Back to Journals » Drug Design, Development and Therapy » Volume 14
The Expression and Therapeutic Potential of Checkpoint Kinase 2 in Laryngeal Squamous Cell Carcinoma
Authors Tian Y, Wang Y, Xu S, Guan C, Zhang Q, Li W
Received 8 January 2020
Accepted for publication 12 June 2020
Published 6 July 2020 Volume 2020:14 Pages 2613—2622
DOI https://doi.org/10.2147/DDDT.S245267
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Manfred Ogris
Ying Tian,1 Yan Wang,1 Shan Xu,1 Chao Guan,1 Qingfu Zhang,2 Wei Li1
1Department of Otorhinolaryngology, The First Affiliated Hospital of China Medical University, Shenyang, People’s Republic of China; 2Department of Pathology, The First Affiliated Hospital and College of Basic Medical Sciences of China Medical University, Shenyang, People’s Republic of China
Correspondence: Wei Li
Department of Otorhinolaryngology, The First Affiliated Hospital of China Medical University, 155, Nanjing North Street, Heping District, Shenyang, Liaoning Province, People’s Republic of China
Tel +86-13604077678
Email [email protected]
Introduction: Laryngeal squamous cell carcinoma (LSCC) is the most common histological subtype of laryngeal cancer. The involved molecular mechanisms and suitable therapeutic targets for LSCC still need to be further investigated. Checkpoint kinase 2 (CHK2) participates in several cellular physiology pathways and plays a role in tumor progression. However, the roles of CHK2 in LSCC remain unclear.
Methods: mRNA expression data were obtained from The Cancer Genome Atlas (TCGA) database, and bioinformatic analysis was performed. Western blot and immunohistochemical analyses were conducted to detect protein expression. MTS assays were performed to examine cell growth of LSCC-derived cell lines.
Results: In the present study, we found that both active form of CHK2 and total CHK2 protein expressions were up-regulated in LSCC tissues. Positive expression of CHK2 was closely associated with advanced clinical features and poor prognosis. Moreover, potential CHK2-involving bioprocesses and signaling pathways were analyzed. In addition, repressed proliferation of LSCC cells was induced by CHK2 inhibitor.
Discussion: Taken together, our findings elucidated that CHK2 may act as an oncogenic factor in LSCC, suggesting a potential target for clinical treatment.
Keywords: checkpoint kinase 2, laryngeal squamous cell carcinoma, BML-277, tumor growth
Introduction
Laryngeal cancer is one of the most common malignant tumors of the head and neck region. The most common histological subtype of laryngeal cancer is referred to as laryngeal squamous cell carcinoma (LSCC).1 It was estimated that 177,422 new cases were diagnosed, with approximately 94,771 patients dying from laryngeal cancer in 2018.2 Although surgery is an integral part of treatment for localized laryngeal cancer, nonsurgical options like radiation and systemic therapy have emerged as viable options.3 There are several risk factors implicated in the pathogenesis of laryngeal cancer, including tobacco smoking,4 alcohol consumption,5 dietary factors,6 and human papillomavirus (HPV) infections,7 but the involved molecular mechanisms and suitable therapeutic targets still need further investigation.
In humans, checkpoint kinase 2 (CHK2), which is encoded by CHEK2, is a serine/threonine kinase of 543 residues and participates in series of signaling networks. CHK2 plays multiple roles in nuclear DNA damage repair (DDR). DNA double-strand breaks induce the activation of ATM serine/threonine kinase, which subsequently phosphorylates CHK2 on T68 and results in dimerization and activation of CHK2.8 As a serine/threonine kinase, activated CHK2 phosphorylates its substrates, including BRCA1, E2F1, p53, CDC25A, and CDC25C, to induce the appropriate cellular response.9 In addition, CHK2 mediates stabilization of the FOXM1 transcription factor to stimulate expression of DNA repair genes.10 Besides DDR, CHK2 is also engaged in DNA structure modification and cell cycle progression in normal cellular physiology.
CHK2 also plays a part in tumor progression. CHK2 can prevent tumor progression by averting genomic instability through DNA repair and, if this is not possible, by causing the cell to senesce or die.11 In Hep-2 laryngeal carcinoma cells, down-regulation of Pak4 shows antitumor effects by activation of the ATM/CHK1/2/p53 pathway.12 But, more than a tumor suppressor in the classical sense, increasing evidence supports that CHK2 appears to function like a cancer susceptibility gene.13 For instance, CHK2 is aberrantly and constitutively activated in invasive urinary bladder carcinomas.14 CHK2 overexpresses and mislocalizes within mitotic structures, which enhances chromosomal instability and hepatocellular carcinoma progression.15
The roles of CHK2 in LSCC remain unclear. Previous data showed that the activated CHK2 (T68-phosphorylated CHK2) expression status is a putative indicator of oral cancer risk, and suggested that early therapeutic exploitation of DDR may be of potential benefit in the prevention of oral squamous cell carcinoma, which is another malignant tumor of the head and neck region.16,17 However, a recent study indicated there is no significant relation between CHK2 expression and clinicopathological data or survival in oral squamous cell carcinoma.18 In this research, our data showed that both active form of CHK2 and total CHK2 protein expressions were higher in LSCC tissues than in matched adjacent nontumor tissues. Further, positive expression of CHK2 was closely associated with advanced clinical features and poor prognosis. Moreover, potential CHK2-involving bioprocesses and signaling pathways were analyzed from The Cancer Genome Atlas (TCGA) database. In addition, repressed proliferation of LSCC cells was induced by CHK2 inhibitor. Taken together, our findings elucidated that CHK2 may act as an oncogenic factor in LSCC, suggesting a potential target for clinical treatment.
Materials and Methods
Tissue Samples
This study was approved by the Human Research Ethics Committee of The First Affiliated Hospital of China Medical University. All patients understood the purpose of this research and signed the informed consent. Nineteen pairs of fresh LSCC tissues and matched adjacent nontumor tissues for Western blot analysis were obtained from laryngeal cancer patients who underwent resection surgery in The First Affiliated Hospital of China Medical University in 2017. The tissues were immediately snap-frozen in liquid nitrogen and stored at −80 °C until protein extraction. Another 64 pairs of formalin-fixed, paraffin-embedded sections were obtained from laryngeal cancer patients who had resection surgery in The First Affiliated Hospital of China Medical University from 2007 to 2012, and the patients were followed up till July 2017. All patients were not pretreated before surgery. The clinicopathological data are listed in Table 1.
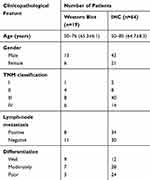 |
Table 1 The Clinicopathological Parameters of Patients |
Cell Culture
Two human LSCC lines (Hep-2 and Tu212) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS; Gibco) at 37 °C with 5% CO2.
Inhibitor and siRNA
BML-277 was purchased from Selleck (S8632), dissolved in dimethyl sulfoxide (DMSO), and stored at −80 °C in small aliquots. siRNAs were obtained from Sigma, and were transfected by jet-PRIME reagent (Polyplus) following the manufacturer’s instructions. The sequence of CHK2-siRNA is 5′-CCUGUGGAGAGGUAAAGCU-3′ and of control-siRNA is 5′-UUCUCCGAACGUGUCACGU-3′.
Western Blot Analysis
Tissue proteins were prepared by extraction with RIPA lysate. Protein concentration was determined with the BCA protein assay kit, and approximately 25 μg of total protein was loaded. GAPDH was used as internal control. Goat anti-mouse/rabbit IgG (HRP) was obtained from Bethyl Laboratories. Antibodies against CHK2 (#6334, 1:1000 dilution) and T68-phosphorylated CHK2 (#2197, 1:1000 dilution) were obtained from Cell Signaling Technology. Antibody against GAPDH was obtained from KangChen Bio-tech Inc.
Immunohistochemical (IHC) Analysis
The IHC experiment was performed with the following procedures. Tissue sections were dewaxed, rehydrated, eliminated of endogenous peroxidase activity, and boiled for antigen retrieval, followed by incubation with indicated CHK2 antibody (#6334, Cell Signaling Technology, IHC antibody validation, 1:400 dilution) overnight at 4 °C. The sections were then incubated with streptavidin-peroxidase-conjugated secondary antibodies. The signals were visualized with diaminobenzidine and the nuclei were counterstained with hematoxylin (ZSGB-bio).
CHK2 expression was scored as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. The frequency of positive cells was defined as follows: 0, 0%; 1, less than 25%; 2, 26% to 50%; 3, 51% to 75%; and 4, greater than 75%. The final score, which is valued between 0 and 12, was determined by multiplying the staining intensity score by the positive cell frequency score.
RNA Expression Profiles and Bioinformatic Analysis
The mRNA expression data of CHEK2 in 519 head and neck squamous cell carcinoma (HNSC) cancer samples and 44 normal samples were obtained from TCGA database using Gene Expression Profiling Interactive Analysis (GEPIA).19 A subset of 499 patients with survival data was analyzed by Kaplan–Meier analysis. Correlated genes of CHEK2 in RNA expression profiles of HNSC cancer samples and 184 esophageal carcinoma (ESCA) cancer samples were obtained from TCGA database by LinkedOmics.20 With thresholds of statistical correlation (+0.5 and −0.4 in HNSC, +0.5 and −0.35 in ESCA), we screened the top correlated genes of CHEK2. Based on the overlap of correlated genes in both HNSC and ESCA samples, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted via Metascape.21
MTS Assay
Cells (5x103 cells/well) were seeded into 96-well plates and cultured overnight. Then, cells were transfected with siRNA or treated with different concentrations of CHK2 inhibitor for indicated times. Cell viability was determined using the CellTiter 96 aqueous one solution cell proliferation assay (Promega).
Statistical Analysis
Statistical analysis was performed using SPSS 17.0 software. The differences between groups were assessed by Student’s t-test. The survival rates were evaluated by the Kaplan–Meier method and tested by the log-rank test. For IHC, the chi-square test was used to determine the significant difference between immunohistochemical expression of CHK2 and clinicopathological features. P<0.05 was regarded as statistically significant.
Results
CHK2 Expression Is Increased in Human LSCC
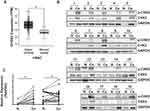
First, a publicly available cohort of HNSC from TCGA database was used to validate CHEK2 expression levels. Compared with normal tissues, we found a significant increased mRNA expression of CHEK2 in HNSC tissues (Figure 1A). We then explored the active form of CHK2 protein (T68-phosphorylated CHK2, hereafter abbreviated as p-CHK2) and total CHK2 protein expressions in 19 pairs of LSCC tissues and matched adjacent nontumor tissues by Western blot analysis (Figure 1B). The protein expressions relative to GAPDH showed that p-CHK2 expression in 18 LSCC tissues (94.7%, except sample 9) and CHK2 expression in 14 LSCC tissues (73.7%, except sample 9, 10, 11, 14, and 19) were significantly increased, compared with that in the matched adjacent noncancerous tissues (P<0.05, Figure 1C), indicating both p-CHK2 and CHK2 levels were up-regulated in LSCC. These data suggested that CHK2 was overexpressed in HNSC and LSCC tissues.
The Clinical Relevance of CHK2 in LSCC
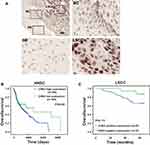
We further detected the expression of CHK2 in 64 pairs of LSCC tissues and matched adjacent nontumor tissues by the IHC method. Nuclear staining of CHK2 was detected in cells (Figure 2A). In accordance with the results shown in Figure 1, positive expression of CHK2 was found in 49 (76.6%) LSCC tissues, but only 19 (29.7%) adjacent nontumor tissues were shown with positive expression of CHK2 (Figure 2A). Moreover, the expression of CHK2 in different clinicopathological features of LSCC was evaluated. Although no significant association between CHK2 expression and age (P=1), gender (P=0.1142) or clinical stage (P=1) was observed, a higher rate of positive-CHK2 expression was significantly associated with positive lymph-node metastasis (P=0.0363) and poor differentiation (P=0.0343) (Table 2).
 |
Table 2 Relation Between the Clinicopathological Features and CHK2 Expression in LSCC |
In addition, Kaplan–Meier analysis showed that high CHK2 expression was associated with poor overall survival rate in HNSC patients (P=0.038, Figure 2B). In our cohort of 64 cases, positive expression of CHK2 in LSCC patients also contributed to bad prognosis, although there was no statistical difference (P=0.113, Figure 2C). Taken together, these data suggested positive CHK2 expression may have an association with lymph-node metastasis, poor differentiation, and poor prognosis in LSCC patients.
Analysis of Potential CHK2 Involving Bioprocesses and Signaling Pathways
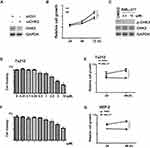
The same as LSCC, the most common histological subtype of esophageal carcinoma (ESCA) is also referred to as squamous cell carcinoma.22 To reveal the underlying mechanisms of CHK2 function in squamous cell carcinoma, we further analyzed RNA-Sequence data of HNSC and ESCA from TCGA database. We analyzed the genes whose expressions were correlated with CHEK2 by LinkedOmics.20 We then focused on the top correlated genes with CHEK2 using the values of statistical correlation as cutoffs. There were 121 positively-correlated genes and 297 negatively-correlated genes in HNSC, while 105 positively-correlated genes and 184 negatively-correlated genes were found in ESCA (listed in Supplementary Tables S1–S4). Further, there were 37 overlapped positively-correlated genes and 25 overlapped negatively-correlated genes between HNSC and ESCA (listed in Supplementary Tables S5 and S6). GO (BP) and KEGG enrichment analysis of those overlapped genes were revealed by Metascape.21 As shown in Figure 3A, the positively-correlated genes appeared to be involved in cell cycle, DNA conformation, DNA repair, and so on. The negatively-correlated genes were involved in Th17 cell differentiation, cell junction organization, vesicle organization, and so on (Figure 3B). These data suggested that CHK2 may be active in these bioprocesses and signaling pathways in the pathogenesis and development of squamous cell carcinoma.
CHK2 Inhibitor BML-277 Reduced LSCC Cell Proliferation
Given that CHK2 plays a role in LSCC development, we then evaluated the effect of CHK2 on cell growth in Tu212 cells. Transient CHK2 knockdown by siRNA was performed, and CHK2 expression was significantly attenuated (Figure 4A). Further, knockdown of CHK2 suppressed the cell growth ability of Tu212 cells, which also suggested an oncogenic potential of CHK2 (Figure 4B). We then sought to investigate the effects of CHK2 inhibitor on LSCC cells. BML-277 is an ATP competitive CHK2 kinase inhibitor, which binds the ATP binding site of CHK2.23 We evaluated the active and inactive forms of CHK2 expression under BML-277 treatment. The results showed that the level of p-CHK2 was decreased under BML-277 treatment, while total CHK2 expression was not obviously changed (Figure 4C), suggesting that BML-277 repressed CHK2 activity. Moreover, we measured cell proliferation by the MTS method. In Tu212 and HEP-2 cells, BML-277 inhibited proliferation of cells in a dose-dependent manner, with maximum suppression observed at 10 μM (Figure 4D and F). Further, Tu212 and HEP-2 cells were treated with 5 μM BML-277 for different time intervals, and cell viability was inhibited in a time-dependent manner (Figure 4E and G). These data suggested that CHK2 inhibition had an anti-growth effect on LSCC cells.
Discussion
Distinct oncogenic signaling pathways have been associated with LSCC, and finding new molecular markers for early detection and treatment in LSCC progression is worthy and important.24 Here, we report that CHK2, which is a key mediator of the DNA damage checkpoint in DDR, may act as an oncogenic gene and administration of a CHK2 inhibitor shows antitumor function in LSCC. Our data validate the potential therapeutic targeting of CHK2 in LSCC, which provides a basis for further experimental and clinical research.
CHK2 demonstrated both tumor-suppressor functions25 and the capacity to promote tumor and cell survival. Overexpression, phosphorylation and mislocalization within the mitotic spindle of CHK2 disrupt the sensitive balance of mitotic proteins, which in turn undermines faithful chromosome segregation and contributes to lagging chromosomes during HCC progression.15 In line with those reports, our data found the expression levels of CHK2 protein were up-regulated in LSCC tissues (Figure 1). Remarkably, significant differences were discovered between positive and negative CHK2 patients within lymph-node metastasis and differentiation (Table 2). More importantly, positive expression of CHK2 in LSCC patients may contribute to bad prognosis (Figure 2). Our data suggested that CHK2 might act as an oncogenic factor in the progression of LSCC; nevertheless, further research studies are required to clarify the functions of CHK2 in vivo.
Using bioprocess and signaling pathway analyses, the biological functions of CHK2 in LSCC were predicted (Figure 3). These bioprocesses and pathways are known to be involved in both LSCC progression and CHK2 function, for example, cell cycle and DNA repair.9,26,27 But further experimental research is required for better understanding the key mechanisms responsible for the processes.
CHK2 mutations have been detected with high incidence in a subset of cancers, including breast, prostate, ovarian, colorectal, kidney, thyroid, and bladder cancers and leukemias.28 Mice harboring the mutant CHEK2 allele are not only at an elevated risk for the development of cancer but also this risk can be further increased as a result of environmental exposure.29 In LSCC, it is not known whether CHK2 protein is mutated or its mutation rate, which will necessitate further investigation. Also, the phosphorylation status of CHK2 in LSCC is another valuable issue, which will make it necessary to further evaluate.
A previous study illustrated that CHK2 is overexpressed in diffuse large B-cell lymphoma (DLBCL) and addressed the therapeutic efficacy of combining ERK and CHK2 inhibitors in the setting of DLBCL.30 Our data make it possible to consider the role that CHK2 plays in tumorigenesis, which indicates CHK2 may serve as a target of cancer therapy. BML-277 is an ATP-competitive inhibitor of CHK2.23 In our study, inhibition of CHK2 by BML-277 significantly inhibited LSCC cell viabilities in vitro (Figure 4). These results prove the possible role of CHK2 in tumorigenesis that consequently functions as a therapeutic target in LSCC. Given that the observed growth inhibition by BML-277 in both lines is modest, future research studies evaluating the interaction between BML-277 and chemotherapy or radiation response are required.
Everything considered, our study indicated that CHK2 expression is up-regulated in LSCC, which associates with advanced clinical features and poor prognosis. Further, inhibition of CHK2 could lead to suppression of proliferation in LSCC cells. Our report provides an insight to support a previously unknown role of CHK2 in LSCC progression and suggests a potential role in the treatment of LSCC. The small sample size, as well as the short-term follow-up, may impose some limitations in our study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Cossu AM, Mosca L, Zappavigna S, et al. Long non-coding RNAs as important biomarkers in laryngeal cancer and other head and neck tumours. Int J Mol Sci. 2019;20(14):3444. doi:10.3390/ijms20143444
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Steuer CE, El-Deiry M, Parks JR, Higgins KA, Saba NF. An update on larynx cancer. CA Cancer J Clin. 2017;67(1):31–50. doi:10.3322/caac.21386
4. Kuper H, Boffetta P, Adami HO. Tobacco use and cancer causation: association by tumour type. J Intern Med. 2002;252(3):206–224. doi:10.1046/j.1365-2796.2002.01022.x
5. Boffetta P, Hashibe M. Alcohol and cancer. Lancet Oncol. 2006;7(2):149–156. doi:10.1016/S1470-2045(06)70577-0
6. Garavello W, Lucenteforte E, Bosetti C, et al. Diet diversity and the risk of laryngeal cancer: a case-control study from Italy and Switzerland. Oral Oncol. 2009;45(1):85–89. doi:10.1016/j.oraloncology.2008.02.011
7. Kobayashi K, Hisamatsu K, Suzui N, Hara A, Tomita H, Miyazaki T. A review of HPV-related head and neck cancer. J Clin Med. 2018;7(9):241. doi:10.3390/jcm7090241
8. Pommier Y, Weinstein JN, Aladjem MI, Kohn KW. Chk2 molecular interaction map and rationale for Chk2 inhibitors. Clin Cancer Res. 2006;12(9):2657–2661. doi:10.1158/1078-0432.CCR-06-0743
9. Zannini L, Delia D, Buscemi G. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol. 2014;6(6):442–457. doi:10.1093/jmcb/mju045
10. Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27(3):1007–1016. doi:10.1128/MCB.01068-06
11. Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy - two sides of the same coin? Nat Rev Cancer. 2007;7(12):925–936. doi:10.1038/nrc2251
12. Sun X, Liu B, Wang J, Li J, Ji WY. Inhibition of p21-activated kinase 4 expression suppresses the proliferation of Hep-2 laryngeal carcinoma cells via activation of the ATM/Chk1/2/p53 pathway. Int J Oncol. 2013;42(2):683–689. doi:10.3892/ijo.2012.1718
13. Cybulski C, Gorski B, Huzarski T, et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75(6):1131–1135. doi:10.1086/426403
14. Bartkova J, Guldberg P, Gronbaek K, et al. Aberrations of the Chk2 tumour suppressor in advanced urinary bladder cancer. Oncogene. 2004;23(52):8545–8551. doi:10.1038/sj.onc.1207878
15. Carloni V, Lulli M, Madiai S, et al. CHK2 overexpression and mislocalisation within mitotic structures enhances chromosomal instability and hepatocellular carcinoma progression. Gut. 2018;67(2):348–361. doi:10.1136/gutjnl-2016-313114
16. Yoon AJ, Shen J, Santella RM, Zegarelli DJ, Chen R, Weinstein IB. Activated checkpoint kinase 2 expression and risk for oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16(12):2768–2772. doi:10.1158/1055-9965.EPI-07-0659
17. Chou SJ, Alawi F. Expression of DNA damage response biomarkers during oral carcinogenesis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2011;111(3):346–353. doi:10.1016/j.tripleo.2010.10.032
18. Cardoso SCS, Duarte A, de Almeida LY, León JE, Ribeiro-Silva A. Immunohistochemistry analysis of checkpoint kinase 2 in oral squamous cell carcinoma. Appl Cancer Res. 2020;40(1):1–6. doi:10.1186/s41241-020-00085-y
19. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
20. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. doi:10.1093/nar/gkx1090
21. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
22. Fatehi Hassanabad A, Chehade R, Breadner D, Raphael J. Esophageal carcinoma: towards targeted therapies. Cell Oncol. 2019.
23. Arienti KL, Brunmark A, Axe FU, et al. Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J Med Chem. 2005;48(6):1873–1885. doi:10.1021/jm0495935
24. Wick CC, Rezaee RP, Wang T, et al. Use of concurrent chemoradiation in advanced staged (T4) laryngeal cancer. Am J Otolaryngol. 2017;38(1):72–76. doi:10.1016/j.amjoto.2016.10.001
25. Li J, Williams BL, Haire LF, et al. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell. 2002;9(5):1045–1054. doi:10.1016/S1097-2765(02)00527-0
26. Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3(5):421–429. doi:10.1016/S1535-6108(03)00110-7
27. Sun H, Wang Y, Wang Z, Meng J, Qi Z, Yang G. Aurora-A controls cancer cell radio- and chemoresistance via ATM/Chk2-mediated DNA repair networks. Biochim Biophys Acta. 2014;1843(5):934–944. doi:10.1016/j.bbamcr.2014.01.019
28. Wu X, Webster SR, Chen J. Characterization of tumor-associated Chk2 mutations. J Biol Chem. 2001;276(4):2971–2974. doi:10.1074/jbc.M009727200
29. Bahassi EM, Robbins SB, Yin M, et al. Mice with the CHEK2*1100delC SNP are predisposed to cancer with a strong gender bias. Proc Natl Acad Sci U S A. 2009;106(40):17111–17116. doi:10.1073/pnas.0909237106
30. Dai B, Zhao XF, Mazan-Mamczarz K, et al. Functional and molecular interactions between ERK and CHK2 in diffuse large B-cell lymphoma. Nat Commun. 2011;2:402. doi:10.1038/ncomms1404
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.