Back to Journals » OncoTargets and Therapy » Volume 10
The combination therapy of salinomycin and gefitinib using poly(D,L-lactic-co-glycolic acid)-poly(ethylene glycol) nanoparticles for targeting both lung cancer stem cells and cancer cells
Authors Zhang Y, Zhang Q, Sun J, Liu H, Li Q
Received 4 May 2017
Accepted for publication 22 August 2017
Published 27 November 2017 Volume 2017:10 Pages 5653—5666
DOI https://doi.org/10.2147/OTT.S141083
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Faris Farassati
Yu Zhang,1,* Qi Zhang,1,* Jing Sun,2 Huijie Liu,1 Qingfeng Li1
1Department of Oncology, Xiangyang Central Hospital, The Affiliated Hospital of Hubei College of Arts and Science, Xiangyang 441000, Hubei, China; 2International Joint Cancer Institute, Second Military Medical University, Shanghai 200433, China
*These authors contributed equally to this work
Purpose: Lung cancer (LC) is the leading cause of cancer death worldwide. Evidences suggest that both LC cancer stem cells (CSCs) and cancer cells are supposed to be eliminated to achieve superior treatment effect against LC. Salinomycin could eradiate CSCs in various types of cancers, and gefitinib is a first-line therapy in LC. The purpose of the present study was to develop salinomycin-loaded nanoparticles (salinomycin-NPs) combined with gefitinib-loaded nanoparticles (gefitinib-NPs) to eradicate both LC CSCs and cancer cells.
Methods: Salinomycin and gefitinib were encapsulated separately by poly(D,L-lactic-co-glycolic acid)-poly(ethylene glycol) nanoparticles by the emulsion/solvent evaporation approach. The anti-LC activity of salinomycin-NPs and gefitinib-NPs was investigated.
Results: Salinomycin-NPs and gefitinib-NPs are of ~140 nm in size, high drug encapsulation efficacy and sustained release of drugs. CD133+ LC CSCs showed the characteristics of CSCs, including significantly enhanced stem cell gene expression, tumorsphere formation ability, and tumorigenicity in mice. Both salinomycin and salinomycin-NPs are capable of selectively inhibiting LC CSCs, as reflected by their enhanced cytotoxic effects toward CD133+ LC CSCs and ability to reduce tumorsphere formation in LC cell lines, whereas gefitinib and gefitinib-NPs could significantly inhibit LC cells. Salinomycin-NPs and salinomycin could reduce the population of LC CSCs in the tumors in vivo. It is noteworthy that salinomycin-NPs combined with gefitinib-NPs inhibited the growth of tumors more efficiently compared with salinomycin combined with gefitinib or single salinomycin-NPs or gefitinib-NPs.
Conclusion: Salinomycin-NPs combined with gefitinib-NPs represent a potential approach for LC by inhibiting both LC CSCs and cancer cells.
Keywords: cancer stem cells, lung cancer, nanoparticles, salinomycin, gefitinib
Introduction
For several decades, lung cancer (LC) is the leading cause of cancer death worldwide. The American Cancer Society predicts that, in 2017, in the USA, LC is the leading cause of cancer-related death among both men and women.1 The Chinese National Cancer Center showed that, in 2015, in China, LC is the most commonly diagnosed cancer and also the primary cause of death from cancer in men aged ≥75 years.2 Thus, the treatment of LC is an urgent task for public health. Despite the recent advances that have been achieved in the therapy of LC, the overall survival and relapse-free survival have been significantly hampered because of the recurrence, metastasis, and drug resistance of LC.3–5 One of the critical reasons for reduced survival and treatment failure is the cancer stem cells (CSCs) in LC.6,7 CSCs are cancer cells with the features of normal stem cells and are responsible for initiation, recurrence, and metastasis of cancer.8 They have unique characteristics that make them resistant to chemotherapy radiotherapy.8 Thus, destruction of LC CSCs is critical to the effective therapy of LC, thus obtaining better therapeutic efficacy against LC and prolonged survival of patients with LC.
Cluster of differentiation 133 (CD133) is regarded as a stem cell marker for both cancer and healthy tissues. To date, cancer cells expressing CD133 have been confirmed to be stem cells in pancreatic cancer, liver cancer, colon cancer, and other types of solid tumors, suggesting that CD133 is a common biomarker of CSCs.9,10 Recently, numerous studies have validated CD133 is a stem cell marker for LC.11 For example, Bertolin et al and Eramo et al have demonstrated that CD133+ LC cells exhibit a stronger potential than CD133− LC cells in the proliferation, differentiation, self-renewal, and tumorigenicity.12,13
Using a series of high-throughput screening, several old chemotherapy drugs have been identified to show potent anti-CSC activity.9 For example, salinomycin has shown potent therapeutic efficacy against various kinds of CSCs in breast cancer, liver cancer, and other cancers.14–17 Researchers have found that salinomycin exerts its potent therapeutic efficacy against CSCs via a series of mechanisms including the interference of the Wnt/β-catenin signaling and ATP-binding cassette transporters and autophagy blockade.14,17 Taken together, these studies indicate that salinomycin represents a promising agent against CSCs.
To the best of our knowledge, the therapeutic efficacy of salinomycin against LC CSCs has been rarely investigated. So far, there have been only two research papers that have investigated the anti-CSC activity of salinomycin in LC. In these two studies, salinomycin has been shown to be able to decrease the proportion of CSCs and tumorsphere forming ability in LC cell lines including murine LLC and human H460, H1299, and A549 cells.18,19 Thus, it is necessary to further validate the therapeutic efficacy of salinomycin against LC CSCs. There is another important issue on salinomycin to be solved before its application in clinic. Due to its poor solubility in water, salinomycin was generally dissolved in ethanol and administered by intraperitoneal injection.15 Nanoparticles could be utilized to increase the solubility and biodistribution of salinomycin, leading to the superior therapeutic effect of salinomycin.19,20 Poly(D,L-lactic-co-glycolic acid) (PLGA) nanoparticles have superior biocompatibility and safety in human beings and thus receive extensive attentions in the research of nanomedicines. Generally, PLGA nanoparticles are modified with poly(ethylene glycol) (PEG) to increase their circulation time in vivo. The passive targeting of nanoparticles to tumors could be greatly improved by PEGylation by enhancing permeability and retention (EPR) effects.21
It has been reported that, to maintain an equilibrium, differentiated cancer cells could be converted to stem-like cells.22–24 Thus, considering the conversion of cancer cells to CSCs, it is generally accepted that both CSCs and cancer cells must be eliminated to improve cancer therapeutic efficacy.24 Thus, the combined therapy consisting of conventional chemotherapy drug with an anti-CSC reagent could offer a promising strategy for eliminating both cancer cells and CSCs. Many successful combination therapies have been raised, such as salinomycin combined with doxorubicin.19,25,26 Both CSCs and cancer cells are eliminated by the combination therapy in these studies.24–26
We hereby developed salinomycin-loaded PLGA-PEG nanoparticles (salinomycin-NPs) to target LC CSCs. Gefitinib is an epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor, and it was approved by the US Food and Drug Administration (FDA) in 2015 to treat non–small cell lung cancer (NSCLC).27,28 It was loaded in PLGA-PEG nanoparticles (gefitinib-NPs) to target LC cells. A549 and A431, two human LC cell lines, were chosen in this study. The antitumor activity and mechanism of salinomycin-NPs and gefitinib-NPs were investigated in vitro and in vivo.
Materials and methods
Reagents
Methoxy poly(ethylene glycol)-b-poly(lactic-co-glycolic acid) copolymers (Mw ~3,000:20,000 Da, PLGA-PEG) was purchased from Polyscitech (Akina, IN, USA). Salinomycin, gefitinib, basic fibroblast growth factor (bFGF), and epidermal growth factor (EGF) were provided by Sigma-Aldrich Co. (St Louis, MO, USA). The CD133 MicroBead Kit was provided from Miltenyi Biotec. (Bergisch Gladbach, Germany). Cell Counting Kit-8 (CCK-8) was provided by Dojindo Laboratories (Kumamoto, Japan). Anti-CD133-fluorescein isothiocyanate (CD133-FITC) rat monoclonal antibody was purchased from BD Biosciences (San Jose, CA, USA). Roswell Park Memorial Institute 1640 (RPMI 1640) and fetal bovine serum (FBS) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Primers of the stemness genes were provided by Invitrogen (Shanghai, China). B27 and insulin-transferrin-selenium (ITS) were purchased from Thermo Fisher Scientific. All the other reagents used were of analytical grade and were used without further purification.
Cell culture
Human LC cell lines A549 (ATCC® CCL185™) and A431 (ATCC® CRL1555™) were purchased from American Type Culture Collection (Manassas, VA, USA). A549 and A431 cells were cultured in RPMI 1640 medium with L-glutamine supplemented with 10%FBS, and 0.1 mM MEM Non-Essential Amino Acids Solution. The cells were incubated at 37°C in a humidified atmosphere of 5%CO2-95%air.
RT-PCR
The RNA was extracted with Trizol reagent (Thermo Fisher Scientific). Using the Reverse Transcription System kit (Promega Corporation, Fitchburg, WI, USA), the first-strand complementary DNA (cDNA) was reverse transcribed. RT-PCR was carried out with a Light Cycler (Hoffman-La Roche Ltd., Basel, Switzerland). The mRNA expression of genes normalized against β-actin was expressed as 2CΔΔCT, where ΔCt = Ct,target − Ct,β-actin and ΔΔCt = Ct,experiment condition − ΔCt,control condition.16
The analysis of CD133 expression of the cells
The LC cells were collected by trypsin digestion and treated with 1 μg/mL CD133-FITC antibodies for 30 minutes at 4°C. The cells were then washed and suspended in phosphate-buffered saline (PBS, pH 7.4). The CD133 expression of the cells was analyzed by flow cytometry (Becton Dickinson, San Jose, CA, USA).
CD133+ cell isolation
With magnetic-activated cell sorting, the CD133+ cells were isolated according to the manufacturer’s protocols. Briefly, the cell suspension was added to 20 μL CD133 microbeads and incubated for 20 minutes at 4°C. With washing, uncombined microbeads were removed from cells. Then, 0.5 mL PBE (PBS supplemented with 0.5% bovine serum albumin and 5 mM ethylenediaminetetraacetic acid) was added to the cells and mixed. The cells were separated by a magnetic separation column. The CD133+ cells retained by the column were eluted with PBE and collected. The purity of the CD133+ cells was determined by flow cytometry, as described in the “The analysis of CD133 expression of the cells” section.
Development of salinomycin-loaded and gefitinib-loaded PLGA nanoparticles
The salinomycin-NPs and gefitinib-NPs were developed as described below. After 30 mg PLGA-PEG and 5 mg salinomycin or 10 mg gefitinib were dissolved in 2 mL dichloromethane, the drug solution was added to 4 mL aqueous solution containing 1.5% sodium cholate. Using a probe sonicator, the mixture was sonicated for 45 seconds at 150 W output. The emulsion was added to 20 mL of 0.5% sodium cholate under magnetic stirring before evaporating dichloromethane with a rotary evaporator. Free drug was removed by ultrafiltration using Amicon® Ultra-15 centrifugal filter devices (100,000 NMWL; EMD Millipore, Billerica, MA, USA). The nanoparticles were collected by centrifugation at 20,000× g at 4°C for 1 hour and resuspended in 1 mL PBS (pH 7.4).
Characteristics of nanoparticles
With a dynamic light scattering detector (Zetasizer; Nano-ZS, Malvern, UK), the size and zeta potential of nanoparticles were measured. The morphology of nanoparticles was evaluated by transmission electron microscopy (TEM) (H-600, Hitachi Ltd., Tokyo, Japan) after being stained with 2% phosphotungstic acid.
The drug encapsulation efficacy and loading efficacy
The drug loading of nanoparticles was measured by high-performance liquid chromatography (HPLC).16 Briefly, 10 mg of lyophilized nanoparticles was completely dissolved in dichloromethane and was analyzed by HPLC (L-2000; Hitachi Ltd.). A reverse phase C-18 column (Diamonsil, 250×4.6 mm, 5 μm) was used. The mobile phase for salinomycin was acetonitrile/deionized water/tetrahydrofuran/phosphoric acid (85/10/5/0.01, v/v) with a flow rate at 1.5 mL/min. The mobile phase for gefitinib was 0.02 M dipotassium hydrogen orthophosphate/methanol (10/90, v/v) with a flow rate at 1 mL/min. For salinomycin, the detection wavelength and column temperature were set at 210 nm and 30°C, respectively, and for gefitinib, the detection wavelength and column temperature were set at 246 nm and 30°C, respectively. The injection volume was 20 μL. Before analysis, all the samples were filtered through a syringe filter. The drug loading = MD/MN ×100%. MD and MN were defined as the mass of loaded drugs and nanoparticles, respectively. The drug encapsulation efficiency = ML/MT ×100%. ML and MT were the mass of loaded drugs and total drugs, respectively.
In vitro drug release assay
The in vitro drug release was carried out as described below. After the nanoparticles (1 mg/mL) were suspended in a centrifuge tube, the tube was placed in an orbital shaker and vibrated horizontally (80× g, 37°C). The release medium was PBS (pH 7.4) and 10% human plasma. The tubes were taken out at different time points and centrifuged (40 minutes, 12,000 rpm). The supernatant was removed and analyzed by HPLC. The pellet was resuspended in fresh medium and placed back in the orbital shaker.
CCK-8
The cytotoxicity was measured by the CCK-8 assay. Briefly, cells were seeded at a density of 5×103 cells per well in 96-well plates for 12 hours. After that, a series of concentrations of the nanoparticles or free drugs were added to the cells. After 72 hours, the absorbance was read with a microplate reader (MK-3; Thermo Fisher Scientific) at 450/630 nm wavelength. The IC50 values were obtained using the logarithmic curves.
The effect of drugs on the CSC proportion of LC cells
The effect of drugs on the CSC proportion of LC cells was examined as described below. Briefly, LC cells were inoculated at a density of 5×104 per well in 12-well plates and cultured for 12 hours. After that, the cells were incubated with various drugs (salinomycin: 5 μg/mL, gefitinib: 10 μg/mL). After 48 hours, the cells were washed and incubated for 3 days. The proportion of CD133+ cells of the treated cells was analyzed by flow cytometry using the method described in the “The analysis of CD133 expression of the cells” section. Alternatively, the cells were removed from the culture plate and cultured at a density of 200 cells per well in ultra-low adherent 96-well dishes to generate tumorspheres, in stem cell-conditioned culture medium consisting of DMEM-F12 with 20 ng/mL bFGF, 20 ng/mL EGF, 1× B27, and 1× ITS. The tumorsphere number was counted after 5–7 days.
Animal studies
All BALB/c nude mice (male, 18–20 g, 6–8 weeks) were provided by the Shanghai Experimental Animal Center of Chinese Academic of Sciences (Shanghai, China). All protocols were approved by the Committee on Animals of the Second Military Medical University (Shanghai, China). They were carried out in accordance with the guidelines of the Committee on Animal of the Second Military Medical University.
In vivo tumorigenicity assay
The in vivo tumorigenicity assay of LC cells was performed in mice. Briefly, CD133+ or CD133− LC cells (5×102 to 1×106 cells) were collected and mixed with growth factor-reduced matrigel (BD Matrigel™). The cells were inoculated subcutaneously (SC) into the same mice on the left and right sides. During 20 weeks, the tumor formation was monitored.
In vivo antitumor assays
In the antitumor activity, assay was performed as described below. Briefly, on day 0, 1×107 A431 cells were injected SC into BALB/c nude mice. By day 5, the average tumor volume of each mice reached about 50 mm3. From day 5, the mice were treated with the nanoparticles or free drugs (5 mg salinomycin/kg, 30 mg gefitinib/kg, intravenous [IV] injections via the tail vein). The nanoparticles were given via the tail vein. Free salinomycin was dissolved in ethanol, and free gefitinib was dissolved in Tween 80R (Sigma-Aldrich Co.). The free drugs were administered by intraperitoneal injection. The drugs were given once every 3 days for 10 times (on days 5, 8, 11, 14, 17, 20, 23, 24, 27, and 30). The effect of the drugs on the CSC proportion of LC was evaluated on day 33. Briefly, three mice were euthanized, and the tumors were excised. After sterilization by 75% ethanol, the tumors were washed with PBS. After that, the tumors were chopped and digested with 1 mg/mL collagenase I solution for 20 minutes at 37°C. After filtration through a 40-μm cell strainer, the cells were washed with RPMI 1640 medium with 5%FBS and cultured overnight at 37°C. The CSC proportion of the cells was evaluated as described in the “The effect of drugs on the CSC proportion of LC cells” section. The tumor volume = width2 × length/2. The weight of the mice was measured during the whole period.
Statistical analysis
The data were analyzed by the statistic package SPSS 13.0 (SPSS Inc., Chicago, IL, USA). Student’s non-paired t-test was conducted to perform a direct comparison between two groups, and the means of three or more groups were compared with one-way ANOVA with the Dunnett’s post-test. P-value of <0.05 was considered statistically significant. *P<0.05; **P<0.01; ***P<0.001; NS represents not significant (P>0.05).
Results
CD133+ LC cells are regarded as LC CSCs
The CD133 expression of the two LC cell lines (A549 and A431) was analyzed by flow cytometry. In A549 cells, the percentage of CD133+ cells was 5.5%, whereas that in A431 was 6.3%. Though the CD133+ population represents a minor population in the LC cell lines, CD133+ cells were significantly enriched, and the percentage of CD133+ cells in the sorted cells was >98%. Next, a series of approaches have been performed to test whether the CD133+ LC cells had characteristics of LC CSCs. As shown in Figure 1, the mRNA levels of CSC-related genes including SMO, CD133, and OCT4 in CD133+ LC cells were increased significantly than CD133− LC cells (P<0.05). The tumorsphere formation ability represents the self-renewal capability of CSCs, and the increased tumorsphere formation indicates that the proportion of CSCs is increased.19,20 Notably, CD133+ LC cells could form more tumorspheres compared with CD133− LC cells (P<0.001) (Figure 1B).
The tumorigenicity of CD133+ and CD133− LC cells was examined in nude mice (Table 1). When the cell count was ≥2×104, CD133+ A549 cells induced tumors in mice with a 100% incidence. In contrast, CD133− A549 cells induced tumors only with an 67% incidence, even at a cell number of 2×106, suggesting that CD133+ A549 cells have greater tumorigenic potential than CD133− A549 cells. Similar results were obtained with CD133+ and CD133− A431 cells. CD133+ A431 cells induced tumors with a 100% incidence in mice when the cell count was ≥2×104. In contrast, CD133− A431 cells induced tumors only with a 50% incidence, even at a cell count of 1×106, suggesting that CD133+ A431 cells have greater tumorigenic potential than CD133− A549 cells. Taken together, CD133+ LC cells exhibited a stronger potential than CD133− LC cells for self-renewal and tumorigenicity in mice, suggesting that CD133+ LC cells could be regarded as LC CSCs.
Development and characteristics of nanoparticles
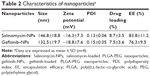
Salinomycin-NPs and gefitinib-NPs were prepared by using the emulsion/solvent evaporation approach (Figure 2). The emulsion/solvent evaporation approach is a simple and common approach to encapsulate hydrophobic drugs in polymeric nanoparticles. The characteristics of salinomycin-NPs and gefitinib-NPs that are summarized in Table 2 were evaluated. The size of the nanoparticles was 130–150 nm with a narrow polydispersity index (PDI) of <0.15, indicating a homogeneous distribution of nanoparticles. Our prepared nanoparticles showed relatively high negative zeta potential of (about −20 mV). The drug loading of our prepared nanoparticles was about 8%, and their encapsulation efficiency was about 80%, suggesting that the emulsion/solvent evaporation method is an effective approach to encapsulate hydrophobic drugs such as salinomycin and gefitinib in polymeric nanoparticles. Taken together, the appropriate size, zeta potential, and drug loading capacity of our prepared nanoparticles suggest that they will be useful drug delivery vehicles.
TEM was taken to examine the morphology of the nanoparticles. As shown in Figure 3A and B, both the nanoparticles showed good uniformity and had smooth surface. Notably, the TEM picture clearly showed a core-shell structure of the nanoparticles. The PEG chain of PLGA-PEG formed a dim ring surrounding the PLGA nanoparticle core.
The in vitro release of salinomycin was examined in PBS and PBS with 10%FBS at 37°C (Figure 3C and D), both the nanoparticles showed slightly faster release in PBS with 10%FBS than in PBS. For salinomycin-NPs, the drug release in PBS with 10%FBS was faster than that in PBS after 4 hours (P>0.05), whereas for gefitinib-NPs, the drug release in PBS with 10%FBS was faster than that in PBS after 8 hours (P>0.05). A fast release of salinomycin and gefitinib (~50%) in the initial 12 hours was observed. The cumulative drug release of the nanoparticles reached ~80% in the following 108 hours. Taken together, both the nanoparticles showed sustained drug release during a long period of 120 hours.
Cytotoxicity toward LC cells
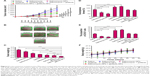
The dose-dependent cytotoxicity of free drugs and nanoparticles was measured on LC cells (Figure 4). NPs, the blank nanoparticles, did not cause significant cytotoxic effects against LC cells, as reflected by the >90% cell viability in the presence of a high concentration of NPs (3,000 μg/mL), indicating that the nanoparticles are rather safe drug carriers for drug delivery. As expected, salinomycin, salinomycin-NPs, gefitinib, and gefitinib-NPs showed concentration-dependent cytotoxic effects on the LC cells. To define the cytotoxic effects of drugs more exactly, the IC50 values of each drug are calculated (Table 3). On A549 cells, salinomycin showed significantly enhanced cytotoxic effect on CD133+ cells than on CD133− cells (5.31±3.22 μg/mL vs 18.39±7.24 μg/mL, P<0.01). In contrast, gefitinib showed a significantly decreased cytotoxic effect on CD133+ cells than on CD133− cells (97.88±22.88 μg/mL vs 38.98±11.74 μg/mL, P<0.05). Similar results were obtained for salinomycin-NPs and gefitinib-NPs on A549 cells. Salinomycin-NPs showed significantly enhanced cytotoxic effect on CD133+ cells than on CD133− cells (8.18±4.46 μg/mL vs 22.32±8.53 μg/mL, P<0.05), whereas gefitinib-NPs showed a significantly decreased cytotoxic effect on CD133+ cells than on CD133− cells (115.03±31.47 μg/mL vs 49.59±17.08 μg/mL, P<0.05). Comparable results were obtained with A431 cells. Thus, both salinomycin and salinomycin-NPs showed increased cytotoxicity against LC CSCs compared with LC cells, whereas gefitinib and gefitinib-NPs have a decreased cytotoxic effect against LC CSCs compared with LC cells. Taken together, salinomycin and salinomycin-NPs preferably target LC CSCs, whereas gefitinib and gefitinib-NPs preferably target LC cells.
The proportion of CSCs in LC cells after treatments
The proportion of CSCs in LC cells after treatment with nanoparticles was evaluated by the tumorsphere formation assay and the proportion of CD133+ cells (Figure 5). Salinomycin and salinomycin-NPs reduced the number of A549 tumorspheres by fourfold (P<0.001, Figure 5A). On the contrary, gefitinib and gefitinib-NPs treatment significantly increased the A549 tumorsphere number (P<0.05), suggesting that salinomycin and salinomycin-NPs reduce but gefitinib and gefitinib-NPs increase the proportion of CSCs in A549 cells. The proportion of CD133+ cells in A549 cells was also measured. As shown in Figure 5B, salinomycin and salinomycin-NPs reduced the proportion of CD133+ cells in A549 cells (P<0.05), whereas gefitinib and gefitinib-NPs increased the proportion (P<0.01) (Figure 5B). However, NPs had no effect on the tumorsphere formation or proportion of CD133+ cells in A549 cells. Similar results were achieved in A431 cells after treatment (Figure 5C and D). Salinomycin and salinomycin-NPs treatment resulted in far fewer tumorspheres (P<0.001) and significantly reduced the proportion of CD133+ cells (P<0.01). Once again, gefitinib and gefitinib-NPs significantly increased the number of tumorspheres (P<0.01) and the proportion of CD133+ cells (P<0.01). Taken together, these results show that salinomycin and salinomycin-NPs could preferably kill LC CSCs, whereas gefitinib and gefitinib-NPs merely increase the proportion of CSCs in LC cells.
The in vivo antitumor assay
The therapeutic efficacy of nanoparticles was evaluated in nude mice bearing A431 xenografts (Figure 6). Strikingly, at the end point (day 35), salinomycin-NP combined with gefitinib-NP treatment achieved the best therapeutic efficacy, resulting in a 94% decrease in tumor volume. On the contrary, gefitinib-NPs and salinomycin combined with gefitinib achieved only 64% and 73% decreases in tumor volume, respectively. Salinomycin, salinomycin-NPs, and gefitinib achieved moderate or minor therapeutic efficacy, resulting in 17%, 44%, and 55% decreases in tumor volume, respectively.
At the end of the treatment (day 35), the tumor volume in the salinomycin-treated mice was increased by 37-folds and that in salinomycin-NP-treated mice was increased by 25-folds, compared with the initial tumor volume (50 mm3). Gefitinib and gefitinib-NPs achieved better therapeutic efficacy compared with salinomycin and salinomycin-NPs, as reflected by the fact that the tumor volume in the gefitinib-treated mice was increased by 20-folds and that in gefitinib-NP-treated mice was increased by 16-folds. The combination therapy achieved the best therapeutic efficacy. Under the treatment of salinomycin combined with gefitinib, the tumor volume was increased only by 12-folds. Notably, the tumor volume in the salinomycin-NPs combined with gefitinib-NP-treated mice did not increase. The mean tumor volume of the salinomycin-NPs combined with gefitinib-NP-treated group was significantly smaller than that of other groups (salinomycin-NPs combined with gefitinib-NPs =130 mm3, salinomycin combined with gefitinib =600 mm3, gefitinib-NPs =799 mm3, gefitinib =1,004 mm3, salinomycin-NPs =1,248 mm3, salinomycin =1,850 mm3, saline =2,234 mm3; salinomycin-NPs combined with gefitinib-NPs vs salinomycin combined with gefitinib: P<0.001, salinomycin-NPs combined with gefitinib-NPs vs gefitinib-NPs: P<0.0001, salinomycin-NPs combined with gefitinib-NPs vs gefitinib: P<0.001, salinomycin-NPs combined with gefitinib-NPs vs salinomycin-NPs: P<0.001, salinomycin-NPs combined with gefitinib-NPs vs salinomycin: P<0.001, salinomycin-NPs combined with gefitinib-NPs vs saline: P<0.001) (Figure 6A and B).
The weights of the excised tumors were measured (Figure 6C). The mean tumor weight of the salinomycin-NPs combined with gefitinib-NP-treated group was significantly lower than that of the other groups (salinomycin-NPs combined with gefitinib-NPs =0.32 g, salinomycin combined with gefitinib =0.56 g, gefitinib-NPs =0.68 g, gefitinib =0.92 g, salinomycin-NPs =1.22 g, salinomycin =1.72 g, saline =2.26 g; salinomycin-NPs combined with gefitinib-NPs vs salinomycin combined with gefitinib: P<0.01, salinomycin-NPs combined with gefitinib-NPs vs gefitinib-NPs: P<0.001, salinomycin-NPs combined with gefitinib-NPs vs gefitinib: P<0.001, salinomycin-NPs combined with gefitinib-NPs vs salinomycin-NPs: P<0.001, salinomycin-NPs combined with gefitinib-NPs vs salinomycin: P<0.001, salinomycin-NPs combined with gefitinib-NPs vs saline: P<0.001) (Figure 6C).
The population of LC CSCs in the excised tumors were evaluated, aiming to investigate the therapeutic efficacy of drugs on LC CSCs in vivo (Figure 6D and E). Consistent with our in vitro results, salinomycin or salinomycin-NPs significantly decreased the percentage of CSCs in the tumors from mice (P<0.01). On the contrary, after the treatment with free gefitinib or gefitinib-NPs, the percentage of CSCs was significantly increased compared with the saline-treated group (P<0.01). Compared with gefitinib or gefitinib-NPs alone, in the tumors treated with salinomycin combined with gefitinib or salinomycin-NPs combined with gefitinib-NPs, the percentage of CSCs was significantly decreased (P<0.01). This result suggested that it is necessary to combine a drug that can kill CSCs when using gefitinib to treat LC. These results demonstrated that the combination therapy with free salinomycin or salinomycin-NPs is effective in reducing the population of CSCs.
The toxicity of all treatments was examined by monitoring the body weight (Figure 6F). The body weight of the mice of all groups gradually increased, without any weight loss, suggesting that mice could well-tolerate all the treatments.
Discussion
LC is a leading cause of cancer death worldwide. Thus, the treatment of LC is an urgent task for public health. One of the critical reasons for reduced survival and treatment failure of LC is the presence of CSCs in LC.6,7 Since it has become generally accepted that both CSCs and cancer cells must be eradicated for successful tumor therapy, it becomes necessary that the CSC-targeting and non-CSC-targeting components should be combined. In this study, we developed a combination therapy of salinomycin-NPs and gefitinib-NPs to kill the LC CSCs while simultaneously eliminating the LC cells, aiming to achieving superior therapeutic efficacy against LC.
In our study, the selection of the two anti-LC drugs is rational and critically important for the superior activity and bright application prospect of our therapy. Gefitinib, the EGFR tyrosine kinase phosphorylation inhibitor, is the first-line treatment for patients with NSCLC. Though gefitinib has shown significant cytotoxic effects against LC cells in our study, it could significantly increase the percentage of LC CSCs, as reflected by the increased number of tumorspheres and CD133+ LC CSCs in the presence of gefitinib. After salinomycin has been found to exert anti-CSCs activity in breast cancer in 2009,15 it has been demonstrated to be an effective anti-CSCs drug in various cancers, suggesting that salinomycin may represent a broad-spectrum reagent against CSCs. Due to the great potential of salinomycin in the treatment of cancer, salinomycin was tested in a small clinical trial and showed partial regression of tumor metastasis, with no severe side effects.14 Thus, salinomycin represents a very promising drug against CSCs. Currently, there have been only two previous reports showing the anti-LC CSC activity of salinomycin.18,19 In our study, salinomycin and salinomycin-NPs have been showed to possess significantly enhanced cytotoxic effects of CD133+ LC cells over CD133− LC cells. Furthermore, both in vitro and in vivo, salinomycin and salinomycin-NPs significantly reduced the proportion of CSCs in LC cells, as reflected by the tumorsphere formation assay and the proportion of CD133+ cells. Significantly, in the tumors treated with salinomycin combined with gefitinib or salinomycin-NPs combined with gefitinib-NPs, the percentage of CSCs was significantly decreased compared with gefitinib or gefitinib-NPs alone (P<0.01), suggesting that it is necessary to combine a drug that could kill CSCs when using gefitinib to treat LC.
Although CSCs have been reported as a new therapeutic target in many cancers, the low viability of CSCs after isolation has severely hampered the investigation of drug therapeutic effects in CSCs. The most common methods of isolation of CSCs include magnetic bead-based or flow cytometry-based cell sorting. Our previous studies have shown that, after fluorescence activated cell sorting, CD133+ LC CSCs tended to die quickly, and this great disadvantage of low cell viability makes it difficult to perform in vitro cell culture and in vivo tumorigenic assay of CD133+ LC CSCs (data not shown). In this study, we isolated CD133+ LC CSCs with good cell viability using magnetic-activated cell sorting from the two LC cell lines A549 and A431. Significantly, the CD133+ LC CSCs were demonstrated to show the characteristics of LC CSCs by stemness gene expression, tumorsphere formation ability, and in vivo tumorigenesis. Thus, our successful isolation of LC CSCs provides a versatile approach for extensive investigation in CSCs.
As we stated before, nanoparticles for eliminating both CSCs and cancer cells could provide a practical solution to radically cure cancer, due to its targeted and controlled drug delivery and preferable pharmacokinetics and biodistribution.9,10,20 We have developed a series of nanoparticles for eliminating both CSCs and cancer cells in melanoma and liver cancer, achieving superior cancer therapeutic efficacy.16,29–31 Our development of salinomycin-loaded nanoliposomes and doxorubicin-loaded nanoliposomes achieved superior therapeutic efficacy against both liver CSCs and cancer cells.29 Although numerous nanoparticles have been developed to eliminate CSCs in many cancers, the nanoparticles developed for targeting LC CSCs have been rarely investigated. The only two nanoparticles developed for targeting LC CSCs are dual drug-loaded biofunctionalized amphiphilic chitosan nanoparticles and miRNA-loaded calcium phosphate nanoparticles.32,33 However, these two nanoparticles have been designed to target only LC CSCs, and their therapeutic effect against LC cells have been never investigated. To the best of our knowledge, this is the first report describing the development of a combination therapy of nanoparticles to eliminate both CSCs and cancer cells in LC, thus providing a very promising approach in the treatment of LC.
The safety of nanoparticles is very critical to their clinic use.34 First, the selection of drug carriers is important for the safety of nanoparticles. PLGA and poly(lactic acid) are FDA-approved and commercially available biodegradable polymers, whereas they are too strong and slow to degrade. PEG is usually used to ameliorate their quality. In this study, we used PLGA-PEG polymer to prepare salinomycin-NPs and gefitinib-NPs. Considering safety is important for the clinical use of nanomedicines and that PLGA and PEG are FDA-approved biodegradable polymers, our prepared nanoparticles are expected to have satisfactory safety.
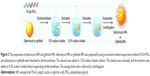
Our combination therapy achieved superior therapeutic efficacy, as reflected by the fact that the mean tumor volume and weight of the salinomycin-NP combined with gefitinib-NP-treated group was significantly lower compared with other groups. The mechanism underlying the superior antitumor activity of the combination therapy could be clarified as described below (Figure 7). In the first step, salinomycin-NPs and gefitinib-NPs could efficiently be accumulated in tumors, due to their long circulation and EPR effect. Then, salinomycin-NPs were bound to and was internalized by LC CSCs. After internalization, salinomycin-NPs release salinomycin and eliminate LC CSCs. Gefitinib-NPs killed the LC cells simultaneously. Thus, both LC CSCs and LC cells are eliminated by the combination of salinomycin-NPs and gefitinib-NPs, resulting in the eradication of LC. On the contrary, a single therapy, either salinomycin-NPs or gefitinib-NPs, only kills LC CSCs or LC cells. After salinomycin-NPs treatment, the remaining LC cells may convert into LC CSCs which initiate the LC recurrence, whereas the remaining LC CSCs will initiate the LC recurrence after gefitinib-NPs treatment.
There are two limitations in this study. First, in addition to CD133, aldehyde dehydrogenase (ALDH) has also been reported to be the marker of lung CSCs.35,36 We need to validate that whether the combination therapy works in lung CSCs isolated by other markers such as ALDH in future studies. Second, more LC cell lines especially the LC cell lines harboring EGFR mutations which are sensitive to gefitinib need to be tested in this study. Since our study focuses on the combination therapy that could eliminate both LC cells and CSCs, the isolation of lung CSCs and tumorsphere formation from LC cell lines should be feasible. However, for two available LC cell lines harboring EGFR mutations (HCC2279 and H3255) maintained in our laboratory, we failed to isolate lung CSCs from them, and both the cell lines could not form tumorspheres. Thus, we currently could not test the combination therapy in the EGFR mutant model. We fully recognize the importance of using LC cell lines of the EGFR mutant model and will test the combination therapy in the EGFR mutant model in the future if possible.
Conclusion
Salinomycin-NPs combined with gefitinib-NPs suppressed tumor growth more effectively than salinomycin combined with gefitinib, with a reduced percentage of LC CSCs in tumors and good safety in vivo. This is the first report describing the development of a combination therapy of nanoparticles to target both CSCs and cancer cells in LC, thus providing a very promising approach in the treatment of LC.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (project number: 81472829) and the Xiangyang Municipal Fund (project number: 20140915).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2017;67(1):7–30. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Hirsch FR, Suda K, Wiens J, Bunn PA Jr. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388(10048):1012–10124. | ||
Karachaliou N, Sosa AE, Barron FB, Gonzalez Cao M, Santarpia M, Rosell R. Pharmacological management of relapsed/refractory NSCLC with chemical drugs. Expert Opin Pharmacother. 2017;18(3):295–304. | ||
Popper HH. Progression and metastasis of lung cancer. Cancer Metastasis Rev. 2016;35(1):75–91. | ||
MacDonagh L, Gray SG, Breen E, et al. Lung cancer stem cells: the root of resistance. Cancer Lett. 2016;372(2):147–156. | ||
Leon G, MacDonagh L, Finn SP, Cuffe S, Barr MP. Cancer stem cells in drug resistant lung cancer: targeting cell surface markers and signaling pathways. Pharmacol Ther. 2016;158:71–90. | ||
Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8(10):806–823. | ||
Gao J, Li W, Guo Y, Feng SS. Nanomedicine strategies for sustained, controlled and targeted treatment of cancer stem cells. Nanomedicine (Lond). 2016;11(24):3261–3282. | ||
Xie F, Xu W, Yin C, Zhang G, Zhong Y, Gao J. Nanomedicine strategies for sustained, controlled, and targeted treatment of cancer stem cells of the digestive system. World J Gastrointest Oncol. 2016;8(10):735–744. | ||
Lundin A, Driscoll B. Lung cancer stem cells: progress and prospects. Cancer Lett. 2013;338(1):89–93. | ||
Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106(38):16281–16286. | ||
Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15(3):504–514. | ||
Naujokat C, Steinhart R. Salinomycin as a drug for targeting human cancer stem cells. J Biomed Biotechnol. 2012;2012:950658. | ||
Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–659. | ||
Mao X, Liu J, Gong Z, et al. iRGD-conjugated DSPE-PEG2000 nanomicelles for targeted delivery of salinomycin for treatment of both liver cancer cells and cancer stem cells. Nanomedicine (Lond). 2015;10(17):2677–2695. | ||
Yue W, Hamaï A, Tonelli G, et al. Inhibition of the autophagic flux by salinomycin in breast cancer stem-like/progenitor cells interferes with their maintenance. Autophagy. 2013;9(5):714–729. | ||
Larzabal L, El-Nikhely N, Redrado M, Seeger W, Savai R, Calvo A. Differential effects of drugs targeting cancer stem cell (CSC) and non-CSC populations on lung primary tumors and metastasis. PLoS One. 2013;8(11):e79798. | ||
Wang Y. Effects of salinomycin on cancer stem cell in human lung adenocarcinoma A549 cells. Med Chem. 2011;7(2):106–111. | ||
Gao J, Feng SS, Guo Y. Nanomedicine for treatment of cancer stem cells. Nanomedicine (Lond). 2014;9(2):181–184. | ||
Otsuka H, Nagasaki Y, Kataoka K. PEGylated nanoparticles for biological and pharmaceutical applications. Adv Drug Deliv Rev. 2003;55(3):403–419. | ||
Gupta PB, Fillmore CM, Jiang G, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. | ||
Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci U S A. 2011;108(4):1397–1402. | ||
Chaffer CL, Brueckmann I, Scheel C, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108(19):7950–7955. | ||
Chen D, Xie F, Sun D, Yin C, Gao J, Zhong Y. Nanomedicine-mediated combination drug therapy in tumor. Open Pharm Sci J. 2017;4:1–10. | ||
Ke XY, Lin Ng VW, Gao SJ, Tong YW, Hedrick JL, Yang YY. Co-delivery of thioridazine and doxorubicin using polymeric micelles for targeting both cancer cells and cancer stem cells. Biomaterials. 2014;35(3):1096–1108. | ||
Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. | ||
Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–13311. | ||
Gong Z, Chen D, Xie F, et al. Co-delivery of salinomycin and doxorubicin using nanoliposomes for targeting both liver cancer cells and cancer stem cells. Nanomedicine (Lond). 2016;11(19):2565–2579. | ||
Xie F, Zhang S, Liu J, et al. Codelivery of salinomycin and chloroquine by liposomes enables synergistic antitumor activity in vitro. Nanomedicine (Lond). 2016;11(14):1831–1846. | ||
Song H, Su X, Yang K, et al. CD20 antibody-conjugated immunoliposomes for targeted chemotherapy of melanoma cancer initiating cells. J Biomed Nanotech. 2015;11(11):1927–1946. | ||
Huang WT, Larsson M, Lee YC, Liu DM, Chiou GY. Dual drug-loaded biofunctionalized amphiphilic chitosan nanoparticles: enhanced synergy between cisplatin and demethoxycurcumin against multidrug-resistant stem-like lung cancer cells. Eur J Pharm Biopharm. 2016;109:165–173. | ||
Zhang J, Zhang C, Hu L, et al. Abnormal expression of mir-21 and mir-95 in cancer stem-like cells is associated with radioresistance of lung cancer. Cancer Invest. 2015;33(5):165–171. | ||
Nystrom AM, Fadeel B. Safety assessment of nanomaterials: implications for nanomedicine. J Control Release. 2012;161(2):403–408. | ||
Sullivan JP, Spinola M, Dodge M, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70(23):9937–9948. | ||
Shao C, Sullivan JP, Girard L, et al. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res. 2014;20(15):4154–4166. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.