. Then, 200–1000 ng DNA was sheared prior to library construction for 150 bp fragments. The NGS libraries were constructed using the IonPlus Fragment Library Kit (Life Technologies, Carlsbad, CA). Next, ligation was performed with barcode adapters, followed by ligated fragments amplification, library purification and concentration. Next-generation sequencing was performed on the Ion Proton instrument (Life Technologies) and tested by the CapitalBio Corporation (Beijing, China). Data were analyzed by the Torrent Suite 4.4.3 and 5.0.4 (Life Technologies) using optimized parameters: minimal depth 300×, detection threshold of 2% and 1% for hotspots. Variant call files from the variant caller were loaded on a galaxy platform and annotated using the Safir2report tool. The sequencing results were then compared with the standard human genome data to obtain the mutation sequence information of the samples to be tested. According to the Human Genome Variation Society (HGVS) guidelines, the genetic variations in this study, were named using the following reference sequences: NM_007294.4 (BRCA1) and NM_000059.4 (BRCA2).
Statistical Analyses
SPSS statistical software version 21.0 was used for data analyses. Continuous variable data are represented by mean ±SD, and analyzed using Student’s t-test or the Mann–Whitney U-test. The Chi-square test was applied for analyzing the categorical variables, which were then presented as percentages. P<0.05 was considered to indicate statistical significance.
Results
Population Characteristics
A total of 195 patients with ovarian cancer (all women) were included in the present study. There were 14 women (7.2%) <35 years of age, 54 cases (27.7%) between 35 and 50 years of age, and 127 women (65.1%) >50 years of age. None of the 195 patients had a family history of breast or ovarian cancer. The findings show that ovarian cancer mainly occurs in people >50 years of age. Some studies have observed that the risk of ovarian cancer decreases with the number of pregnancies.23,24 We intended to analyze the relationship between the number of pregnancies and BRCA mutations in the patients with ovarian cancer. Accordingly, 8 women (4.1%) never had a pregnancy, 94 women (48.2%) had 1–3 pregnancies, 34 women (17.4%) had 4–5 pregnancies, 14 women (7.2%) had >5 pregnancies, and the remaining 45 (23.1%) were unknown. These results show that the risk of ovarian cancer decreases with the number of pregnancies. The CEA, CA199, AFP, CA125, NLR, LMR, and PLR levels of these patients were 11.56 ± 114.89 ng/mL, 144.34 ± 769.04 U/mL, 225.98 ± 1779.13 ng/mL, 733.15 ± 1553.06 U/mL, 4.22 ± 4.17, 3.61 ± 2.24, and 245.88 ± 169.87, respectively (Table 1).
 |
Table 1 Clinical Characteristics of Ovarian Cancer Patients |
Clinical Features of Patients with Different Types of Ovarian Cancer
There were 29 (14.9%) patients with type 1 ovarian cancer, 119 (61.0%) cases with type 2 ovarian cancer and 47 (24.1%) cases with other ovarian cancer types. The majority of the patients had type 2 ovarian cancer. It was found that 1 patient (3.4%) was <35 years of age, 6 patients (20.7%) were between 35 and 50 years of age, and 22 patients (75.9%) were >50 years of age in the type 1 group. On the other hand, in the type 2 group, 2 patients (1.7%) were <35 years of age, 28 patients (23.5%) were between 35 and 50 years of age, and 89 patients (74.8%) were >50 years of age. In the other types, there were 11 patients (23.4%) <35 years of age, 20 patients (42.6%) between 35 and 50 years of age, and 16 patients (34.0%) >50 years of age. There were significant differences in age distribution among the three groups of patients with ovarian cancer (P < 0.001). The results show that type 1 and type 2 ovarian cancers mainly occur in people >50 years of age, whereas the other types of cancer mainly occur in people in the age group of 35–50 years. Regarding the number of pregnancies, in the group of type 1 ovarian cancer, 17 women (58.6%) had never been pregnant or had 1–3 pregnancies, and 8 women (27.6%) had ≥4 pregnancies. In the type 2 ovarian cancer group, 55 women (46.2%) had never been pregnant or had 1–3 pregnancies, and 32 (26.9%) had ≥4 pregnancies. Although there was no significant difference in the number of pregnancies among the groups, these types of ovarian cancer were predominated by women with ≤3 pregnancies (Table 2). There were significant differences in CEA (P = 0.035) and AFP (P = 0.012) levels among the groups. The CEA level was the highest in patients with type 1 ovarian cancer (62.24 ± 296.74 ng/mL vs 2.87 ± 7.89 and 2.28 ± 2.26 ng/mL), whereas the AFP level was the lowest in patients with type 1 ovarian cancer (2.92 ± 1.50 ng/mL vs 16.83 ±120.70 and 893.17 ± 3565.47 ng/mL) (Table 2).
 |
Table 2 Clinical Characteristics of Participants with Type 1, Type 2, and Other Ovarian Cancer |
The Frequencies and Distributions of the BRCA Gene Mutations
The protein coding region and exon-intron boundary region of the BRCA1 and BRCA2 genes of the patients were sequenced using next-generation sequencing. There were 64 patients (64/195, 32.8%) with BRCA gene mutations, among whom 32 patients (32/64, 50.0%) had BRCA1 gene mutation/mutations, 27 patients (27/64, 42.2%) had BRCA2 gene mutation/mutations, and 5 patients (5/64, 7.8%) had both mutations. The numbers of patients with type 1, type 2, and other types of ovarian cancer who had BRCA mutations were 7, 42, and 15, respectively. The numbers of patients with type 1 ovarian cancer who had BRCA1, BRCA2, and both mutations were 3 (42.9%), 4 (57.1%), and 0(0), respectively. The corresponding numbers of patients with type 2 ovarian cancer and other types of cancer were 23 (54.8%), 15 (35.7%), 4 (9.5%) and 6 (40.0%), 8 (53.3%), 1 (6.7%), respectively. These results show that type 1 ovarian cancer with BRCA mutation mainly involves the BRCA2 gene, while type 2 ovarian cancer with BRCA mutation mainly involves the BRCA1 gene. The frequencies and distributions of BRCA1 and BRCA2 gene mutations are presented in Table 3.
 |
Table 3 The Frequencies and Distributions of BRCA1 and BRCA2 Gene Mutations |
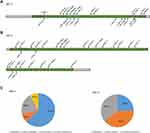
In this study, a total of 52 variants of the BRCA gene were detected. The sequence analysis revealed 22 distinct pathogenic mutations detected in 26 patients, 2 likely pathogenic variants in 2 patients, 12 variants of uncertain significance (VOUS) in 20 patients, and 16 likely benign variants in 24 patients. The mutations were predominantly seen in exons 8, 14, and 17 of the BRCA1 gene and exons 10, 11, 14 and 15 of the BRCA2 gene (Figure 1A and B). Among the BRCA1 gene variants, 15 (62.5%) were pathogenic variants, 2 (8.3%) were likely pathogenic variants, 5 (20.8%) were likely benign variants, and 2 (8.3%) were VOUSs. On the other hand, among the BRCA2 mutations, 7 (25.0%) were pathogenic variants, 11 (39.3%) were likely benign variants, and 10 (35.7%) were VOUSs (Figure 1C).
Table 4 provides detailed information on each patient with BRCA mutation/mutations, including ClinVar information (pathogenic, likely pathogenic, likely benign, and VOUS) pertaining to the BRCA1 and BRCA2 genes. Data on mutations, mutation types, family history of cancer, and number of pregnancies are presented. In this study, 10 patients (15.63%, 10/64) carried the BRCA1 gene c.536 A>T variant, 4 patients (6.25%, 4/64) carried the BRCA1 gene c.2635 G>T variant, 4 patients (6.25%, 4/64) carried the BRCA1 gene c.2566 T>C variant, and 3 patients (4.69%, 3/64) carried the BRCA2 gene c.5785 A>G variant, 3 patients (4.69%, 3/64) carried the BRCA2 gene c.8187 G>T variant. Based on these observations, BRCA1 c.536 A>T might be a founder mutation in this population.
 |  |  |
Table 4 All Mutations and the Characteristics of Ovarian Cancer Patients |
Discussion
Ovarian cancer is one of the most common malignant tumors that threaten women’s health and quality of life and has the highest mortality rate among gynecological tumors.25 BRCA is an important gene that determines the genetic susceptibility to cancer by participating in the regulation of DNA damage and repair, cell growth and apoptosis and by playing an indispensable role in maintaining the genetic stability of cells.26,27 Mutations in the BRCA gene can lead to ovarian cancer. Screening for BRCA gene mutations can effectively assess and predict the risk for ovarian cancer, intervene to reduce the incidence of the disease, and guide precise treatment.
Worldwide, the incidence of BRCA mutations in patients with ovarian cancer is approximately 10%-15%. BRCA mutations are chiefly distributed in Europe and North America, among which the incidence of BRCA1 mutation is significantly higher than that of BRCA2.28 The incidence of BRCA1/2 mutations among Mexican patients with ovarian cancer is approximately 28%, of which BRCA1 constitutes 88% and BRCA2 constitutes 12%.29 Another study on patients with ovarian cancer in Mexico found that the incidence of BRCA1/2 mutations was 33%, of which 66.1% was BRCA1 and 33.9% was BRCA2.30 These differences may be related to the geographical distribution of the selected population. The prevalence of BRCA mutations among Spanish patients with ovarian cancer was 16%, of which the prevalence of BRCA2 mutations was 63%, which is contrary to most studies.31 The prevalence of BRCA mutations in Israeli Arab patients with ovarian cancer was 32%.32 The prevalence of BRCA mutations in Korean patients with ovarian cancer was 24.6%.33 At present, there are several studies based on large samples in China. Shi et al34 performed BRCA1/2 gene detection in 916 patients with epithelial ovarian cancer. The results showed that the incidence of BRCA1/2 gene mutation was 16.7%, of which BRCA1 mutation accounted for 13.1%, BRCA2 mutation accounted for 3.9%, and the simultaneous presence of both mutations accounted for 0.3%. Wu et al35 conducted the BRCA test on 826 patients with ovarian cancer in a multiCenter nationwide study, and the incidence of BRCA1/2 gene mutations was found to be 28.5%, of which the BRCA1 mutation accounted for 20.8% and the BRCA2 mutation accounted for 7.6%. Bu et al36 attempted to detect BRCA mutation in 547 patients with ovarian cancer and found the incidence of BRCA1/2 gene mutations to be 23.6%, of which BRCA1 mutation accounted for 15.4% and BRCA2 mutation accounted for 8.2%. The proportion of BRCA mutations was 5.41% in breast and ovarian cancer in a Hakka population.37 The proportion of patients with BRCA mutation in Hong Kong was 15.3%.38 In this study, the incidence of BRCA1/2 gene mutations was 32.8% in patients with ovarian cancer, of which BRCA1 mutation accounted for 50.0%, BRCA2 mutation accounted for 42.2%, and both mutations accounted for 7.8%. In the future, multicenter BRCA gene mutation studies should be conducted in China with a larger sample size by adopting unified standards so as to create a BRCA gene mutation database consistent with the characteristics of the Chinese population.
To the best of our knowledge, this study is the largest on BRCA1 and BRCA2 gene sequencing in patients with ovarian cancer for mutation screening analysis in the Chinese Hakka population. According to researches, mutations in the BRCA1 gene are concentrated in exons 8, 11, 22, and 24 and mutations in the BRCA2 gene are concentrated in exons 10, 11, 14, and 21 in Mainland China populations.39–43 Our results suggested that mutations in the BRCA1 gene mainly occur in exons 8, 14, and 17, whereas mutations in the BRCA2 gene chiefly occur in exons 10, 11, 14, and 15. The findings indicate that the BRCA genes have different mutation hotspots in different ethnic groups and regions.
In addition, some variants were present at higher frequencies when compared with the other variants. It was observed that 10 patients (15.63%, 10/64) carried the BRCA1 gene c.536 A>T variant, 4 patients (6.25%, 4/64) carried the BRCA1 gene c.2635 G>T variant, 4 patients (6.25%, 4/64) carried the BRCA1 gene c.2566 T>C variant, and 3 patients (4.69%, 3/64) carried the BRCA2 gene c.5785 A>G variant, 3 patients (4.69%, 3/64) carried the BRCA2 gene c.8187 G>T variant. Whether these mutations are founder and hotspot mutations in the patients with ovarian cancer in the Chinese Hakka population remains to be confirmed. Founder mutations in the BRCA genes have been reported in many nations and ethnic groups worldwide. It has been documented that BRCA1 c. 5470_5477del8 is a BRCA1 founder mutation of ovarian cancer in the Chinese population;34,44 this mutation may be the BRCA1 founder mutation unique to Asians.45 The mutation BRCA2 c.3109 C>T is a founder mutation in the Southern Chinese population.46 Another study showed that the most common mutation was BRCA1 ex9-12del, a Mexican founder mutation.30 Mutational data show that the most frequently recorded BRCA1 c.5266dupC mutation is the founder mutation in Italian,47 Northeastern Romanian,48 and Turkish populations.49 Slavic BRCA1 and BRCA2 founder mutations include BRCA1 c.5266dupC, BRCA1 c.4034delA, and BRCA1 c.68_69delAG.50 BRCA1 c.4136_4137delCT and c.1140dupG represent the founder mutations in the Middle Eastern population.51 BRCA2 c.3922G>T is a founder mutation in the Puerto Rican population.52 BRCA1 c.5266dupC and c.181T>G are founder mutations in the Polish population.53 BRCA1 c.798_799delTT might be a founder mutation in the North African population.54 BRCA1 c.3319G>T might be a founder mutation in the Western Danish population.55 In the present study, the BRCA1 c.536 A>T mutation was observed which might be considered a founder mutation in this ovarian cancer population. The recurrent BRCA1 mutation reported herein has rarely been observed in other ethnic groups.
In general, the mutation frequency of the BRCA1/2 gene in the Hakka patients with ovarian cancer in Southern China is different from that in other ethnic groups. Moreover, differences exist in the exon regions where the mutations occur. This study provides a basis and serves as a reference for clinical counseling and for devising individualized prevention and treatment strategies to combat ovarian cancer. Identifying founder and recurrent mutations is an important way to improve genetic counseling because molecular testing can target the founder and recurrent mutations, thereby enabling faster and less expensive testing. As the frequency of founder mutations increases, molecular testing can analyze a large number of cases and provide accurate information on the relationship between the patient’s mutation status and disease risk, thereby improving disease management. Owing to the small sample size of this study, the distributions of the BRCA gene mutations among the patients with ovarian cancer in the Hakka population of Southern China have not been entirely revealed. The distributions of the BRCA gene mutations in different populations, and the relationship between mutation status and disease risk and pathological features need to be investigated further.
In this population, the significance of identifying the founder mutations mainly lies in reducing the cost of population screening. Genetic screening can be performed by first focusing on the founder mutation. In this manner, genetic screening can be easily implemented in the Hakka population. Although this study has shed light on the founder BRCA1 mutation in the Hakka population, we cannot rule out the possibility that other founder BRCA mutations may exist in the larger patient population. However, given the economic advantages of genetic screening, we believe that this study would pave the way for future studies in the Hakka population.
Conclusions
In this study, the BRCA gene mutations were found to account for a certain proportion of the patients with ovarian cancer in the Hakka population of Southern China. The BRCA1 c.536 A>T mutation was detected among in 10/64 (15.63%) of the individuals with BRCA mutation/mutations in the cohort and can, therefore, be considered a founder mutation in this ovarian cancer population. This recurrent BRCA1 mutation has rarely been observed in other ethnic groups. Understanding the frequency of BRCA1 and BRCA2 gene mutations in the Hakka patients with ovarian cancer will provide valuable data for clinical consultation and for devising individualized therapeutic strategies for patients with ovarian cancer.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics Approval and Consent to Participate
This study was conducted on the basis of the Declaration of Helsinki, and was supported by the Ethics Committee of the Meizhou People’s Hospital.
Acknowledgments
The author would like to thank other colleagues whom were not listed in the authorship of Center for Precision Medicine, Meizhou People’s Hospital (Huangtang Hospital), Meizhou Academy of Medical Sciences for their helpful comments on the manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Guangdong Provincial Key Laboratory of Precision Medicine and Clinical Translation Research of Hakka Population (Grant No.: 2018B030322003), the Science and Technology Program of Meizhou (Grant No.: 2019B0202001), and Key Scientific and Technological Project of Meizhou People’s Hospital (Grant No.: MPHKSTP-20190102).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Momenimovahed Z, Tiznobaik A, Taheri S, Salehiniya H. Ovarian cancer in the world: epidemiology and risk factors. Int J Womens Health. 2019;11:287–299. doi:10.2147/IJWH.S197604
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Stewart C, Ralyea C, Lockwood S. Ovarian cancer: an integrated review. Semin Oncol Nurs. 2019;35(2):151–156. doi:10.1016/j.soncn.2019.02.001
4. Marcišauskas S, Ulfenborg B, Kristjansdottir B, Waldemarson S, Sundfeldt K. Univariate and classification analysis reveals potential diagnostic biomarkers for early stage ovarian cancer type 1 and type 2. J Proteomics. 2019;196:57–68. doi:10.1016/j.jprot.2019.01.017
5. Terada KY, Ahn HJ, Kessel B. Differences in risk for type 1 and type 2 ovarian cancer in a large cancer screening trial. J Gynecol Oncol. 2016;27(3):e25. doi:10.3802/jgo.2016.27.e25
6. Song H, Dicks E, Ramus SJ, et al. Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol. 2015;33(26):2901–2907. doi:10.1200/JCO.2015.61.2408
7. Alenezi WM, Fierheller CT, Recio N, Tonin PN. Literature review of BARD1 as a cancer predisposing gene with a focus on breast and ovarian cancers. Genes. 2020;11(8):856. doi:10.3390/genes11080856
8. Gu J, Tang Y, Liu Y, et al. Murine double minute 2 siRNA and wild-type p53 gene therapy enhances sensitivity of the SKOV3/DDP ovarian cancer cell line to cisplatin chemotherapy in vitro and in vivo. Cancer Lett. 2014;343(2):200–209. doi:10.1016/j.canlet.2013.10.011
9. Ignacio RMC, Dong YL, Kabir SM, et al. CXCR2 is a negative regulator of p21 in p53-dependent and independent manner via Akt-mediated Mdm2 in ovarian cancer. Oncotarget. 2018;9(11):9751–9765. doi:10.18632/oncotarget.24231
10. Huber D, Seitz S, Kast K, Emons G, Ortmann O. Use of fertility treatments in BRCA1/2 mutation carriers and risk for ovarian and breast cancer: a systematic review. Arch Gynecol Obstet. 2020;302(3):715–720. doi:10.1007/s00404-020-05690-4
11. Hall JM, Lee MK, Newman B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250(4988):1684–1689. doi:10.1126/science.2270482
12. Deng CX, Brodie SG. Roles of BRCA1 and its interacting proteins. Bioessays. 2015;22(8):728–737. doi:10.1002/1521-1878(200008)22:8<728::AID-BIES6>3.0.CO;2-B
13. Gudas JM, Li T, Nguyen H, Jensen D, Cowan KH, Cowan KH. Cell cycle regulation of BRCA1 messenger RNA in human breast epithelial cells. Cell Growth Differ. 1996;7(6):717–723.
14. Kumaravel S. Breast cancer gene 1 (BRCA1): role in cell cycle regulation and DNA repair–perhaps through transcription. J Cell Biochem. 2003;88(6):1084–1091. doi:10.1002/jcb.10469
15. Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408(6811):429–432. doi:10.1038/35044000
16. Davies AA, Masson JY, Mcilwraith MJ, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7(2):273–282. doi:10.1016/s1097-2765(01)00175-7
17. Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene. J Biol Chem. 1997;272(51):31941–31944. doi:10.1074/jbc.272.51.31941
18. Castilla LH, Couch FJ, Erdos MR, et al. Mutations in the BRCA1 gene in families with early-onset breast and ovarian cancer. Nat Genet. 1994;8(4):387. doi:10.1038/ng1294-387
19. Wooster R, Neuhausen SL, Mangion J, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265(5181):2088–2090. doi:10.1126/science.8091231
20. Bhaskaran SP, Chandratre K, Gupta H, et al. Germline variation in BRCA1/2 is highly ethnic-specific: evidence from over 30,000 Chinese hereditary breast and ovarian cancer patients. Int J Cancer. 2019;145(4):962–973. doi:10.1002/ijc.32176
21. Armstrong N, Ryder S, Forbes C, Ross J, Quek RG. A systematic review of the international prevalence of BRCA mutation in breast cancer. Clin Epidemiol. 2019;11:543–561. doi:10.2147/CLEP.S206949
22. Wang WZ, Wang CY, Cheng YT, et al. Tracing the origins of Hakka and Chaoshanese by mitochondrial DNA analysis. Am J Phys Anthropol. 2010;141(1):124–130. doi:10.1002/ajpa.21124
23. Park JS, Lee ST, Han JW, Kim TI, Nam EJ, Park HS. Difference in risk of breast and ovarian cancer according to putative functional domain regions in Korean BRCA1/2 mutation carriers. Clin Breast Cancer. 2018;18(5):362–373.e361. doi:10.1016/j.clbc.2018.02.007
24. Lee AW, Rosenzweig S, Wiensch A, et al. Expanding our understanding of ovarian cancer risk: the role of incomplete pregnancies. J Natl Cancer Inst. 2021;113(3):301–308. doi:10.1093/jnci/djaa099
25. Kuroki L, Guntupalli SR. Treatment of epithelial ovarian cancer. BMJ. 2020;371:m3773. doi:10.1136/bmj.m3773
26. Petsalaki E, Zachos G. DNA damage response proteins regulating mitotic cell division: double agents preserving genome stability. FEBS J. 2020;287(9):1700–1721. doi:10.1111/febs.15240
27. Gorodetska I, Kozeretska I, Dubrovska A. BRCA genes: the role in genome stability, cancer stemness and therapy resistance. J Cancer. 2019;10(9):2109–2127. doi:10.7150/jca.30410
28. Manchana T, Phoolcharoen N, Tantbirojn P. BRCA mutation in high grade epithelial ovarian cancers. Gynecol Oncol Rep. 2019;29:102–105. doi:10.1016/j.gore.2019.07.007
29. Villarreal-Garza C, Alvarez-Gómez RM, Pérez-Plasencia C, et al. Significant clinical impact of recurrent BRCA1 and BRCA2 mutations in Mexico. Cancer. 2015;121(3):372–378. doi:10.1002/cncr.29058
30. Gallardo-Rincón D, Álvarez-gómez RM, Montes-Servín E, et al. Clinical evaluation of BRCA1/2 mutation in Mexican ovarian cancer patients. Transl Oncol. 2020;13(2):212–220. doi:10.1016/j.tranon.2019.11.003
31. Ruiz de Sabando A, Urrutia Lafuente E, García-Amigot F, et al. Genetic and clinical characterization of BRCA-associated hereditary breast and ovarian cancer in Navarra (Spain). BMC Cancer. 2019;19(1):1145. doi:10.1186/s12885-019-6277-x
32. Bernstein-Molho R, Barnes-Kedar I, Ludman MD, et al. The yield of full BRCA1/2 genotyping in Israeli Arab high-risk breast/ovarian cancer patients. Breast Cancer Res Treat. 2019;178(1):231–237. doi:10.1007/s10549-019-05379-6
33. Eoh KJ, Park JS, Park HS, et al. BRCA1 and BRCA2 mutation predictions using the BRCAPRO and Myriad models in Korean ovarian cancer patients. Gynecol Oncol. 2017;145(1):137–141. doi:10.1016/j.ygyno.2017.01.026
34. Shi T, Wang P, Xie C, et al. BRCA1 and BRCA2 mutations in ovarian cancer patients from China: ethnic-related mutations in BRCA1 associated with an increased risk of ovarian cancer. Int J Cancer. 2017;140(9):2051–2059. doi:10.1002/ijc.30633
35. Wu X, Wu L, Kong B, et al. The first nationwide multicenter prevalence study of germline BRCA1 and BRCA2 mutations in Chinese ovarian cancer patients. Int J Gynecol Cancer. 2017;27(8):1650–1657. doi:10.1097/IGC.0000000000001065
36. Bu H, Chen J, Li Q, et al. BRCA mutation frequency and clinical features of ovarian cancer patients: a report from a Chinese study group. J Obstet Gynaecol Res. 2019;45(11):2267–2274. doi:10.1111/jog.14090
37. Wu H, Wang Q, Guo X, et al. Frequency of BRCA1 and BRCA2 mutations in individuals with breast and ovarian cancer in a Chinese Hakka population using next-generation sequencing. Hum Hered. 2019;84(4–5):160–169. doi:10.1159/000505268
38. Ava K, Ng EKO, Wong CLP, et al. Identification of BRCA1/2 founder mutations in Southern Chinese breast cancer patients using gene sequencing and high resolution DNA melting analysis. PLoS One. 2012;7(9):e43994. doi:10.1371/journal.pone.0043994
39. Juan Z, Renguang P, Zhiyuan P, et al. Prevalence and characterization of BRCA1 and BRCA2 germline mutations in Chinese women with familial breast cancer. Breast Cancer Res Treat. 2012;132(2):421–428. doi:10.1007/s10549-011-1596-x
40. Song CG, Hu Z, Wu J, et al. The prevalence of BRCA1 and BRCA2 mutations in eastern Chinese women with breast cancer. J Cancer Res Clin Oncol. 2006;132(10):617–626. doi:10.1007/s00432-006-0105-9
41. Li WF, Hu Z, Rao NY, et al. The prevalence of BRCA1 and BRCA2 germline mutations in high-risk breast cancer patients of Chinese Han nationality: two recurrent mutations were identified. Breast Cancer Res Treat. 2008;110(1):99–109. doi:10.1007/s10549-007-9708-3
42. Suter NM, Ray RM, Hu YW, et al. BRCA1 and BRCA2 mutations in women from Shanghai China. Cancer Epidemiol Biomarkers Prev. 2004;13(2):181–189. doi:10.1158/1055-9965.epi-03-0196
43. Xu J, Wang B, Li R, Wang Y, Zhang S, Zhang S. Clinical implications for BRCA gene mutation in breast cancer. Mol Biol Rep. 2012;39(3):3097–3102. doi:10.1007/s11033-011-1073-y
44. Kim YC, Zhao L, Zhang H, et al. Prevalence and spectrum of BRCA germline variants in mainland Chinese familial breast and ovarian cancer patients. Oncotarget. 2016;7(8):9600–9612. doi:10.18632/oncotarget.7144
45. Jang JH, Lee JE, Kwon MJ, et al. Spectra of BRCA1 and BRCA2 mutations in Korean patients with breast cancer: the importance of whole-gene sequencing. J Hum Genet. 2012;57(3):212–215. doi:10.1038/jhg.2011.139
46. Kwong A, Wong LP, Wong HN, et al. A BRCA2 founder mutation and seven novel deleterious BRCA mutations in southern Chinese women with breast and ovarian cancer. Breast Cancer Res Treat. 2009;117(3):683–686. doi:10.1007/s10549-009-0385-2
47. Artioli G, Giannone G, Valabrega G, et al. Characteristics and outcome of BRCA mutated epithelial ovarian cancer patients in Italy: a retrospective multicenter study (MITO 21). Gynecol Oncol. 2021;161(3):755–761. doi:10.1016/j.ygyno.2021.04.014
48. Negură L, Duşa CP, Balmuş MI, et al. BRCA1 5382insC founder mutation has not a significative recurrent presence in Northeastern Romanian cancer patients. Rom J Morphol Embryol. 2015;56(2):379–385.
49. Gun-Bilgic D, Aydin-Gumus A, Bilgic A, Cam FS. Mutations of BRCA1/2 genes in the West of Turkey and genotype-phenotype correlations. Clin Lab. 2022;68(1). doi:10.7754/Clin.Lab.2021.210425
50. Sokolenko AP, Sokolova TN, Ni VI, et al. Frequency and spectrum of founder and non-founder BRCA1 and BRCA2 mutations in a large series of Russian breast cancer and ovarian cancer patients. Breast Cancer Res Treat. 2020;184(1):229–235. doi:10.1007/s10549-020-05827-8
51. Siraj AK, Bu R, Iqbal K. Prevalence, spectrum, and founder effect of BRCA1 and BRCA2 mutations in epithelial ovarian cancer from the Middle East. Hum Mutat. 2019;40(6):729–733. doi:10.1002/humu.23736
52. Diaz-Zabala HJ, Ortiz AP, Garland L, et al. A recurrent BRCA2 mutation explains the majority of hereditary breast and ovarian cancer syndrome cases in Puerto Rico. Cancers (Basel). 2018;10(11):419. doi:10.3390/cancers10110419
53. Perkowska M, BroZek I, Wysocka B, et al. BRCA1 and BRCA2 mutation analysis in breast-ovarian cancer families from northeastern Poland. Hum Mutat. 2003;21(5):553–554. doi:10.1002/humu.9139
54. Laraqui A, Uhrhammer N, Rhaffouli HE, et al. BRCA genetic screening in Middle Eastern and North African: mutational spectrum and founder BRCA1 mutation (c.798_799delTT) in North African. Dis Markers. 2015;2015:194293. doi:10.1155/2015/194293
55. Nielsen HR, Nilbert M, Petersen J, et al. BRCA1/BRCA2 founder mutations and cancer risks: impact in the western Danish population. Fam Cancer. 2016;15(4):507–512. doi:10.1007/s10689-016-9875-7
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.