Back to Journals » Infection and Drug Resistance » Volume 14
The Escherichia coli Sequence Type 131 Harboring Extended-Spectrum Beta-Lactamases and Carbapenemases Genes from Poultry Birds
Authors Ilyas S, Rasool MH, Arshed MJ, Qamar MU , Aslam B, Almatroudi A , Khurshid M
Received 13 January 2021
Accepted for publication 19 February 2021
Published 2 March 2021 Volume 2021:14 Pages 805—813
DOI https://doi.org/10.2147/IDR.S296219
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Sana Ilyas,1 Muhammad Hidayat Rasool,1 Muhammad Javed Arshed,2 Muhammad Usman Qamar,1 Bilal Aslam,1 Ahmad Almatroudi,3 Mohsin Khurshid1
1Department of Microbiology, Government College University, Faisalabad, Pakistan; 2National Veterinary Laboratory, National Agriculture Research Council, Islamabad, Pakistan; 3Department of Medical Laboratories, College of Applied Medical Sciences, Qassim University, Buraydah, Saudi Arabia
Correspondence: Muhammad Hidayat Rasool
Department of Microbiology, Government College University, Faisalabad 38000, Pakistan
Tel +92 41-9203023
Email [email protected]
Background and Aim: The extended-spectrum beta-lactamases (ESBLs), as well as carbapenemases, are considered as the foremost resistance determinants throughout the world. However, the relevant data especially related to the sequence types of ESBL and carbapenemases producing Escherichia coli from the poultry is limited from Pakistan. Here, we present the data on the genetic diversity of E. coli strains isolated from the poultry birds from the poultry farms located in Islamabad, Pakistan, and the underlying resistance mechanisms to beta-lactam agents.
Methods: Of 250 broilers from 25 different farms (10 birds from each farm), the cecal samples were obtained and analyzed for the presence of ESBLs producing E. coli (ESBL-Ec) as well as carbapenemases producing E. coli (CPEc) strains using selective agar for ESBL and carbapenemases screening. The susceptibility profiling of the ESBL-Ec and CPEc isolates was evaluated followed by multi-locus sequence typing.
Results: A total of 119 strains were positive for ESBL production whereas 37 strains were found positive to produce carbapenemases in addition to ESBLs. The MLST analysis has shown a diversity of isolates as the E. coli isolates from poultry birds correspond to a total of 16 sequence types (STs). The ST131 (22/48, 46%) followed by ST8051 (10/48, 21%) were the main STs in this study. The blaCTX-M gene was detected in all the poultry E. coli strains whereas the blaTEM was found in 45.5% of strains. The blaVIM was found in all 37 CPEc isolates whereas the blaNDM and blaIMP were found in 31/37 (83.8%) and 16/37 (43.2%) CPEc isolates respectively.
Conclusion: The overall results have shown the prevalence of diverse genotypes among the ESBL-Ec and carbapenemase-producing E. coli (CPEC) from poultry. Furthermore, the study documents poultry birds as a persisting reservoir of extensively antimicrobial-resistant E. coli ST131 in Pakistan, suggesting a potential threat to public health.
Keywords: MLST, E. coli, poultry, Pakistan, AMR, ESBL
Introduction
The extended-spectrum beta-lactamases (ESBLs) as well as carbapenemases are increasingly being spreading to members of the Enterobacteriaceae family, which results in limiting the therapeutic options for the management of infections caused by these organisms.1–3 Escherichia coli strains that harbor these enzymes have been reported to colonize healthy human beings as well as animals such as poultry birds.4 It is widely accepted that the increased and indiscriminate use of antimicrobial drugs for humans and animals is the main driving force for the increasing colonization and infections caused by ESBL producing E. coli strains.5–7
The gut colonization by the ESBL producing E. coli (ESBL-Ec) and carbapenemase-producing E. coli (CPEC), is an important feature for the spread of these multidrug resistant (MDR) bacteria among livestock especially poultry.4,8 Studies have widely reported the presence of ESBL-producing E. coli (ESBL-EC) strains in the food-producing animals, especially poultry, which poses a serious threat to public health.9,10 ESBL producing MDR strains have also been reported from European countries such as Netherlands and Germany from poultry birds despite the controlled use of antibiotics in poultry farming.11,12 The ESBL-Ec in poultry farms has also been reported from Asian countries, such as China, Japan, Thailand, and Vietnam, although, the prevalence of ESBL-Ec strains varied with the sampling strategy and the study catchment area.13–16 Moreover, the clonal dissemination of ESBL-Ec strains among the poultry and surrounding environments, as well as poultry-human transmission, has also been reported.16,17 Despite the carbapenems are not approved for use in livestock according to the Animal Medicinal Drug Use Clarification Act (1994) in the United States and similar guidelines in many other countries. However, studies have demonstrated the spread of E. coli harboring these resistance determinists among pets or livestock and to exposed humans.18,19
In Pakistan, the few published studies have confirmed the occurrence of ESBL-Ec isolates from poultry.20,21 Most studies are limited to a single large poultry farm and no data is available regarding the sequence types of ESBLs and carbapenemases producing E. coli. The extent of the spread of ESBLs and carbapenemase among poultry birds can be helpful to determine the implication of these potential reservoirs as a source of transmission of such MDR strains to humans. Therefore, it is important to screen the poultry for the colonization of ESBLs and carbapenemase-producing strains in poultry birds. Therefore, the present study was designed to explore the occurrence of the sequence types ESBLs and carbapenemase-producing E. coli from the poultry farms of the capital region of Pakistan.
Materials and Methods
Ethical Approval
The research proposal was approved by the Ethical Review Committee (ERC), Government College University, Faisalabad, Pakistan, with reference number GCUF/ERC/17/16. Furthermore, the prior consent to collect poultry samples was taken from farm owners individually.
Samples and Bacterial Strains
The current study was conducted in Islamabad, the capital territory of Pakistan, between February 2018 to June 2019 at 25 poultry farms in and around Islamabad, Pakistan. The poultry farms were having around 500–1500 chickens on each farm and infrastructure was comparable in all the poultry farms. The 250 chicken ceca samples (10 samples from each farm) were collected and transported to the laboratory within 2 hours.
Screening for ESBL and Carbapenemases Producing E. coli
The cecal material (1 gram) was suspended in 9 mL sterile saline (0.9%) and mixed by gentle vortex for 30–45 seconds. A small amount (100μL) of this cecal saline suspension was inoculated on the commercial HiCrome™ ESBL Agar Base (HiMedia, India) and MacConkey agar comprising cefotaxime (4 μg/mL) for screening the ESBL producing E. coli. For the screening of carbapenem-resistant E. coli, the MacConkey agar supplemented with imipenem (1 μg/mL) was used. The plates were aerobically incubated at 37°C for 24 hours.
The isolated colonies were sub-cultured on nutrient agar for phenotypic identification, preservation, and subsequent antimicrobial susceptibility testing. The species-level identification was done by standard microbiological methods and further confirmed by the API 20E kit (BioMérieux, France). All isolates including the ESBL-Ec and CPEC were preserved in brain heart infusion (BHI) broth and glycerol in cryovials and stored at −20°C.
Phenotypic Detection of ESBL and Carbapenemase Production
For the detection of ESBL production, the double-disc synergy test was performed for all isolates using the combination disc test (CDT) according to the CLSI guidelines. Briefly, the test discs containing cephalosporin a (cefotaxime and ceftazidime) alone and the combination of cephalosporins in combination with clavulanic acid were applied on the surface of Muller Hinton agar (Oxoid, UK). The ESBL production was considered positive in case, the zone of inhibition for the cephalosporin-clavulanic acid disc is ≥ 5 mm larger than the zone of the cephalosporin disc alone. The isolates were further screened for the carbapenemase production by the modified Hodge test as described previously.22
Antibiotic Susceptibility Testing
The antibiotic susceptibility testing of the ESBL positive isolates was performed by the Kirby Bauer disc diffusion method as per CLSI guidelines and interpreted accordingly.23 The following antimicrobial agents were used; cefuroxime, ceftazidime, cefotaxime, cefixime, ceftriaxone, cefepime, ampicillin-clavulanic acid, and piperacillin-tazobactam, imipenem and meropenem, aztreonam, ciprofloxacin, amikacin and gentamicin, and trimethoprim-sulfamethoxazole. The E. coli (ATCC® 25922) and K. pneumoniae (ATCC® 700603) were used for quality control. The antimicrobial disks used in the study were obtained from Oxoid (Oxoid, UK).
The minimal inhibitory concentrations (MICs) of cefuroxime, cefotaxime, cefepime as well as imipenem, and colistin were determined by the broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI, 2018) guidelines. Pseudomonas aeruginosa (ATCC® 27853) and E. coli (ATCC® 25922) and were used for quality control.
Molecular Characterization of β-Lactamase Encoding Genes
All the 156 isolates were also screened for the ESBL encoding genes including blaCTX-M, blaTEM, and blaSHV genes. The isolates positive for blaCTX-M were further screened for the blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9, and blaCTX-M-25. The presence of class B beta-lactamases such as blaIMP, blaVIM, and blaNDM was also detected by PCR assays for all the 156 isolates. The primers (Macrogen, South Korea) were used and the expected size of PCR products for these enzymes are shown in Table 1. For the DNA extraction, a commercially available DNA extraction kit FavorPrep™ (FAVORGEN Biotech Corporation, Taiwan) was used. The PCR reactions (30 μL) was used using 15 μL 2X DreamTaq Green master mix (Thermo Fisher Scientific, Massachusetts, USA), 1 μL of each primer (forward and reverse) were used and 1 μL of the template DNA was used. The PCR water was used to make a total volume of 30 μL. The agarose gel (1.5%) was used to visualize the amplicons after electrophoresis at 100V for 30–45 minutes. All the amplified samples were sent for Sanger sequencing from Macrogen, Inc (South Korea) The obtained sequences were aligned and compared with the sequences in the GenBank database using the NCBI BLAST tool.
 |
Table 1 Primers and Annealing Temperature for the Amplification of ESBLs and Carbapenemases Genes |
Multi-Locus Sequence Typing (MLST)
For the MLST, seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) were amplified and sequenced according to the conditions provided by the EnteroBase Database.24 The obtained PCR products were sent for sequencing to Macrogen (South Korea). The raw sequences were edited using the ChromasPro software (Technelysium, Australia). The sequences were aligned using the ClustalW algorithm (MEGA7 software). The allele number was assigned to each gene locus and the sequence types (STs) were determined according to the allelic profiles for each of the isolates.
Results
Prevalence of ESBL and Carbapenemases Producing E. coli
A total of 156 (62.4%) strains were positive for ESBL production. Among these 156 strains, 119 strains were positive for ESBLs only whereas a total of 37 (14.8%) strains were found positive for carbapenemases production in addition to ESBL production.
Antibiotic Resistance Profile
The data obtained from the antibiotic sensitivity testing obtained by disc diffusion tests have been shown in Table 2. The ESBL-Ec isolates showed the highest resistance to sulfamethoxazole-trimethoprim (83%) followed by ciprofloxacin (74.4%) and aminoglycosides ie gentamicin (45%) and amikacin (44%). 37 (23.7%) isolates were found resistant to carbapenems (imipenem and meropenem). All (n=156) ESBL-Ec were found susceptible to colistin.
 |
Table 2 Overall Distribution of MICs of Various β-Lactams Agents Against Escherichia coli (n=156) Strains |
Diversity of Beta-Lactamases Genes
All one hundred and fifty-six isolates were screened for the ESBL gene and all were found positive for the blaCTX-M gene. Overall, the blaCTX-M-1 was found in 131 isolates followed by blaCTX-M-9 which was found in 27 isolates. The blaCTX-M-2 and blaCTX-M-9 were found from 19 isolates. The blaTEM was the second most common ESBL gene which was found in 71 isolates, whereas the blaSHV was found in 28 isolates. Many of the strains were found to co-harbor multiple ESBL genes. Among the carbapenemases genes, the blaVIM was the most common gene and was found in all the 37 carbapenem-resistant isolates whereas the blaNDM gene was found in 31/37 (83.8%) isolates and blaIMP were found in 16/37 (43.2%) isolates.
Sequence Types of ESBL-Ec
The analysis of MLST results has shown that the 48 E. coli strains were classified into 17 different sequence types (STs) including the 10 already reported STs and seven new STs. The ST131 (22/48, 46%) was the most common ST, followed by ST8051 (10/48, 21%). The ST8900 and ST2847 accounted for 4%, whereas the STs including ST8420, ST8431, ST2741, ST3499, and ST6293 accounted for 2% (1/48) (Table 3). The ST131 was found to harbor the ESBL (blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9 blaTEM, blaSHV) and carbapenemases (blaVIM and blaNDM) genes. The ST8051 was harboring the blaCTX-M-1, blaSHV, blaIMP, and blaVIM gene combinations.
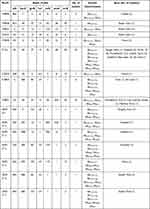 |
Table 3 Distribution of Various Genetic Determinants Among Different Escherichia coli Sequence Types (STs) Isolated from Poultry in Islamabad, Pakistan |
Discussion
Antimicrobial resistance is a serious global issue; however, in developing countries such as Pakistan, the indiscriminate use of antimicrobials especially in veterinary settings has worsened the situation.25–27 The emergence of ESBLs and carbapenemases among the Enterobacteriaceae family in the food-producing animals can impact the consumer’s health as there is growing evidence supporting the hypothesis that the animals can be a source of antimicrobial resistance and can be involved in the spread of such resistant bacterial strains. Recent studies have highlighted the similarity between the E. coli strains isolated from the poultry and humans in terms of antimicrobial resistance determinants, which suggests the transmission of these strains from poultry to humans.28,29
In this study, 62.4% (156/250) of the chicken cecal samples were found to harbor the ESBL producing E. coli. This is comparable to previous studies showing a varying level of ESBL phenotypes in poultry birds. In a study from Punjab, India a total of 1,556 Escherichia coli isolates were obtained from 18 poultry farms and have reported the prevalence of ESBL-producing E. coli strains as 87% in broilers while 42% in layers.30 The incidence of ESBL-Ec in poultry as reported in the previous studies was relatively less, ie, 30% in a study from Bangladesh31 and 10.7% in a report in France.32
In our study, the ESBL-Ec strains were resistant to ciprofloxacin (74.4%), sulfamethoxazole-trimethoprim (83%), gentamicin (45%), and amikacin (44%). The susceptibility profiles of the E. coli isolates from the poultry are quite variable. A recent study from Ethiopia has shown that the E. coli isolates were 100% susceptible to fluoroquinolones and sulfamethoxazole-trimethoprim, whereas the susceptibility to gentamicin, streptomycin, and kanamycin was 93%, 85%, and 75% respectively.33 However, a few studies have reported highly resistant strains of E. coli which were resistant to 100 resistant to nalidixic acid while the resistance to kanamycin and streptomycin was 77% and 67% respectively.34 These variations may be attributed to the differences in the use of antimicrobial agents in different regions in humans and poultry or due to the presence of diverse clones of E. coli.
The 37 isolates were found resistant to carbapenems (imipenem and meropenem). A study from China has recently isolated a blaNDM-5 producing E. coli strain ST156 from a poultry farm. Which showed resistance to twenty-seven antimicrobial agents including carbapenems.35 In a study comprising of 220 samples of chicken litter, chicken feed, intestinal content, and water, the Escherichia species were the most prevalent (39%) bacterial species. A total of 42 carbapenem-resistant isolates were found.36 In a systematic review published in 2018, the prevalence of CRE was reported from the livestock. The prevalence was low in the European countries including the UK, Netherlands, and Switzerland, ie, <1%. However, the prevalence was higher in the Asian countries including China, India, Lebanon, and Algeria.19
In the present study, we have found that ST131 was the most prevalent sequence type (ST) among the E. coli strains subjected to MLST ie 46% (22/48). The previous studies have shown that the ST131 E. coli have been frequently isolated in the clinical settings and frequently harboring the blaCTX-M gene with multidrug-resistant phenotypes.37 This ST is also identified in various animal reservoirs, which shows that this clone has not only adapted to humans but other reservoirs as well, including animals.37,38 It is also suggested that due to the acquisition of multiple virulence factors, the ST131 is also involved in the increase of E. coli carrying the blaCTX-M genes responsible for ESBL phenotypes.37
The studies that have indicated the presence of ST131 in the food animals are quite limited. In a study from Italy, only one E. coli ST131 isolates that was not producing ESBL was identified among a total of 101 E. coli isolates in healthy turkeys and chickens.39 One E. coli ST131 isolates harboring blaCTXM-1 was found among 22 ESBL producers in a total collection of 1378 E. coli strains from different animals mainly poultry, pigs, and cattle.40 A single E. coli ST131 isolate was found in a pig with a gastrointestinal infection. A study reported the ESBL producing E. coli ST131 carrying blaCTX-M-9 from the poultry birds in different countries from the samples collected at different periods.41 In contrast to these findings, many studies indicated the absence of E. coli ST131 from broilers, turkey sheep, cattle, and pigs.42–45
The ST8900 has recently been reported among the avian origin colistin-resistant E. coli in Shandong Province, China harboring the mcr-1 gene in addition to blaCTXM and blaTEM genes.46 The E. coli ST2847 have also been reported from Faisalabad, Pakistan from healthy broilers and was found resistant to colistin and positive for mcr-1 gene.47 In contrast to these studies, both STs (ST8900 and ST2847) in our study were susceptible to colistin although positive for ESBL phenotypes and harboring the blaCTXM and blaTEM genes as found in the previous reports.
Conclusion
The molecular characterization and MLST of ESBL and carbapenemases producing Escherichia coli have helped to establish the genetic determinants among various sequence types for the first time in poultry settings in Pakistan. The disparities in the distribution of genetic determinists among various strains as seen in our study compared to the other reports may be attributed to the variable practices of poultry farming and the use of antimicrobial agents. It is important to find the presence of ST131 from poultry birds as this ST was previously reported from the clinical settings in most of the studies. The further characterization of ESBL producing E. coli from poultry, human as well as environmental sources in Pakistan is essential to understand the transmission dynamics of resistance determinants. The control strategies are necessary to prevent the dissemination of ESBL-Ec and CRE in animals, humans, and foods.
Acknowledgments
We are thankful to Dr. Amir Bin Zahoor (Director General), Dr. Muhammad Abubaker (Senior Scientific Officer), Dr. Shumaila Manzoor (Laboratory Technologist), and Mr. Eid Nawaz Khan (Laboratory Technologist) for their guidance and support.
Funding
This work was supported by the World Health Organization for Global Tricycle project on ESBL positive E. coli and partially by the grant from the National Veterinary Laboratory, Ministry of Food and Security.
Disclosure
Ms Sana Ilyas reports grants from WHO, during the conduct of the study. The authors have declared no conflict of interest.
References
1. Doi Y, Iovleva A, Bonomo RA. The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J Travel Med. 2017;24(suppl_1):S44–s51. doi:10.1093/jtm/taw102
2. Saeed M, Rasool MH, Rasheed F, et al. Extended-spectrum beta-lactamases producing extensively drug-resistant Salmonella Typhi in Punjab, Pakistan. J Infect Dev Ctries. 2020;14(2):169–176. doi:10.3855/jidc.12049
3. Rasheed F, Saeed M, Alikhan NF, et al. Emergence of Resistance to Fluoroquinolones and Third-Generation Cephalosporins in Salmonella Typhi in Lahore, Pakistan. Microorganisms. 2020;8:9. doi:10.3390/microorganisms8091336
4. Carattoli A. Animal reservoirs for extended spectrum beta-lactamase producers. Clin Microbiol Infect. 2008;14(Suppl 1):117–123. doi:10.1111/j.1469-0691.2007.01851.x
5. Hawkey PM. Prevalence and clonality of extended-spectrum beta-lactamases in Asia. Clin Microbiol Infect. 2008;14(Suppl 1):159–165. doi:10.1111/j.1469-0691.2007.01855.x
6. Nguyen VT, Jamrozy D, Matamoros S, et al. Limited contribution of non-intensive chicken farming to ESBL-producing Escherichia coli colonization in humans in Vietnam: an epidemiological and genomic analysis. J Antimicrob Chemother. 2019;74(3):561–570. doi:10.1093/jac/dky506
7. Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–1658.
8. Ojer-Usoz E, González D, Vitas AI, et al. Prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Sci. 2013;93(2):316–321. doi:10.1016/j.meatsci.2012.09.009
9. Seiffert SN, Hilty M, Perreten V, Endimiani A. Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: an emerging problem for human health? Drug Resist Updat. 2013;16(1–2):22–45.
10. Saliu EM, Vahjen W, Zentek J. Types and prevalence of extended-spectrum beta-lactamase producing Enterobacteriaceae in poultry. Anim Health Res Rev. 2017;18(1):46–57. doi:10.1017/S1466252317000020
11. Reich F, Atanassova V, Klein G. Extended-spectrum β-lactamase- and AmpC-producing enterobacteria in healthy broiler chickens, Germany. Emerg Infect Dis. 2013;19(8):1253–1259. doi:10.3201/eid1908.120879
12. Huijbers PM, Graat EA, Haenen AP, et al. Extended-spectrum and AmpC β-lactamase-producing Escherichia coli in broilers and people living and/or working on broiler farms: prevalence, risk factors and molecular characteristics. J Antimicrob Chemother. 2014;69(10):2669–2675. doi:10.1093/jac/dku178
13. Tansawai U, Walsh TR, Niumsup PR. Extended spectrum ß-lactamase-producing Escherichia coli among backyard poultry farms, farmers, and environments in Thailand. Poult Sci. 2019;98(6):2622–2631. doi:10.3382/ps/pez009
14. Li S, Zhao M, Liu J, Zhou Y, Miao Z. Prevalence and antibiotic resistance profiles of extended-spectrum β-lactamase-producing escherichia coli isolated from healthy broilers in Shandong Province, China. J Food Prot. 2016;79(7):1169–1173. doi:10.4315/0362-028X.JFP-16-025
15. Ueda S, Ngan BT, Huong BT, Hirai I, Tuyen le D, Yamamoto Y. Limited transmission of bla(CTX-M-9)-type-positive Escherichia coli between humans and poultry in Vietnam. Antimicrob Agents Chemother. 2015;59(6):3574–3577. doi:10.1128/AAC.00517-15
16. Kameyama M, Chuma T, Yabata J, Tominaga K, Iwata H, Okamoto K. Prevalence and epidemiological relationship of CMY-2 AmpC β-lactamase and CTX-M extended-spectrum β-lactamase-producing Escherichia coli isolates from broiler farms in Japan. J Vet Med Sci. 2013;75(8):1009–1015. doi:10.1292/jvms.12-0453
17. Ma J, Liu JH, Lv L, et al. Characterization of extended-spectrum β-lactamase genes found among Escherichia coli isolates from duck and environmental samples obtained on a duck farm. Appl Environ Microbiol. 2012;78(10):3668–3673. doi:10.1128/AEM.07507-11
18. Poirel L, Stephan R, Perreten V, Nordmann P. The carbapenemase threat in the animal world: the wrong culprit. J Antimicrob Chemother. 2014;69(7):2007–2008. doi:10.1093/jac/dku054
19. Köck R, Daniels-Haardt I, Becker K, et al. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect. 2018;24(12):1241–1250. doi:10.1016/j.cmi.2018.04.004
20. Umair M, Mohsin M, Ali Q, et al. Prevalence and genetic relatedness of extended spectrum-β-lactamase-producing escherichia coli among humans, cattle, and Poultry in Pakistan. Microb Drug Resist. 2019;25(9):1374–1381. doi:10.1089/mdr.2018.0450
21. Ahmad K, Khattak F, Ali A, et al. Carbapenemases and extended-spectrum β-lactamase–producing multidrug-resistant escherichia coli isolated from retail chicken in Peshawar: first report from Pakistan. J Food Prot. 2018;81(8):1339–1345. doi:10.4315/0362-028X.JFP-18-045
22. Amjad A, Mirza I, Abbasi S, Farwa U, Malik N, Zia F. Modified Hodge test: a simple and effective test for detection of carbapenemase production. Iran J Microbiol. 2011;3(4):189–193.
23. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
24. Wirth T, Falush D, Lan R, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60(5):1136–1151. doi:10.1111/j.1365-2958.2006.05172.x
25. Baloch Z, Aslam B, Muzammil S, Khurshid M, Rasool MH, Ma K. Selection inversion: a probable tool against antibiotic resistance. Infect Drug Resist. 2018;11:1903–1905.
26. Khurshid M, Rasool MH, Ashfaq UA, Aslam B, Waseem M. Emergence of ISAba1 harboring carbapenem-resistant Acinetobacter baumannii isolates in Pakistan. Future Microbiol. 2017;12:1261–1269. doi:10.2217/fmb-2017-0080
27. Aslam B, Chaudhry TH, Arshad MI, et al. The First bla(KPC) Harboring Klebsiella pneumoniae ST258 Strain Isolated in Pakistan. Microb Drug Resist. 2020;26(7):783–786. doi:10.1089/mdr.2019.0420
28. Liebana E, Carattoli A, Coque TM, et al. Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin Infect Dis. 2013;56(7):1030–1037. doi:10.1093/cid/cis1043
29. Lazarus B, Paterson DL, Mollinger JL, Rogers BA. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis. 2015;60(3):439–452. doi:10.1093/cid/ciu785
30. Brower CH, Mandal S, Hayer S, et al. The prevalence of extended-spectrum beta-lactamase-producing multidrug-resistant Escherichia Coli in poultry chickens and variation according to farming practices in Punjab, India. Environ Health Perspect. 2017;125(7):077015. doi:10.1289/EHP292
31. Hasan B, Sandegren L, Melhus A, et al. Antimicrobial drug-resistant Escherichia coli in wild birds and free-range poultry, Bangladesh. Emerg Infect Dis. 2012;18(12):2055–2058. doi:10.3201/eid1812.120513
32. Girlich D, Poirel L, Carattoli A, et al. Extended-spectrum beta-lactamase CTX-M-1 in Escherichia coli isolates from healthy poultry in France. Appl Environ Microbiol. 2007;73(14):4681–4685. doi:10.1128/AEM.02491-06
33. Sarba EJ, Kelbesa KA, Bayu MD, Gebremedhin EZ, Borena BM, Teshale A. Identification and antimicrobial susceptibility profile of Escherichia coli isolated from backyard chicken in and around ambo, Central Ethiopia. BMC Vet Res. 2019;15(1):85. doi:10.1186/s12917-019-1830-z
34. Salehi TZ, Bonab SF. Antibiotics susceptibility pattern of Escherichia coli strains isolated from chickens with colisepticemia in Tabriz province, Iran. Int J Poultry Sci. 2006;5(7):677–684. doi:10.3923/ijps.2006.677.684
35. Tang B, Chang J, Cao L, et al. Characterization of an NDM-5 carbapenemase-producing Escherichia coli ST156 isolate from a poultry farm in Zhejiang, China. BMC Microbiol. 2019;19(1):82. doi:10.1186/s12866-019-1454-2
36. Elmanama AA, Al-Reefi MR, Shamali MA, Hemaid HI. Carbapenem-resistant Gram-negative bacteria isolated from poultry samples: a cross-sectional study. The Lancet. 2019;393:S21. doi:10.1016/S0140-6736(19)30607-5
37. Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565–591. doi:10.1128/CMR.00116-14
38. Platell JL, Johnson JR, Cobbold RN, Trott DJ. Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet Microbiol. 2011;153(1–2):99–108. doi:10.1016/j.vetmic.2011.05.007
39. Giufrè M, Graziani C, Accogli M, Luzzi I, Busani L, Cerquetti M. Escherichia coli of human and avian origin: detection of clonal groups associated with fluoroquinolone and multidrug resistance in Italy. J Antimicrob Chemother. 2012;67(4):860–867. doi:10.1093/jac/dkr565
40. Schink AK, Kadlec K, Kaspar H, Mankertz J, Schwarz S. Analysis of extended-spectrum-β-lactamase-producing Escherichia coli isolates collected in the GERM-Vet monitoring programme. J Antimicrob Chemother. 2013;68(8):1741–1749. doi:10.1093/jac/dkt123
41. Mora A, Herrera A, Mamani R, et al. Recent emergence of clonal group O25b: K1:H4-B2-ST131ibeA strains among Escherichia coli poultry isolates, including CTX-M-9-producing strains, and comparison with clinical human isolates. Appl Environ Microbiol. 2010;76(21):6991–6997. doi:10.1128/AEM.01112-10
42. Wu G, Day MJ, Mafura MTet al. Comparative analysis of ESBL-positive Escherichia coli isolates from animals and humans from the UK, The Netherlands and Germany. PLoS One. 2013;8(9):e75392.
43. Randall LP, Clouting C, Horton RAet al. Prevalence of Escherichia coli carrying extended-spectrum β-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J Antimicrob Chemother. 2011;66(1):86–95. doi:10.1093/jac/dkq396
44. Dahmen S, Métayer V, Gay E, Madec JY, Haenni M. Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. Vet Microbiol. 2013;162(2–4):793–799. doi:10.1016/j.vetmic.2012.10.015
45. Hordijk J, Wagenaar JA, van de Giessen A, et al. Increasing prevalence and diversity of ESBL/AmpC-type β-lactamase genes in Escherichia coli isolated from veal calves from 1997 to 2010. J Antimicrob Chemother. 2013;68(9):1970–1973. doi:10.1093/jac/dkt132
46. Zhao X, Liu Z, Zhang Y, Yuan X, Hu M, Liu Y. Prevalence and molecular characteristics of avian-origin mcr-1-Harboring Escherichia coli in Shandong Province, China. Front Microbiol. 2020;11:255. doi:10.3389/fmicb.2020.00255
47. Lv J, Mohsin M, Lei S, et al. Discovery of a mcr-1-bearing plasmid in commensal colistin-resistant Escherichia coli from healthy broilers in Faisalabad, Pakistan. Virulence. 2018;9(1):994–999. doi:10.1080/21505594.2018.1462060
48. Fang H, Lundberg C, Olsson-Liljequist B, et al. Molecular epidemiological analysis of Escherichia coli isolates producing extended-spectrum β-lactamases for identification of nosocomial outbreaks in Stockholm, Sweden. J Clin Microbiol. 2004;42(12):5917–5920. doi:10.1128/JCM.42.12.5917-5920.2004
49. Monstein Ö, Nilsson M, Nilsson M, Dornbusch K, Nilsson L. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX‐M genes in Enterobacteriaceae. APMIS. 2007;115(12):1400–1408. doi:10.1111/j.1600-0463.2007.00722.x
50. Boyd DA, Tyler S, Christianson S, et al. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother. 2004;48(10):3758–3764. doi:10.1128/AAC.48.10.3758-3764.2004
51. Woodford N, Ellington MJ, Coelho JM, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–353. doi:10.1016/j.ijantimicag.2006.01.004
52. Fallah F, Noori M, Hashemi A, et al. Prevalence of blaNDM, blaPER, blaVEB, blaIMP, and blaVIM Genes among Acinetobacter baumannii Isolated from Two Hospitals of Tehran, Iran. Scientifica. 2014;2014:245162. doi:10.1155/2014/245162
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
