Back to Journals » Journal of Asthma and Allergy » Volume 16
The Assessment of TLR1 Gene Polymorphism Association with the Risk of Allergic Rhinitis in the Chinese Han Population from Northern China
Authors Han H, Lian P, Chen H, Shamsi BH, Liu Y, Niu Y
Received 18 May 2023
Accepted for publication 7 September 2023
Published 18 September 2023 Volume 2023:16 Pages 979—986
DOI https://doi.org/10.2147/JAA.S421939
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Luis Garcia-Marcos
Hui Han,1 Penggang Lian,2 Haiyuan Chen,3 Bilal Haider Shamsi,4 Yonglin Liu,4 Yongliang Niu5
1Clinical Laboratory, Shenmu Hospital, the Affiliated Shenmu Hospital of Northwest University, Shenmu, 719300, People’s Republic of China; 2Otorhinolaryngologic Department, Shenmu Hospital, the Affiliated Shenmu Hospital of Northwest University, Shenmu, 719300, People’s Republic of China; 3Information Department, Shenmu Hospital, the Affiliated Shenmu Hospital of Northwest University, Shenmu, 719300, People’s Republic of China; 4Department of Science and Education, Shenmu Hospital, the Affiliated Shenmu Hospital of Northwest University, Shenmu, 719300, People’s Republic of China; 5Department of Respiratory and Critical Care Medicine, Shenmu Hospital, the Affiliated Shenmu Hospital of Northwest University, Shenmu, 719300, People’s Republic of China
Correspondence: Yongliang Niu, Tel/Fax +86-13892234287, Email [email protected]
Background: Environmental factors and genetic predisposition can influence the occurrence and development of AR. Toll-like receptor 1 (TLR1) belongs to the TLR receptor family, which plays a fundamental role in the activation of innate immunity. This study aimed to explore the association between TLR1 genetic loci and AR susceptibility in the Han Chinese from northern China.
Methods: Genotyping of three SNPs in the TLR1 has proceeded using the Agena MassARRAY platform. Odds ratio (OR) and 95% confidence interval (CI) were used to assess the correlation between candidate SNPs and AR susceptibility. Using FPRP (false-positive report probability analysis) to detect whether the positive results are noteworthy findings. The SNP-SNP interactions were detected by multifactor dimensionality reduction (MDR).
Results: TLR1-rs72493538 (Allele “G”: OR=0.77, p = 0.034) and -rs76600635 (Allele “G”: OR=0.75, p = 0.024) were associated with reducing the risk of AR among Han Chinese in northern China. In addition, we found evidence that TLR1-rs72493538 (males, participants with aging > 43 years, or coming from the wind-blown sand region) and -rs76600635 (males, participants with BMI ≤ 24 kg/m2, or coming from the wind-blown sand region) were associated with AR risk in stratified analyses. FPRP showed that all positive results are noteworthy findings. MDR analysis showed that a two-loci genetic model composed of rs72493538 and rs76600635 can be chosen as the best genetic model to predict the risk of AR.
Conclusion: TLR1-rs72493538 and -rs76600635 have a close association with reducing the risk of AR.
Keywords: allergic rhinitis, TLR1, susceptibility, case-control study
Introduction
Allergic rhinitis (AR) is a chronic non-infectious disease characterized by occasional nasal congestion, nasal itching, sneezing, and runny nose, which can occur at all stages.1 Epidemiological surveys in recent years have shown that the incidence of AR in the United States is 10%-20%, and the global prevalence is about 10%-25%.2 AR not only triggers a significant decline in the quality of life but also imposes a heavy social and economic burden on individuals, families, communities, and countries.3 Therefore, it is necessary to investigate the etiology of AR.
Previous studies have demonstrated that environmental factors (air pollutants, aeroallergens) and genetic predisposition promote the occurrence and development of AR.4 The heritability of AR has been estimated to be over 0.65, indicating a strong genetic component in AR.5 Moreover, Noguchi et al have proposed that when both parents have allergies, the prevalence of AR in children is as high as 75%; when one parent has allergies, the prevalence is 50%.6 In addition, single nucleotide polymorphisms (SNPs) are important genetic factors for AR.7,8 Numerous investigations have indicated that SNPs in ADAM33, IL-18, FOXP3, and IL13 have been implicated in susceptibility to AR.9–11
Toll-like receptor 1 (TLR1) belongs to the TLR receptor family which plays a fundamental role in the activation of innate immunity. It has been reported that TLRs are involved in the pathogenesis of AR.12,13 Renkonen et al have found the protein expression levels of TLR1 and TLR6 are down-regulated in the allergic group after allergen stimulation.14 Another study has also revealed that the expression levels of TLR2 and TLR4 are increased in patients with AR compared with controls.15 Aoi et al have indicated that TLR2 signaling participates in the development of AR through regulating OK-432-induced anti-T helper 2 (Th2) immunity.16 Moreover, the allele TLR4-rs1927911 contributes to a higher risk of AR in children.17 In general, these findings emphasize the critical impact of TLRs on AR development. However, there are relatively few studies on the correlation between TLR1 and allergic rhinitis. Based on the above studies, it is necessary to further explore the TLR1 polymorphism related to AR susceptibility, which will provide a theoretical basis for the study of the mechanism of TLR1 in the AR and then further supplement the mechanism research of TLR family genes in allergic rhinitis.
The purpose of this study is to explore the relationship between TLR1 polymorphism and AR risk, which is helpful to further understand the pathogenesis of AR at the genetic level. This study will provide a new research direction and theoretical basis for the early prevention and treatment of AR and the development of individualized treatment programs.
Materials and Methods
Study Population
A total of 982 patients with AR and 984 healthy controls were recruited from Daliuta Town, Jinjie Town, Langanbao Town, Hejiachuan Town, and central urban area (including Linzhou Street and Yingbin Road Street) of Shenmu city, Shaanxi province, China. AR patients were diagnosed based on clinical features (sneezing, nasal congestion, nasal itching, and runny nose), clinical examination, and serum-specific IgE detection. The inclusion criteria for patients were positive history of AR, positive physical examination, and positive serum IgE. Patients with possible chronic systemic disorder, atopic or allergic disease, family history of asthma, airway diseases, malignant disease, auto-immune disease, any diffuse dermatitis, or respiratory tract infection before recruitment were excluded from the study. The inclusion criteria for controls were no history of AR and no treatment with antihistamine drugs.
The Ethics Committee of Shenmu Hospital approved the study. The experimental procedures of this study were in accordance with Declaration of Helsinki and informed consent was signed by all subjects.
Selection of Candidate SNPs and Genotyping
Through an online tool, we found that the physical position of TLR1 was on Chromosome 4: 38,790,677–38,856,817 (e!Ensmbl Human GRCh38.p13: https://asia.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000174125;r=4:38790677-38856817). There were 33,914 variants in TLR1. In order to narrow the research scope, we conducted the following screening steps: First, we used “Ensembl Tools: VCF to PED conversion window” to download TLR1 gene variants files (CHB and CHS populations). Then, putting the above files into Haploview software to further narrow the scope of the study after setting specific conditions (MAF> 0.05, Min Genotype > 75%, Tagger r2 > 0.8, and HWE> 0.01). Finally, rs72493538, rs76600635, and rs17616434 of TLR1 were randomly selected as the candidate genetic variants for this study.
We collected venous blood from each subject and stored it in EDTA-treated tubes. Genomic DNA was isolated by an extraction kit (GoldMag Co. Ltd, Xi’an, China) and its concentration was quantified by NanoDrop 2000. Genotyping of SNPs was proceeded using the Agena MassARRAY platform (Agena Bioscience, USA). All primers were designed through the MassARRAY platform (https://support.agenabio.com/s/online-tools). All primer sequences can be found in Supplemental Table 1. Agena Typer 4.0 software was utilized to analyze and manage data.
Data Analysis
We performed Student’s t-test to compare differences in age and BMI between the case and control groups. In addition, the gender distribution and p-values for Hardy-Weinberg equilibrium (HWE) were assessed by χ2 test. Odds ratio (OR) and 95% confidence interval (CI) were used to assess the correlation between SNPs and AR susceptibility. Then, performing false-positive report probability analysis (FPRP) to detect whether the positive results obtained in this study are noteworthy findings.18 The SNP-SNP interactions were detected by multifactor dimensionality reduction (MDR). A p-value of 0.05 or less was considered statistically significant.
Results
Characteristics of Subjects
There were 982 AR patients (357 males and 625 females) and 984 healthy controls (344 males and 640 females) in this study. The mean ages of the case group and the control group were 42.43 ± 10.39 years and 43.62 ± 8.05 years, respectively (Table 1).
 |
Table 1 Characteristics of the AR Patients and Healthy Controls |
Rs72493538, rs76600635, and rs17616434 in TLR1 were selected as candidate SNPs. The HWE P-values of these SNPs were all greater than 0.05 (Table 2).
 |
Table 2 Basic Information of the SNPs in TLR1 Gene |
AR Risk Assessment
The results demonstrated (Table 3) that Allele “G” (OR=0.77; p = 0.034) and heterozygous genotype “AG” (OR=0.70; p = 0.011) of TLR1-rs72493538 were protective genetic factors of susceptibility to AR. And TLR1-rs72493538 has a significant association with decreasing risk of AR under dominant (p = 0.019), and additive (p = 0.044) genetic models. Similarly, Allele “G” (OR=0.75; p = 0.024) and heterozygous genotype “AG” (OR=0.71; p = 0.014) of TLR1-rs76600635 were protective genetic factors of susceptibility to AR. And TLR1-rs76600635 has a significant association with decreasing risk of AR under dominant (p = 0.017), and additive (p = 0.028) genetic models. No evidence has been found that TLR1-rs17616434 was associated with susceptibility to AR in overall analysis.
 |
Table 3 Associations Between TLR1 Polymorphisms and AR Susceptibility |
Next, we conducted a stratified analysis based on gender, age, BMI, and region. Figure 1 showed the positive results obtained in stratified analysis. TLR1-rs72493538 had significant associations with susceptibility of AR in males (Allele: OR = 0.34, p < 0.0001; Codominant: OR = 0.30, p < 0.0001; Dominant: OR = 0.31, p < 0.0001; Additive: OR = 0.34, p < 0.0001), participants with aging > 43 years old (Codominant: OR = 0.66, p = 0.030; Dominant: OR = 0.67, p = 0.032; Additive: OR = 0.69, p = 0.039), or coming from wind beach region (Allele: OR = 0.57, p = 0.038). Similarly, TLR1-rs76600635 was significant associated with susceptibility of AR in males (Allele: OR = 0.41, p = 0.001; Codominant: OR = 0.40, p = 0.001; Dominant: OR = 0.40, p = 0.001; Additive: OR = 0.42, p = 0.001), participants with BMI ≤ 24 kg/m2 (Allele: OR = 0.66, p = 0.027; Codominant: OR = 0.65, p = 0.031; Dominant: OR = 0.64, p = 0.025; Additive: OR = 0.65, p = 0.024), or coming from wind beach region (Allele: OR = 0.52, p = 0.048; Additive: OR = 0.56, p = 0.048).
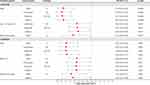 |
Figure 1 Positive results about TLR1-rs72493538 and -rs76600635 found in stratified analysis are shown in forest map. |
TLR1-rs17616434 was not associated with susceptibility to AR in all stratified analyses. There was no association between all candidate genetic loci (rs72493538, rs76600635, and rs17616434) and susceptibility to AR in females, participants with aging ≤ 43 years old, BMI > 24 kg/m2, or coming from loess hilly region.
In addition, FPRP analysis showed (Supplemental Table 2) that all positive results found in this study are noteworthy at a probability level of 0.25 and FPRP cutoff value of 0.2.
MDR Analysis
MDR analysis showed (Table 4) that two-loci genetic model composed of rs72493538 and rs76600635 can be chose as the best model to predict the risk of AR, which with the highest Bal. Acc. Testing (0.536) and perfect CVC (10/10). The Fruchterman-Region of SNP-SNP interactions is shown in Figure 2.
 |
Table 4 SNP-SNP Interactions of TLR1 Gene Analyzed by MDR Method |
Discussion
In this research, we found TLR1-rs72493538 and –rs76600635 reduced the susceptibility to AR among Han Chinese in northern China. In addition, we found evidence that TLR1-rs72493538 and -rs76600635 were associated with AR risk in stratified analyses. These results highlight the importance of TLR1 polymorphisms in AR development and may provide potential biological targets for the treatment of AR.
Koponen and his colleagues have confirmed that TLR1-rs5743618 is involved in the development of allergy and asthma.19 Furthermore, it has been reported that TLR1-rs76600635 is correlated with poor outcomes of melioidosis in Thais populations.20 A targeted sequencing study has identified TLR1-rs72493538 as a novel genetic variant associated with asthma risk in the Chinese Han population.21 TLR1-rs76600635 has been reported to be significantly associated with the occurrence, presentation and adverse drug reactions of tuberculosis in adults in western China.22 Based on the above researches, it can be concluded that TLR1 gene polymorphism may play a potential role in the occurrence and development of nose-related diseases or allergic diseases. All the above studies were conducted on populations with other genetic backgrounds. However, there are relatively few studies on the association between TLR1 gene polymorphism and disease risk in the Chinese population. Our study is the first to investigate the association between TLR1 gene polymorphism and susceptibility to allergic rhinitis in the Chinese population, and noteworthy results have been found. It is worth noting that this study is the first to find that TLR1-rs72493538 and -rs76600635 are associated with the susceptibility to AR in the northern Chinese Han population.
Previous study has reported that TLR1-rs17616434 is associated with susceptibility to asthma and rhinoconjunctivitis in a cohort of Sicilian children,23 but not with the genetic susceptibility of milk allergy in Chinese children.24 More importantly, no evidence was found in our study that TLR1-rs17616434 is associated with AR susceptibility. We speculate that why TLR1-rs17616434 is not associated with allergic diseases in Chinese population may be influenced by the genetic background of the study population, which further indicates that genetic loci play different genetic roles in different populations. In our study, TLR1-rs72493538 has a significant association with AR risk in males, participants with aging > 43 years, or coming from wind-blown sand region. Similarly, TLR1-rs76600635 is significantly associated with the decreasing risk of AR in males, participants with BMI ≤ 24 kg/m2, or coming from wind-blown sand region. Studies have found that the clinical symptoms of allergic rhinitis vary with age and gender.25 In general, the symptoms of allergic rhinitis will slowly improve with age, and the skin test reactivity tends to weaken.26 Environment is also one of the important factors in the pathogenesis of AR.27 Different regions of China may have different environmental factors, climate factors and economic levels, which will cause prevalence of AR may be different. More importantly, many epidemiological studies have found significant differences in AR incidence in different regions of China.28,29 Combined with previous studies and the results of this study, it is necessary to identify the genetic loci associated with AR susceptibility in a specific population, which will provide valuable reference for individualized treatment and diagnosis.
The TLR1 gene is located on chromosome 4p14 and contains seven exons. It plays a critical role in the innate immune system and inflammatory diseases.30 Some research has illustrated that TLR1 participates in the development of nasal diseases.14,31 For example, Kaczmarek et al have observed an upregulated expression of TLR1 in nasal polyps of patients with chronic rhinosinusitis, further suggesting an important role of TLR1 in the formation and maintenance of nasal polyp.31 Moreover, another study has reported that the protein expression level of TLR1 is decreased in nasal epithelium of AR patients.14 Combined with the results of previous studies and this study, we hypothesized that TLR1-rs72493538 and rs76600635 might affect the AR susceptibility of the Han population from northern China by affecting the expression level of TLR1 in AR patients. However, the above is only a speculation, and further design function studies for TLR1-rs72493538 and -rs76600635 will help to better understand the specific molecular mechanism of the above SNPs in AR. In any case, this study provides a new idea for studying the pathogenesis of AR.
However, this study has some limitations that must be faced. Firstly, the research scope is further expanded in the following research to further verify the results of this study. Secondly, further designing functional studies based on the positive results of this study will help to better understand the mechanism of candidate SNPs in the development of AR. In any case, this study is the first to find TRL1 genetic loci associated with reducing AR risk in the Han population in northern China, which will provide a new idea for clinical monitoring of AR risk, as well as early prevention and treatment of AR.
Conclusions
TLR1-rs72493538 and -rs76600635 have closely association with reducing risk of AR. Our study provides a theoretical basis for the treatment and prevention of AR.
Abbreviations
TLR1, Toll-like receptor 1; SNPs, single nucleotide polymorphisms; HWE, Hardy-Weinberg equilibrium; OR, odds ratio; 95% CI, 95% confidence interval; MDR, multifactor dimensionality reduction.
Data Sharing Statement
All data generated or analyzed during this study are included in this manuscript.
Ethics Statement
This research received approval from the Shenmu Hospital, and conformed to the Declaration of Helsinki.
Consent to Participate
Informed consent was obtained from each participant in recruitment after a full understanding of our research.
Acknowledgments
We sincerely thank all participants in this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by Natural Science Foundation of Shaanxi Province (2021SF-075), Science and Technology Plan Project of Yulin City (YF-2020-191) and Shenmu Municipal Government Scientific Research Project (2019) No.5.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Moh’d Al-Rawashdeh B, Sada Alhanjori A, Ali E, Zihlif M. Association of IL-4 polymorphisms with allergic rhinitis in Jordanian population. Medicina. 2020;56(4):179. doi:10.3390/medicina56040179
2. Mims JW. Epidemiology of allergic rhinitis. Int Forum Allergy Rhinol. 2014;4(Suppl 2):S18–20. doi:10.1002/alr.21385
3. Bousquet J, Anto JM, Bachert C, et al. Allergic rhinitis. Nat Rev Dis Primers. 2020;6(1):95. doi:10.1038/s41572-020-00227-0
4. Meng Y, Wang C, Zhang L. Recent developments and highlights in allergic rhinitis. Allergy. 2019;74(12):2320–2328. doi:10.1111/all.14067
5. Choi BY, Han M, Kwak JW, Kim TH. Genetics and Epigenetics in Allergic Rhinitis. Genes. 2021;12(12):2004. doi:10.3390/genes12122004
6. Noguchi E, Shibasaki M, Arinami T, et al. Evidence for linkage between asthma/atopy in childhood and chromosome 5q31-q33 in a Japanese population. Am J Respir Crit Care Med. 1997;156(5):1390–1393. doi:10.1164/ajrccm.156.5.9702084
7. Ashley SE, Tan HT, Vuillermin P, et al. The skin barrier function gene SPINK5 is associated with challenge-proven IgE-mediated food allergy in infants. Allergy. 2017;72(9):1356–1364. doi:10.1111/all.13143
8. Suzuki H, Makino Y, Nagata M, et al. A rare variant in CYP27A1 and its association with atopic dermatitis with high serum total IgE. Allergy. 2016;71(10):1486–1489. doi:10.1111/all.12950
9. Xu Y, Zhang JX. ADAM33 polymorphisms and susceptibility to allergic rhinitis: a meta-analysis. Eur Archi Oto-Rhino-Laryngology. 2015;272(3):597–605. doi:10.1007/s00405-014-3130-3
10. Tharabenjasin P, Pabalan N, Jarjanazi H, Poachanukoon O. Influence of polymorphisms in the interleukin-18 gene on allergic rhinitis: a meta-analysis. Int Arch Allergy Immunol. 2020;181(5):375–384. doi:10.1159/000506010
11. Tang L, Chen Y, Xiang Q, Xiang J, Tang Y, Li J. The association between IL18, FOXP3 and IL13 genes polymorphisms and risk of allergic rhinitis: a meta-analysis. Inflammation Res. 2020;69(9):911–923. doi:10.1007/s00011-020-01368-4
12. Kirtland ME, Tsitoura DC, Durham SR, Shamji MH. Toll-Like Receptor Agonists as Adjuvants for Allergen Immunotherapy. Front Immunol. 2020;11:599083. doi:10.3389/fimmu.2020.599083
13. Larsson O, Sunnergren O, Bachert C, Kumlien Georén S, Cardell LO. The SP-TLR axis, which locally primes the nasal mucosa, is impeded in patients with allergic rhinitis. Clin Transl Allergy. 2021;11(1):e12009. doi:10.1002/clt2.12009
14. Renkonen J, Toppila-Salmi S, Joenväärä S, et al. Expression of Toll-like receptors in nasal epithelium in allergic rhinitis. APMIS. 2015;123(8):716–725. doi:10.1111/apm.12408
15. Cui XY, Chen X, Yu CJ, et al. Increased expression of toll-like receptors 2 and 4 and related cytokines in persistent allergic rhinitis. Otolaryngology--head and neck surgery. Official j Am Acad Otolaryngol Head Neck Surgery. 2015;152(2):233–238.
16. Aoi N, Morikura I, Fuchiwaki T, Yamada T, Prokopakis E, Kawauchi H. OK-432 Administration Inhibits Murine Allergic Rhinitis at the Induction Phase, through the Macrophage Activation with TLR2 Signaling Pathway. Med sci. 2018;6(4):107. doi:10.3390/medsci6040107
17. Fuertes E, Brauer M, MacIntyre E, et al. Childhood allergic rhinitis, traffic-related air pollution, and variability in the GSTP1, TNF, TLR2, and TLR4 genes: results from the TAG Study. J Allergy Clin Immunol. 2013;132(2):342–52.e2. doi:10.1016/j.jaci.2013.03.007
18. Dervieux T. Methotrexate pharmacogenomics in rheumatoid arthritis: introducing false-positive report probability. Rheumatology. 2009;48(6):597–598. doi:10.1093/rheumatology/kep060
19. Koponen P, Vuononvirta J, Nuolivirta K, Helminen M, He Q, Korppi M. The association of genetic variants in toll-like receptor 2 subfamily with allergy and asthma after hospitalization for bronchiolitis in infancy. Pediatr Infect Dis J. 2014;33(5):463–466. doi:10.1097/INF.0000000000000253
20. Wright SW, Emond MJ, Lovelace-Macon L, et al. Exonic sequencing identifies TLR1 genetic variation associated with mortality in Thais with melioidosis. Em Microbes Infections. 2019;8(1):282–290. doi:10.1080/22221751.2019.1575172
21. Liu J, Deng Y, Yu B, et al. Targeted resequencing showing novel common and rare genetic variants increases the risk of asthma in the Chinese Han population. J Clin Lab Anal. 2021;35(6):e23813. doi:10.1002/jcla.23813
22. Peng W, Chen H, Zhao Z, et al. TLR1 polymorphisms are significantly associated with the occurrence, presentation and drug-adverse reactions of tuberculosis in Western Chinese adults. Oncotarget. 2018;9(2):1691–1704. doi:10.18632/oncotarget.23067
23. Sottile G, Ferrante G, Torregrossa M, et al. An association analysis to identify genetic variants linked to asthma and rhino-conjunctivitis in a cohort of Sicilian children. Ital J Pediatr. 2019;45(1):16. doi:10.1186/s13052-019-0603-4
24. Hou L, Ma Z, Chao S, et al. Genetic susceptibility to cow’s milk allergy in Chinese children. Asia Pac J Clin Nutr. 2022;31(1):147–155. doi:10.6133/apjcn.202203_31(1).0016
25. Hong SN, Won JY, Nam EC, et al. Clinical Manifestations of Allergic Rhinitis by Age and Gender: a 12-Year Single-Center Study. Ann Otol Rhinol Laryngol. 2020;129(9):910–917. doi:10.1177/0003489420921197
26. Schoenwetter WF. Allergic rhinitis: epidemiology and natural history. Allergy Asthma Proc. 2000;21(1):1–6. doi:10.2500/108854100778248971
27. Meng Y, Wang C, Zhang L. Advances and novel developments in allergic rhinitis. Allergy. 2020;75(12):3069–3076. doi:10.1111/all.14586
28. Zhang L, Han D, Huang D, et al. Prevalence of self-reported allergic rhinitis in eleven major cities in China. Int Arch Allergy Immunol. 2009;149(1):47–57. doi:10.1159/000176306
29. Wang XY, Ma TT, Wang XY, et al. Prevalence of pollen-induced allergic rhinitis with high pollen exposure in grasslands of northern China. Allergy. 2018;73(6):1232–1243. doi:10.1111/all.13388
30. Vijay K. Toll-like receptors in immunity and inflammatory diseases: past, present, and future. Int Immunopharmacol. 2018;59:391–412. doi:10.1016/j.intimp.2018.03.002
31. Kaczmarek M, Banaszewski J, Leszczyńska M, et al. High frequency of macrophages expressing elevated level of CD80, PD-Ls and TLR1 in nasal polyps of CRS patients. Immunobiology. 2019;224(1):154–162. doi:10.1016/j.imbio.2018.09.004
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

