Back to Journals » Infection and Drug Resistance » Volume 15
The Application of the CRISPR-Cas System in Antibiotic Resistance
Authors Tao S , Chen H, Li N, Liang W
Received 14 April 2022
Accepted for publication 17 June 2022
Published 2 August 2022 Volume 2022:15 Pages 4155—4168
DOI https://doi.org/10.2147/IDR.S370869
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Shuan Tao,1,2 Huimin Chen,1 Na Li,3 Wei Liang2
1School of Medical, Jiangsu University, Zhenjiang, Jiangsu Province, 212013, People’s Republic of China; 2Lianyungang Clinical College of Jiangsu University, Lianyungang, Jiangsu Province, 222023, People’s Republic of China; 3Bengbu Medical College, Bengbu, Anhui Province, 233030, People’s Republic of China
Correspondence: Wei Liang, Lianyungang Clinical College of Jiangsu University, No. 161. Xingfu Road, Haizhou District, Lianyungang, Jiangsu Province, 222023, People’s Republic of China, Tel/Fax +86-51885213100 ; Tel/Fax +86 15351883016, Email [email protected]
Abstract: The emergence and global epidemic of antimicrobial resistance (AMR) poses a serious threat to global public health in recent years. AMR genes are shared between bacterial pathogens mainly via horizontal gene transfer (HGT) on mobile genetic elements (MGEs), thereby accelerating the spread of antimicrobial resistance (AMR) and increasing the burden of drug resistance. There is an urgent need to develop new strategies to control bacterial infections and the spread of antimicrobial resistance. The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) are an RNA-guided adaptive immune system in prokaryotes that recognizes and defends against invasive genetic elements such as phages and plasmids. Because of its specifically target and cleave DNA sequences encoding antibiotic resistance genes, CRISPR/Cas system has been developed into a new gene-editing tool for the prevention and control of bacterial drug resistance. CRISPR-Cas plays a potentially important role in controlling horizontal gene transfer and limiting the spread of antibiotic resistance. In this review, we will introduce the structure and working mechanism of CRISPR-Cas systems, followed by delivery strategies, and then focus on the relationship between antimicrobial resistance and CRISPR-Cas. Moreover, the challenges and prospects of this research field are discussed, thereby providing a reference for the prevention and control of the spread of antibiotic resistance.
Keywords: antibiotic resistance, CRISPR-Cas, horizontal gene transfer
Introduction
In recent years, antibiotic resistance has intensified due to the extensive use of antibiotics.1 Antimicrobial resistance genes (ARGs) may be carried on the bacterial chromosome, and mobile genetic elements (MGEs) such as plasmid, and transposons. The resistance mechanisms mainly include the production of inactivated enzymes and modified enzymes (β-lactamases, aminoglycoside modifying enzymes), changes in the target site of the drug, the alteration of membrane permeability, increased active efflux pump system expression, formation of the bacterial biofilm (BBF).2,3 BBF (adaptive resistance) serves as a diffusion barrier to limit the access of antibiotics to the bacterial cells.4 Loss of efficacy in antibiotics due to antibiotic resistance in bacteria is an urgent threat to the success of microbial infection therapy. The spread of antibiotic-resistant bacteria poses a substantial threat to morbidity and mortality worldwide.5,6 ARGs can spread through vertically inherited chromosomal mutations and horizontal gene transfer (HGT) mediated by MGEs among bacteria.7 The acquisition of ARGs mediated by HGT is the main reason for the spread of drug resistance.8 HGT is mainly driven by conjugation, transduction, and natural transformation.9 MGEs share their genetic elements carrying resistance genes with other non-resistant bacterial species via HGT, which promoted the accumulation and dissemination of ARGs in Gram-negative and Gram-positive bacteria.10,11 To treat bacterial infections and prevent and control the spread of antibiotic resistance, the research and development of new agents with therapeutic potential have attracted more attention worldwide.
Bacteria have evolved multiple defense mechanisms under long-term selection pressure, such as restriction-modification systems and CRISPR-Cas system, which act as innate immune and adaptive systems in bacteria, and significantly affect the spread of antibiotic resistance genes and phage infection.12–14 CRISPR-Cas systems are considered as barriers to HGT in bacteria.15 CRISPR-Cas system can be used to re-sensitize drug-resistant bacteria to antibiotics by specifically eliminating the plasmids carrying antibiotic resistance genes.16 CRISPR has been highlighted as an important tool to prevent and control the spread of bacterial resistance.
In this review, we describe the structure and working mechanism of the CRISPR-Cas system, introduce the delivery method, elucidate the relationship between antimicrobial resistance and CRISPR-Cas, discuss the status of clinical challenges, which aims at providing new ideas for the prevention and control of bacterial resistance. Overall, CRISPR-Cas systems have different effects on antibiotic resistance in different bacteria. The CRISPR-Cas system plays an important role in limiting the spread of antibiotic resistance.
The Structure and Working Mechanism of the CRISPR-Cas System
The CRISPR system is composed of a CRISPR array and a set of CRISPR-associated (Cas) genes.17 The CRISPR array consists of short repeat sequences separated by spacer sequences.18 The CRISPR array is typically preceded by an A-T-rich leader sequence containing a promoter used to initiate transcription of the repeat and spacer sequences19,20 (see Figure 1).
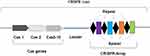 |
Figure 1 The structure of CRISPR system. |
The CRISPR-Cas system is mainly divided into two categories according to its constituent proteins and modes of action.21,22 Class 1 utilizes multi-protein effector complexes to degrade nucleic acids, and can be further divided into type I, type III, and type IV; Class 2 uses single-protein effector complexes to degrade nucleic acids, which can be further divided into type II, type V and type VI.23,24 Types II CRISPR-Cas systems are more widely studied and applied to eliminate resistance genes due to their relatively simple structure.25 In recent years, Type I CRISPR-Cas systems have also been developed as genetic manipulation tools to eliminate resistance genes.26
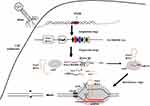
The bacterial defense mechanism of the CRISPR/Cas systems includes three stages.14,27 (i) Adaptation stage: acquisition of spacer sequences.28,29 (ii) Expression stage: generation of the crRNA and Cas protein;24 (iii) Interference stage: crRNA-guided nucleic acid-targeted cleavage.30,31 The mature crRNA binds to Cas protein to form a nucleic acid-protein complex, which can recognize sequences complementary to crRNA in nucleic acid and exert endonuclease activity to degrade the nucleic acid near the recognition site32 (see Figure 2). This specificity allows the CRISPR system to be developed to eliminate specific drug resistance genes, thus controlling horizontal gene transfer and limiting the spread of antibiotic resistance.33
Immune Recognition and Defense Mechanism of I and II CRISPR-Cas System
Type I CRISPR-Cas system is a typical representative of Class 1, and its marker protein is the Cas3 protein containing the phosphohydrolase domain and the helicase domain.34 Type I CRISPR systems rely on multiple proteins working together in a complex called Cascade to target DNA.35 After binding to DNA, Cascade recruits the Cas3 protein to cut DNA.36 The adaptation process of the type I CRISPR-Cas system is mediated by Cas1 and Cas2, which integrate the exogenous DNA segment between its leader sequence and the first repeat of CRISPR to form a new spacer. At the expression stage, the Cas protein processed the pre-CRISPR RNA (pre-crRNA) into mature crRNA and binds into a complex with a group of Cas proteins (cascade), which guides the Cas3 protein to cleavage foreign target sequences.37
Class 2 type II CRISPR-Cas9 system has been most widely adapted for immune defense and gene editing.38 Cas9 contains a unique active site of RuvC at the amino-terminal end and HNH2 in the middle of the protein, playing a role in crRNA maturation and double-stranded DNA shear.39 At the same time as pre-crRNA is transcribed, trans-activating crRNA (tracrRNA) that has sequence complementarity to CRISPR repeats is also transcribed and stimulates Cas9 and double-stranded RNA-specific RNase III nuclease to process pre-crRNA into mature crRNA.12,40 After processing, crRNA, tracrRNA, and Cas9 form a complex, which recognizes and binds to the MGE genome.41,42 Then the complementary protospacer strand is cleaved by the HNH endonuclease domain of Cas9, and the non-complementary strand is cleaved by the RuvC endonuclease domain of Cas9, generating a double-stranded DNA break (DSB) in the invading MGE.43 The cleavage site of CRISPR/Cas9 is located in the NGG site of the PAM region (Protospacer Adjacent Motif) adjacent to the downstream of the crRNA complementary sequence, and cleaves nucleotides on the nearby target sequence to generate a double-strand break (DSB)44–46 (See Figure 2).
Delivery Strategies for CRISPR-Cas Systems
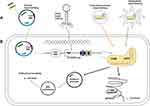
The CRISPR-Cas system has potential applications in the prevention and control of the spread of bacterial resistance caused by drug resistance genes. The direct application of the CRISPR-Cas system is to design the genomic gRNA targeting drug-resistant genes or resistant bacteria to guide the CRISPR-Cas system to cleave the targeted sequences, thereby restoring their antibiotic sensitivity or killing bacteria.45 The efficient delivery of the CRISPR-Cas system into microorganisms is the main problem to be solved by using this system to prevent and control bacterial drug resistance.47 Based on a brief introduction of the working mechanism of the CRISPR-Cas system, we will elucidate the progress of reducing resistance genes and resistance plasmids using the CRISPR-Cas system with plasmids, extracellular vesicles, phage, and nanoparticles as carriers (see Figure 3A).
Plasmid Vector-Based Delivery of CRISPR-Cas Systems
Integrating the sequences of the CRISPR-Cas system or some of its components into a plasmid vector enables the CRISPR-Cas system to target and cut drug-resistant genes, thereby reducing the resistance of drug-resistant bacteria to antibiotics.48
Conjugation is an important mode of bacterial gene transfer that enhances the spread of resistance genes.49,50 CRISPR-Cas encoding delivery plasmids can reduce the occurrence of antibiotic resistance in Enterococcal populations in a sequence-specific manner.51 Moreover, the introduction of a plasmid vector carrying the pCasCure system into carbapenem-resistant Enterobacteriaceae could remove carbapenemase resistance genes such as blaNDM and blaKPC, re-sensitize the resistant bacteria to carbapenems, and have good therapeutic effects on clinical isolation of carbapenem-resistant Enterobacteriaceae.52 The recipient bacteria with the conjugation plasmid can further transmit the CRISPR-Cas system to other recipient bacteria, which greatly expands the application scope of using the CRISPR-Cas system to reduce drug resistance genes.53 In addition, the targeted antimicrobial plasmids (TAPs) can be used to carry the CRISPR-Cas system for efficient transfer to Escherichia coli and the highly related Gram-negative Enterobacteriaceae, re-sensitize the recipient cells carrying pOXA48 and prevent the spread of drug resistance.54 Dong et al55 constructed the conjugative CRISPR/Cas9 system targeting the mobile colistin resistance gene (mcr-1) in Escherichia coli, this engineered CRISPR/Cas9 system can not only eliminate drug-resistant plasmids and re-sensitize to antibiotics but also make the recipient cell acquire immunity against mcr-1. The recombinant plasmid pMBLcas9-sgRNA was able to transfer into clinically isolated E. coli carrying different MCR-1 plasmids with a conjugation efficiency of about 10–1, successfully eliminating the multidrug resistance plasmids.56
Delivery efficiency is the main factor limiting the clinical application of the CRISPR-Cas system.57 Improving plasmid conjugation efficiency can expand the application prospect of the CRISPR-Cas system in preventing and controlling drug resistance gene transfer.58 The pheromone-responsive plasmid (PRP) in Enterococcus faecalis has a higher conjugative transfer efficiency than other plasmids.59 pPD1 plasmid has been used to transfer CRISPR/Cas systems to the Gram-positive Enterococcus faecalis. Studies have shown that the pheromone response plasmid (PRP) in Enterococcus faecalis synthesizes a protein adhesin under the induction of pheromone secreted by the recipient bacteria without the conjugation plasmid, and promoting the aggregation of the donor and recipient bacteria, thereby increasing the conjugation efficiency of pheromone-responsive plasmids.60
Phage Vector-Based Delivery of CRISPR-Cas Systems
Compared with plasmid vectors, phage vectors not only have a stronger ability to infect host bacteria but also can carry larger DNA fragments, which can be introduced into the CRISPR-Cas system by encoding multiple proteins.61 In addition, the nucleic acid encapsulated by the phage protein is relatively stable and not easily degraded.62 The advantage of phage naturally targeting bacteria makes it a preferred delivery tool for researchers.63
Using bacteriophage as a vector, the sgRNA and Cas9 designed based on the conserved sequences of β-lactamase mutants were introduced into the target strain, which successfully inactivated more than 200 mutant pathogenic bacteria and re-sensitized to β-lactamase.64 In addition, a type I CRISPR-Cas3 system containing six Cas genes and multiple spacers targeting ndm-1 and ctx-M-15 resistance genes were packaged using lysogenic phages,65 which could selectively destroy drug-resistant plasmids and restore the susceptibility of host bacteria to multiple antibiotics.66 Bikard et al67 used phagemids to carry the CRISPR-Cas9 system to target the resistance gene of Staphylococcus aureus, and found that it can destroy the plasmid containing the target gene, and if the target gene is on the chromosome, it will cause bacterial death. Research68 indicated that a new mild phage packaging CRISPR-Cas9 system can deliver the CRISPR-Cas9 system and target bacterial drug resistance plasmids. CRISPR-Cas9 wrapped by phages can mediate the degradation of DNA of antibiotic resistance genes (ARG) on plasmids but does not cause cell death. However, the CRISPR-Cas13a wrapped by the phage showed a strong bactericidal activity after the recognition of the target ARG on the plasmid.69
Extracellular Vesicles Vector-Based Delivery of CRISPR-Cas Systems
Membrane vesicles (MV) are the lipid membrane nanoparticles first found in Gram-negative bacteria that are also known as outer membrane vesicles (OMV).70 Bacterial outer membrane vesicles (OMVs) are vesicle-like structures secreted into the extracellular compartment of Gram-negative bacteria during growth.71,72 OMVs can act as delivery systems for antibiotic resistance genes, virulence genes, or plasmids because these components are protected from DNases when present in the vesicle lumen, thereby facilitating the horizontal gene transfer of DNA.73,74 Recently, OMVs have been explored as vectors for delivering Cas9 ribonucleoprotein (RNP) for gene editing and gene regulatory purposes.74 It has been shown that OMVs secreted by Escherichia coli can act as carriers for the CRISPR-Cas9 system targeting Streptococcus agalactiae, achieving efficient and specific clearance of Streptococcus agalactiae in a mixed culture manner.75 However, the lack of a mechanism to enrich RNP into membrane vesicles limits the efficiency of membrane vesicles as an RNP delivery tool.74,76
Nanoparticle-Based Delivery of CRISPR-Cas
Nanoparticles are small in size and have a strong ability to penetrate biological membranes.77 After the CRISPR-Cas9 system is encapsulated by nanomaterials, CRISPR-Cas9 is not easily degraded and can be highly efficient and targeted transported.78 Many studies have been conducted on the application of nanoparticles as carriers for antibiotics, such as polymeric nanoparticles and nanocrystals, zwitterion amino lipid (ZALs) nanoparticles, exosome-liposome mixed nanoparticles, and cationic lipid nanoparticles, nanocages et al.79 Kang et al52 assembled Cas effector proteins and guide RNAs into branched polyethyleneimine nanomaterials, which can be efficiently delivered into methicillin-resistant Staphylococcus aureus (MRSA) for more efficient editing of bacterial genomes targeting mecA. More studies investigated the role of nanotechnology-based CRISPR-Cas9 delivery systems in controlling and preventing antimicrobial resistance.80 Although great progress has been made in optimizing the CRISPR-Cas system with nanoparticles for the treatment of bacterial resistance, further research is needed to achieve its safety and efficacy.81
CRISPR-Cas System’s Role in the Development of Antibiotic Resistance
Relationship Between CRISPR/Cas and the Spread of Antimicrobial Resistance
The emergence and spread of antimicrobial resistance represents a threat to human health.82 Antimicrobial resistance is mainly transmitted by HGT, leading to the spread of bacterial drug resistance.83 CRISPR-Cas systems serve as genome defense systems that can defend against invading exogenous genetic material.84 The structure and function of the CRISPR-Cas system are related to bacterial resistance. The CRISPR-Cas system can prevent the spread of plasmids and phages harboring antibiotic resistance genes, thus limiting the HGT of antibiotic resistance genes mediated by these mobile genetic elements16,85 (see Figures 2 and 3B).
The ubiquitous CRISPR-Cas system in prokaryotes can be used to prevent and control the spread of antibiotic resistance.86 Aydin et al87 found that the type I CRISPR system in Escherichia coli may interfere with the bacteria acquiring the drug-resistant plasmids and maintain the sensitivity of the strain. A study by Price et al59 demonstrated that CRISPR-Cas from mammalian intestinal flora can block the spread of antibiotic resistance plasmids in the mouse intestinal colonization model. Wang et al88 show a negative correlation between the acquisition of antibiotic-resistant genes (ARGs) and the presence of CRISPR/Cas. It is worth noting that pathogens with CRISPR-Cas systems were less likely to carry antibiotic resistance genes than those lacking this defense system.89 However, recent studies have shown that the CRISPR systems are sometimes missed or inactivated and may not be an effective barrier to plasmid and drug resistance spread.90 Therefore, the analysis of the relationship between the CRISPR-Cas syetem and antibiotic resistance will help to better understand the mechanism of bacterial resistance and provide new directions for the prevention and treatment of bacterial resistance.
CRISPR-Cas System’s Role in the Spread of Antibiotic Resistance in Gram-Negative Bacteria
Gram-negative bacteria have a highly restrictive permeability barrier, and their low permeability of the outer membrane is a major cause of resistance to many antibiotics.91 Infections caused by multiple drug-resistant (MDR) and widely drug-resistant (XDR) Gram-negative bacteria (GNB) have become a major challenge to public health.92 The emergence and prevalence of drug-resistance genes such as bla KPC, blaNDM, blaVIM, and MCR-1 suggested that new therapeutic options for the treatment of MDR Gram-negative bacterial infections are urgently needed.93,94 Previous studies have shown that the CRISPR-Cas system has been used to limit the spread of drug-resistant Gram-negative bacteria.
Carbapenems are the preferred treatment for severe infections caused by MDR and XDR Gram-negative bacteria (GNB).95 Of particular concern is an increasing proportion of drug-resistant strains that are generated in Gram-positive bacteria such as extended-spectrum beta-lactamase (ESBL) and carbapenemase-producing Enterobacteriaceae (CPE). Hence, new treatment and prevention strategies are urgently required.96 CRISPR-Cas9 technology can effectively eliminate carbapenem-resistant plasmids in pathogens and restore pathogen susceptibility to carbapenems.97 The constructed CRISPR-Cas9 system was introduced into ESBL-producing Escherichia coli and the results indicated that this system can restore sensitivity to antimicrobial agents in the form of clearing resistance plasmids in ESBLs.64 CRISPR-Cas9-mediated plasmid-curing system (pCasCure) can effectively remove carbapenemase genes (bla KPC, blaNDM, and blaOXA-48) and plasmids (bla KPC-harboring IncFIIK-pKpQIL and the blaNDM-harboring IncX3 plasmid, et al) in CRE isolates, thereby resensitizing CRE to carbapenems.97,98 A study carried out by Kim et al,64 where indicated that the CRISPR-Cas9 system targeting ESBLs-resistant plasmids was introduced into ESBLs-producing Escherichia coli constructed by transformation experiments (ESBLs-producing Klebsiella pneumoniae isolated from patients as plasmid donors and Escherichia coli as plasmid recipients) and the results showed that the system could restore susceptibility to antimicrobials through eliminating ESBLs resistance plasmid. Wan et al99 showed that horizontally transferable colistin resistance genes on MCR-1 plasmids can be knocked out using the CRISPR-Cas9 system.
CRISPR -Cas9 can simultaneously target multiple genes in the same single cell. The study by Vad-Nielsen et al100 simultaneously targeted ten genomic loci and inhibited multiple endogenous genes by constructing a golden gate assembly of the CRISPR gRNA expression array. Further research achieved the simultaneous knockout of multiple β-lactamases with similar sequences in Escherichia coli by CRISPR-Cas, thus solving the problem of high diversity of β-lactamases to a certain extent.101 It has been well reported in the literature that the CRISPR-Cas9 system simultaneously targets two super drug-resistant genes (MCR-1 and blaNDM-1), so that these two drug-resistant genes can be eliminated at the same time, ensuring the efficacy of carbapenem and colistin antibiotics.102
CRISPR-Cas System’s Role in the Spread of Antibiotic Resistance in Gram-Positive Bacteria
Gram-positive bacteria have thick cell walls, which can cause clinical infections of various diseases.103 The emergence of drug-resistant Gram-positive bacteria has raised concerns, and methicillin-sensitive Staphylococcus aureus (MRSA), vancomycin-resistant Enterococci (VRE), and highly resistant strains to Streptococcus penicillin pneumoniae infections are a growing concern to human health.104 As a gene defense system, CRISPR-Cas has been widely used to combat drug resistance in Gram-positive bacteria.
Enterococcus faecalis is a Gram-positive opportunistic pathogen that is the leading cause of nosocomial infections. Enterococcal infections are recognized as a serious public health threat, and the rise in antibiotic resistance makes these infections particularly difficult to treat.105,106 Genetic analysis showed that CRISPR-Cas is a potent barrier to the horizontal acquisition of antibiotic resistance in E. faecalis.59 The literature suggested an inverse relationship between the occurrence of the type II CRISPR-Cas system and antibiotic resistance in E. faecalis.107 E. faecalis isolates generally possess only the orphan CRISPR2.108 Most multidrug-resistant E. faecalis strains generally lack the functional CRISPR1-Cas or CRISPR3-Cas systems. The presence of the CRISPR-Cas system is associated with antibiotic sensitivity and lack of virulence features.109 In Enterococcus faecalis, several studies110,111 have revealed that the absence of antibiotic resistance was associated with the presence of CRISPR3. Differences in CRISPR1 loci have been reported among E. faecalis species, which may lead to the variability of CRISPR activity against antibiotic resistance genes in different species.112 A study carried out by Rodrigues et al51 where indicated that conjugative delivery of CRISPR-Cas9 can reduce the occurrence of antibiotic resistance in the Enterococcal population in a sequence-specific manner. Price et al12 showed that orphan CRISPR2 loci requires the presence of CRISPR1-Cas derived from Enterococcus faecalis for genomic defense against MGE. As demonstrated by Gholizadeh et al113 in studies with Enterococcus faecalis where they present a study showing that CRISPR-Cas can prevent the acquisition of some corresponding pathogenic factors in some isolates. In a study by Wu and coworkers found that the CRISPR-Cas9 systems targets the tetracycline resistance gene (tetM) and erythromycin resistance gene (ermB), respectively, successfully reducing antibiotic resistance to E. faecalis in vitro and in vivo.102 These findings suggest that CRISPR-Cas systems can limit the spread of drug resistance in Gram-positive bacteria.
CRISPR-Cas Systems’ Role in Bacterial Biofilms
Bacterial pathogens often form biofilms, which helps them to resist different threats within the host.114 Bacterial biofilms can increase the possibility of bacterial drug resistance and horizontal gene transfer.115 The changes in the permeability of bacterial membranes can affect bacterial resistance.116 The integrity of the bacterial envelope plays an important role in mediating antibiotic resistance and evasion of some inflammation caused by membrane-targeted antibiotics.117 Studies found that the CRISPR-Cas system enhanced the envelope integrity by regulating bacterial lipoproteins, and ultimately cells provided the ability to resist membrane damage caused by antibiotics.81 The CRISPR-Cas system is associated with the biofilm formation and colonization of the host.114 In Pseudomonas aeruginosa, the type I-F CRISPR-Cas system has been revealed to inhibit biofilm formation by crRNA-guided targeting and destroying of prophage DNA.118 Also, Zegans et al119 demonstrated that the CRISPR system is required for bacteriophage DMS3-dependent inhibition of biofilm formation in Pseudomonas aeruginosa. In addition, studies by Sampson et al117 revealed that the CRISPR-Cas endonuclease gene cas9 along with tracrRNA and ScaRNA represses the expression of bacterial lipoprotein (BLP), thereby affecting envelope integrity-mediated antibiotic resistance and contributing to immune avoidance during infection in Francisella novicida.120,121
CRISPR-Cas System’s Role in Increasing Antibiotic Resistance
CRISPR-Cas systems may have a different effect on antibiotic resistance among different species.16,122 Most studies showed that CRISPR-Cas system involvement in antimicrobial resistance.123 However, several studies demonstrated that there was no significant relationship between the CRISPR-Cas system and antibiotic resistance in Escherichia coli.124 Furthermore, Touchon et al125 showed little effect of CRISPR on the epidemiology of plasmids in E. coli or on the spread of antibiotic-resistant genes. Type I CRISPR-Cas system not only plays a role in mediating anti-antibiotic resistance but also is associated with increased bacterial antibiotic resistance. For example, the I-F CRISPR-Cas system in Pseudomonas aeruginosa removed resistance genes or plasmids, thereby reducing the level of antibiotic resistance of the bacteria.126 However, studies have suggested that the type I-E CRISPR system of Vibrio cholerae may facilitate the acquisition of bacteria carrying β-lactamase resistance genes.127 Moreover, Shabbir et al128 revealed that the CRISPR-Cas system promotes antimicrobial resistance in Campylobacter jejuni through transcriptome analysis. These results indicated that apart from the impact of limiting the spread of antibiotic resistance, the CRISPR-Cas system is associated with increased bacterial antibiotic resistance.
Previous studies indicated that Klebsiella pneumoniae strains with type I CRISPR system have a high number of tetracycline resistance genes, while they were more sensitive to aminoglycoside and β-lactam antibiotics and had fewer associated resistance genes.129 A few resistant strains containing the CRISPR-Cas system were considered to be due to the mutation of the original spacer sequence, the partial or total deletion in the Cas gene cluster, and the presence of anti-CRISPR proteins.
Anti-CRISPR (Acr) Proteins and Antibiotic Resistance
CRISPR-Cas system provides adaptive immunity in bacteria against diverse categories of phage infections and can defend against mobile genetic elements (MGEs).130 Parallelly, many phages and MGEs have evolved anti-CRISPR proteins (Acres) to counteract the CRISPR-Cas system at different stages under the strong selective pressure exerted by CRISPR-Cas immunity.131 Anti-CRISPR proteins (Acres) disable CRISPR-Cas systems with diverse mechanisms. For example, AcrIF1, AcrIIA et al132–134 can block DNA binding sites, and AcrIIC1, AcrIE1, AcrIF3, et al135–137 can block DNA cleavage and even act enzymatically to disable CRISPR-Cas. Previous studies have shown that anti-type I CRISPR-Cas system ACPs such as AcrIE and AcrIF can inhibit the degradation of target DNA by preventing the recruitment of Cas3;137,138 while anti-type II CRISPR-Cas system ACP can prevent from binding to target DNA by binding to Cas9.137,139–141 Meeske et al142 described that listeriaphage (ϕLS46) encoding an anti-CRISPR protein (AcrVIA1) inhibits Cas13a by interacting with the guide-exposed face of Cas13a nuclease that prevents access to the target RNA and the activation of Cas13a RNase function and eventually inactivates the type VI-A CRISPR system. Prior studies have suggested that anti-CRISPR proteins, AcrF1 and AcrF2, inhibit the type I-F CRISPR/Cas system of Pseudomonas aeruginosa by preventing target DNA recognition. Shehreen et al143 indicated that anti-CRISPRs might facilitate the uptake of ARGs in Pseudomonas aeruginosa. Anti-CRISPR (Acr) proteins carried by mobile genetic elements such as phages and conjugative plasmids demonstrate a role in the horizontal transfer of different MGE-encoded traits.139 Anti-CRISPR promotes HGT by inhibiting CRISPR-Cas, and anti-CRISPR genes may be positively associated with antibiotic resistance. However, as pointed out by Stanley et al,144 the phage encoding anti-CRISPRs remain sensitive to CRISPR-Cas, suggesting that anti-CRISPR action may be an imperfect process.
Discussion
Antibiotic resistance is spreading rapidly around the world and poses a critical threat to public health. There is an urgent need for new strategies to control multidrug-resistant (MDR) bacterial infections and the spread of antimicrobial resistance.145 The CRISPR-Cas system can specifically recognize and target the genetic elements carrying drug resistance genes or their transcripts and limit the spread of drug resistance genes, which shows great potential for preventing and controlling bacterial drug resistance.
As a gene-editing tool for adaptive immune defense against foreign nucleic acid invasion, the CRISPR-Cas9 system is more advanced than zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALEN), and other traditional gene-editing tools.146 CRISPR-Cas shows many remarkable features of simple operation high efficiency, simple operation, good knockout effect, and low cytotoxicity.37 Ding et al147 used TALEN and CRISPR-Cas9 to modify the same gene, and the experimental results showed that the efficiencies were 0% to 34% and 51% to 79%, respectively, indicating the high efficiency of CRISPR. In addition, from an experimental point of view, CRISPR is easier to operate than TALEN, because each pair of TALEN needs to be resynthesized, but the design of Guide-RNA in the application of CRISPR technology is much simpler.148 As a gene-editing tool, CRISPR/Cas9 technology, when combined with existing nucleic acid amplification technology, luminescence technology, rapid detection technology, etc., will show huge technical advantages such as high detection sensitivity, specificity, and reduced reaction time. For example, in 2017, Science published a molecular detection platform SHERLOCK based on Cas13a and recombinant polymerase amplification, which can detect Zika virus and dengue virus, identify mutations in circulating tumor DNA, etc., with extremely high sensitivity.149 The specifically targeted site-specific cutting capability of CRISPR-Cas has made some achievements in nucleic acid testing and bacterial typing. These recent studies show that CRISPR technology has broad and rich application prospects in the field of medical laboratory.
In this review, we briefly introduce the structure, and mechanism of the CRISPR-Cas system, and the correlation of the CRISPR-Cas system with the spread of antibiotic resistance. Collectively, the effect of the CRISPR-Cas system on antibiotic resistance varies among different bacteria. The progress of introducing different types of CRISPR-Cas systems into plasmids, extracellular vesicles, phages, and nanoparticle vectors to reduce the level of bacterial drug resistance by targeting drug-resistant genes and plasmids was analyzed. Delivery of the CRISPR-Cas system or some of its components to bacteria still has some problems such as difficulty in the introduction and easy degradation by bacterial intracellular proteases or nucleases. Activation of ubiquitous endogenous CRISPR-Cas systems in bacteria using small molecule compounds or physicochemical factors may provide new directions for controlling the problem of bacterial multiple resistance due to drug resistance gene transfer.
The CRISPR-Cas system can limit the horizontal transfer of drug resistance genes in bacteria, and the application of bacterial genome programming provides important genetic tools for future research in the field of molecular biology. The CRISPR system has attracted more and more attention and become the research object of many scholars. Researchers have proposed and developed various efficient strategies to improve editing efficiency and reduce the off-target rate of CRISPR systems, and some remarkable results have been achieved. It is of great significance to utilize the CRISPR-Cas systems for the rapid detection of pathogenic microorganisms, the inhibition of viral infection, and the prevention and treatment of human infectious diseases. In the future, the CRISPR system will surprise us even more in terms of human health and medical development.
Conclusion
CRISPR-Cas has the application prospect of preventing and controlling horizontal gene transfer and limiting the spread of antibiotic resistance. The CRISPR system can defend against the invasion of exogenous genes and is associated with bacterial drug resistance. Elucidating the structure and regulatory mechanisms of the CRISPR-Cas system and analyzing the relationship between CRISPR-Cas and bacterial drug resistance can help to better understand bacterial drug resistance and provide a new direction for controlling bacterial drug resistance. The CRISPR-Cas system has updated the understanding of bacterial function regulation, providing a new direction for the prevention and control of bacterial drug resistance and a means for effective immunotherapy in the medical industry.
Acknowledgments
Thanks to Jiangsu University and the Department of Laboratory Medicine of the Second People’s Hospital of Lianyungang City, Jiangsu Province, for supporting the work. Special thanks to my tutor Dr. Liang for my guidance.
Funding
This study was supported by grants from the Natural Science Foundation of Jiangsu Province (BK20191210), the fifth phase of the “333 Project” scientific research project in Jiangsu Province (BRA2019248), the Jiangsu Commission of Health (H2018073), and the Subject of Lianyungang Science and Technology Bureau (SF2015).
Disclosure
None of the authors has a conflict of interest to disclose.
References
1. Kim JH, Yu D, Eom SH, et al. Synergistic antibacterial effects of chitosan-caffeic acid conjugate against antibiotic-resistant acne-related bacteria. Mar Drugs. 2017;15(6):167. doi:10.3390/md15060167
2. Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int. 2016;2016:2475067. doi:10.1155/2016/2475067
3. Christaki E, Marcou M, Tofarides A. Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol. 2020;88(1):26–40.
4. Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177–192. doi:10.1016/j.biotechadv.2018.11.013
5. Rostock L, Driller R, Grätz S, et al. Molecular insights into antibiotic resistance - how a binding protein traps albicidin. Nat Commun. 2018;9(1):3095. doi:10.1038/s41467-018-05551-4
6. Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol Rev. 2018;42(1). doi:10.1093/femsre/fux053
7. von Wintersdorff CJ, Penders J, van Niekerk JM, et al. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol. 2016;7:173. doi:10.3389/fmicb.2016.00173
8. Meng M, Li Y, Yao H. Plasmid-mediated transfer of antibiotic resistance genes in soil. Antibiotics. 2022;11(4). doi:10.3390/antibiotics11040525
9. Fogg PCM. Identification and characterization of a direct activator of a gene transfer agent. Nat Commun. 2019;10(1):595. doi:10.1038/s41467-019-08526-1
10. Engelstädter J, Harms K, Johnsen PJ. The evolutionary dynamics of integrons in changing environments. ISME J. 2016;10(6):1296–1307. doi:10.1038/ismej.2015.222
11. Koonin EV, Makarova KS. Mobile genetic elements and evolution of CRISPR-Cas systems: all the way there and back. Genome Biol Evol. 2017;9(10):2812–2825. doi:10.1093/gbe/evx192
12. Price VJ, Huo W, Sharifi A, Palmer KL. CRISPR-Cas and restriction-modification act additively against conjugative antibiotic resistance plasmid transfer in Enterococcus faecalis. mSphere. 2016;1(3). doi:10.1128/mSphere.00064-16
13. Mackow NA, Shen J, Adnan M, et al. CRISPR-Cas influences the acquisition of antibiotic resistance in Klebsiella pneumoniae. PLoS One. 2019;14(11):e0225131. doi:10.1371/journal.pone.0225131
14. Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc Lond B Biol Sci. 2016;371(1707). doi:10.1098/rstb.2015.0496
15. Zheng Z, Zhang Y, Liu Z, et al. The CRISPR-Cas systems were selectively inactivated during evolution of Bacillus cereus group for adaptation to diverse environments. ISME J. 2020;14(6):1479–1493. doi:10.1038/s41396-020-0623-5
16. Gholizadeh P, Ş K, Dao S, et al. How CRISPR-Cas system could be used to combat antimicrobial resistance. Infect Drug Resist. 2020;13:1111–1121. doi:10.2147/IDR.S247271
17. Rath D, Amlinger L, Hoekzema M, Devulapally PR, Lundgren M. Efficient programmable gene silencing by Cascade. Nucleic Acids Res. 2015;43(1):237–246. doi:10.1093/nar/gku1257
18. Hillary VE, Ignacimuthu S, Ceasar SA. Potential of CRISPR/Cas system in the diagnosis of COVID-19 infection. Expert Rev Mol Diagn. 2021;21(11):1179–1189. doi:10.1080/14737159.2021.1970535
19. Hu T, Cui Y, Qu X. Characterization and comparison of CRISPR Loci in Streptococcus thermophilus. Arch Microbiol. 2020;202(4):695–710. doi:10.1007/s00203-019-01780-3
20. Alkhnbashi OS, Shah SA, Garrett RA, et al. Characterizing leader sequences of CRISPR loci. Bioinformatics. 2016;32(17):i576–i585. doi:10.1093/bioinformatics/btw454
21. Nishimasu H, Nureki O. Structures and mechanisms of CRISPR RNA-guided effector nucleases. Curr Opin Struct Biol. 2017;43:68–78. doi:10.1016/j.sbi.2016.11.013
22. Mohanraju P, Makarova KS, Zetsche B, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353(6299):aad5147. doi:10.1126/science.aad5147
23. Shmakov S, Abudayyeh OO, Makarova KS, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60(3):385–397. doi:10.1016/j.molcel.2015.10.008
24. Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13(11):722–736. doi:10.1038/nrmicro3569
25. Kalds P, Zhou S, Cai B, et al. Sheep and goat genome engineering: from random transgenesis to the CRISPR era. Front Genet. 2019;10:750. doi:10.3389/fgene.2019.00750
26. Zhang Q, Doak TG, Ye Y. Expanding the catalog of cas genes with metagenomes. Nucleic Acids Res. 2014;42(4):2448–2459. doi:10.1093/nar/gkt1262
27. Jackson SA, McKenzie RE, Fagerlund RD, et al. CRISPR-Cas: adapting to change. Science. 2017;356(6333). doi:10.1126/science.aal5056.
28. Nuñez JK, Kranzusch PJ, Noeske J, et al. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 2014;21(6):528–534. doi:10.1038/nsmb.2820
29. Nuñez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519(7542):193–198. doi:10.1038/nature14237
30. Plagens A, Richter H, Charpentier E, Randau L. DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol Rev. 2015;39(3):442–463. doi:10.1093/femsre/fuv019
31. Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39(3):428–441. doi:10.1093/femsre/fuv023
32. Le Rhun A, Escalera-Maurer A, Bratovič M, Charpentier E. CRISPR-Cas in Streptococcus pyogenes. RNA Biol. 2019;16(4):380–389. doi:10.1080/15476286.2019.1582974
33. Teng M, Yao Y, Nair V, Luo J. Latest advances of virology research using CRISPR/Cas9-based gene-editing technology and its application to vaccine development. Viruses. 2021;13(5):779. doi:10.3390/v13050779
34. Makarova KS, Zhang F, Koonin EV. SnapShot: class 1 CRISPR-Cas systems. Cell. 2017;168(5):946–946.e1. doi:10.1016/j.cell.2017.02.018
35. Zheng Y, Li J, Wang B, et al. Endogenous type I CRISPR-Cas: from foreign DNA defense to prokaryotic engineering. Front Bioeng Biotechnol. 2020;8:62. doi:10.3389/fbioe.2020.00062
36. Westra ER, van Erp PB, Künne T, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46(5):595–605. doi:10.1016/j.molcel.2012.03.018
37. Liu Z, Dong H, Cui Y, Cong L, Zhang D. Application of different types of CRISPR/Cas-based systems in bacteria. Microb Cell Fact. 2020;19(1):172. doi:10.1186/s12934-020-01431-z
38. Shmakov S, Smargon A, Scott D, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15(3):169–182. doi:10.1038/nrmicro.2016.184
39. Mir A, Edraki A, Lee J, Sontheimer EJ. Type II-C CRISPR-Cas9 biology, mechanism, and application. ACS Chem Biol. 2018;13(2):357–365. doi:10.1021/acschembio.7b00855
40. Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi:10.1016/j.cell.2014.02.001
41. Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109(39):E2579–86. doi:10.1073/pnas.1208507109
42. Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168(1–2):20–36. doi:10.1016/j.cell.2016.10.044
43. Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi:10.1038/nature17946
44. Fu YW, Dai XY, Wang WT, et al. Dynamics and competition of CRISPR-Cas9 ribonucleoproteins and AAV donor-mediated NHEJ, MMEJ and HDR editing. Nucleic Acids Res. 2021;49(2):969–985. doi:10.1093/nar/gkaa1251
45. Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20(8):490–507. doi:10.1038/s41580-019-0131-5
46. Cencic R, Miura H, Malina A, et al. Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage. PLoS One. 2014;9(10):e109213. doi:10.1371/journal.pone.0109213
47. Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234–1257. doi:10.1080/10717544.2018.1474964
48. Cabezón E, Ripoll-Rozada J, Peña A, de la Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev. 2015;39(1):81–95. doi:10.1111/1574-6976.12085
49. Harrison E, Brockhurst MA. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012;20(6):262–267. doi:10.1016/j.tim.2012.04.003
50. Köstlbacher S, Collingro A, Halter T, Domman D, Horn M. Coevolving plasmids drive gene flow and genome plasticity in host-associated intracellular bacteria. Curr Biol. 2021;31(2):346–357.e3. doi:10.1016/j.cub.2020.10.030
51. Rodrigues M, McBride SW, Hullahalli K, Palmer KL, Duerkop BA. Conjugative delivery of CRISPR-Cas9 for the selective depletion of antibiotic-resistant enterococci. Antimicrob Agents Chemother. 2019;63(11). doi:10.1128/AAC.01454-19
52. Kang YK, Kwon K, Ryu JS, et al. Nonviral genome editing based on a polymer-derivatized CRISPR nanocomplex for targeting bacterial pathogens and antibiotic resistance. Bioconjug Chem. 2017;28(4):957–967. doi:10.1021/acs.bioconjchem.6b00676
53. Gama JA, Zilhão R, Dionisio F. Conjugation efficiency depends on intra and intercellular interactions between distinct plasmids: plasmids promote the immigration of other plasmids but repress co-colonizing plasmids. Plasmid. 2017;93:6–16. doi:10.1016/j.plasmid.2017.08.003
54. Reuter A, Hilpert C, Dedieu-Berne A, et al. Targeted-antibacterial-plasmids (TAPs) combining conjugation and CRISPR/Cas systems achieve strain-specific antibacterial activity. Nucleic Acids Res. 2021;49(6):3584–3598. doi:10.1093/nar/gkab126
55. Dong H, Xiang H, Mu D, Wang D, Wang T. Exploiting a conjugative CRISPR/Cas9 system to eliminate plasmid harbouring the mcr-1 gene from Escherichia coli. Int J Antimicrob Agents. 2019;53(1):1–8. doi:10.1016/j.ijantimicag.2018.09.017
56. Wang P, He D, Li B, et al. Eliminating mcr-1-harbouring plasmids in clinical isolates using the CRISPR/Cas9 system. J Antimicrob Chemother. 2019;74(9):2559–2565. doi:10.1093/jac/dkz246
57. Chen G, Cheng D, Chen B. [Development of CRISPR technology and its application in bone and cartilage tissue engineering]. Nan Fang Yi Ke Da Xue Xue Bao. 2019 Dec 30;39(12):1515-1520. Chinese. doi:10.12122/j.issn.1673-4254.2019.12.19
58. Ling X, Xie B, Gao X, et al. Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates. Sci Adv. 2020;6(15):eaaz0051. doi:10.1126/sciadv.aaz0051
59. Price VJ, McBride SW, Hullahalli K, et al. Enterococcus faecalis CRISPR-Cas Is a robust barrier to conjugative antibiotic resistance dissemination in the murine intestine. mSphere. 2019;4(4). doi:10.1128/mSphere.00464-19.
60. Zou J, Tang Z, Yan J, et al. Dissemination of linezolid resistance through sex pheromone plasmid transfer in Enterococcus faecalis. Front Microbiol. 2020;11:1185. doi:10.3389/fmicb.2020.01185
61. Borges AL, Castro B, Govindarajan S, et al. Bacterial alginate regulators and phage homologs repress CRISPR-Cas immunity. Nat Microbiol. 2020;5(5):679–687. doi:10.1038/s41564-020-0691-3
62. Górski A, Międzybrodzki R, Borysowski J, et al. Phage as a modulator of immune responses: practical implications for phage therapy. Adv Virus Res. 2012;83:41–71.
63. Fage C, Lemire N, Moineau S. Delivery of CRISPR-Cas systems using phage-based vectors. Curr Opin Biotechnol. 2021;68:174–180. doi:10.1016/j.copbio.2020.11.012
64. Kim JS, Cho DH, Park M, et al. CRISPR/Cas9-mediated re-sensitization of antibiotic-resistant Escherichia coli harboring extended-spectrum β-Lactamases. J Microbiol Biotechnol. 2016;26(2):394–401. doi:10.4014/jmb.1508.08080
65. Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci U S A. 2015;112(23):7267–7272. doi:10.1073/pnas.1500107112
66. Oechslin F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses. 2018;10(7):351. doi:10.3390/v10070351
67. Bikard D, Euler CW, Jiang W, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32(11):1146–1150. doi:10.1038/nbt.3043
68. Lam KN, Spanogiannopoulos P, Soto-Perez P, et al. Phage-delivered CRISPR-Cas9 for strain-specific depletion and genomic deletions in the gut microbiome. Cell Rep. 2021;37(5):109930. doi:10.1016/j.celrep.2021.109930
69. Kiga K, Tan XE, Ibarra-Chávez R, et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat Commun. 2020;11(1):2934. doi:10.1038/s41467-020-16731-6
70. Gerritzen MJH, Martens DE, Wijffels RH, Stork M. High throughput nanoparticle tracking analysis for monitoring outer membrane vesicle production. J Extracell Vesicles. 2017;6(1):1333883. doi:10.1080/20013078.2017.1333883
71. Collins SM, Brown AC. Bacterial outer membrane vesicles as antibiotic delivery vehicles. Front Immunol. 2021;12:733064. doi:10.3389/fimmu.2021.733064
72. Martora F, Pinto F, Folliero V, et al. Isolation, characterization and analysis of pro-inflammatory potential of Klebsiella pneumoniae outer membrane vesicles. Microb Pathog. 2019;136:103719. doi:10.1016/j.micpath.2019.103719
73. Dell’Annunziata F, Folliero V, Giugliano R, et al. Gene transfer potential of outer membrane vesicles of gram-negative bacteria. Int J Mol Sci. 2021;22(11):5985. doi:10.3390/ijms22115985
74. Yao X, Lyu P, Yoo K, et al. Engineered extracellular vesicles as versatile ribonucleoprotein delivery vehicles for efficient and safe CRISPR genome editing. J Extracell Vesicles. 2021;10(5):e12076. doi:10.1002/jev2.12076
75. Fantappiè L, de Santis M, Chiarot E, et al. Antibody-mediated immunity induced by engineered Escherichia coli OMVs carrying heterologous antigens in their lumen. J Extracell Vesicles. 2014;3(1):24015. doi:10.3402/jev.v3.24015
76. Lu M, Xing H, Yang Z, et al. Recent advances on extracellular vesicles in therapeutic delivery: challenges, solutions, and opportunities. Eur J Pharm Biopharm. 2017;119:381–395. doi:10.1016/j.ejpb.2017.07.010
77. Pudlarz A, Szemraj J. Nanoparticles as carriers of proteins, peptides and other therapeutic molecules. Open Life Sci. 2018;13(1):285–298. doi:10.1515/biol-2018-0035
78. Givens BE, Naguib YW, Geary SM, Devor EJ, Salem AK. Nanoparticle-based delivery of CRISPR/Cas9 genome-editing therapeutics. AAPS J. 2018;20(6):108. doi:10.1208/s12248-018-0267-9
79. Lima R, Del Fiol FS, Balcão VM. Prospects for the use of new technologies to combat multidrug-resistant bacteria. Front Pharmacol. 2019;10:692. doi:10.3389/fphar.2019.00692
80. Xu X, Liu C, Wang Y, et al. Nanotechnology-based delivery of CRISPR/Cas9 for cancer treatment. Adv Drug Deliv Rev. 2021;176:113891. doi:10.1016/j.addr.2021.113891
81. Wan F, Draz MS, Gu M, et al. Novel strategy to combat antibiotic resistance: a sight into the combination of CRISPR/Cas9 and nanoparticles. Pharmaceutics. 2021;13(3):352. doi:10.3390/pharmaceutics13030352
82. Shaw DJ, Hill RE, Simpson N, et al. Examining the role of protein structural dynamics in drug resistance in Mycobacterium tuberculosis. Chem Sci. 2017;8(12):8384–8399. doi:10.1039/C7SC03336B
83. Stevenson C, Hall JP, Harrison E, Wood A, Brockhurst MA. Gene mobility promotes the spread of resistance in bacterial populations. ISME J. 2017;11(8):1930–1932. doi:10.1038/ismej.2017.42
84. Liu T, Liu Z, Ye Q, et al. Coupling transcriptional activation of CRISPR-Cas system and DNA repair genes by Csa3a in sulfolobus islandicus. Nucleic Acids Res. 2017;45(15):8978–8992. doi:10.1093/nar/gkx612
85. Serajian S, Ahmadpour E, Oliveira SMR, Pereira ML, Heidarzadeh S. CRISPR-Cas technology: emerging applications in clinical microbiology and infectious diseases. Pharmaceuticals. 2021;14(11). doi:10.3390/ph14111171
86. Xu Y, Li Z. CRISPR-Cas systems: overview, innovations and applications in human disease research and gene therapy. Comput Struct Biotechnol J. 2020;18:2401–2415. doi:10.1016/j.csbj.2020.08.031
87. Aydin S, Personne Y, Newire E, et al. Presence of Type I-F CRISPR/Cas systems is associated with antimicrobial susceptibility in Escherichia coli. J Antimicrob Chemother. 2017;72(8):2213–2218. doi:10.1093/jac/dkx137
88. Wang G, Song G, Xu Y. Association of CRISPR/Cas system with the drug resistance in Klebsiella pneumoniae. Infect Drug Resist. 2020;13:1929–1935. doi:10.2147/IDR.S253380
89. Pursey E, Dimitriu T, Paganelli FL, Westra ER, van Houte S. CRISPR-Cas is associated with fewer antibiotic resistance genes in bacterial pathogens. Philos Trans R Soc Lond B Biol Sci. 2022;377(1842):20200464. doi:10.1098/rstb.2020.0464
90. Gophna U, Kristensen DM, Wolf YI, et al. No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales. Isme j. 2015;9(9):2021–2027. doi:10.1038/ismej.2015.20
91. Farrag HA, Abdallah N, Shehata MMK, Awad EM. Natural outer membrane permeabilizers boost antibiotic action against irradiated resistant bacteria. J Biomed Sci. 2019;26(1):69. doi:10.1186/s12929-019-0561-6
92. Hawkey PM, Warren RE, Livermore DM, et al. Treatment of infections caused by multidrug-resistant Gram-negative bacteria: report of the British Society for Antimicrobial Chemotherapy/Healthcare Infection Society/British Infection Association Joint Working Party. J Antimicrob Chemother. 2018;73(suppl_3):iii2–iii78. doi:10.1093/jac/dky027
93. Liang WJ, Liu HY, Duan GC, et al. Emergence and mechanism of carbapenem-resistant Escherichia coli in Henan, China, 2014. J Infect Public Health. 2018;11(3):347–351. doi:10.1016/j.jiph.2017.09.020
94. Wang R, Liu Y, Zhang Q, et al. The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: coexistence of mcr-1 and bla(NDM) with low fitness cost. Int J Antimicrob Agents. 2018;51(5):739–744. doi:10.1016/j.ijantimicag.2018.01.023
95. Sheu CC, Chang YT, Lin SY, Chen YH, Hsueh PR. Infections caused by carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Front Microbiol. 2019;10:80. doi:10.3389/fmicb.2019.00080
96. Choi YK, Byeon EJ, Park JJ, Lee J, Seo YB. Antibiotic resistance patterns of Enterobacteriaceae isolated from patients with healthcare-associated infections. Infect Chemother. 2021;53(2):355–363. doi:10.3947/ic.2021.0030
97. Hao M, He Y, Zhang H, et al. CRISPR-Cas9-mediated carbapenemase gene and plasmid curing in carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2020;64(9). doi:10.1128/AAC.00843-20.
98. Wang Y, Wang S, Chen W, et al. CRISPR-Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl Environ Microbiol. 2018;84(23). doi:10.1128/AEM.01834-18.
99. Wan P, Cui S, Ma Z, et al. Reversal of mcr-1-mediated colistin resistance in Escherichia coli by CRISPR-Cas9 system. Infect Drug Resist. 2020;13:1171–1178. doi:10.2147/IDR.S244885
100. Vad-Nielsen J, Lin L, Bolund L, Nielsen AL, Luo Y. Golden gate assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes. Cell Mol Life Sci. 2016;73(22):4315–4325. doi:10.1007/s00018-016-2271-5
101. Feng X, Zhao D, Zhang X, Ding X, Bi C. CRISPR/Cas9 assisted multiplex genome editing technique in Escherichia coli. Biotechnol J. 2018;13(9):e1700604. doi:10.1002/biot.201700604
102. Wu Y, Battalapalli D, Hakeem MJ, et al. Engineered CRISPR-Cas systems for the detection and control of antibiotic-resistant infections. J Nanobiotechnology. 2021;19(1):401. doi:10.1186/s12951-021-01132-8
103. Bloem A, Bax HI, Yusuf E, Verkaik NJ. New-generation antibiotics for treatment of gram-positive infections: a review with focus on Endocarditis and Osteomyelitis. J Clin Med. 2021;10(8):1743. doi:10.3390/jcm10081743
104. Feng Z, Chakraborty D, Dewell SB, Reddy BV, Brady SF. Environmental DNA-encoded antibiotics fasamycins A and B inhibit FabF in type II fatty acid biosynthesis. J Am Chem Soc. 2012;134(6):2981–2987. doi:10.1021/ja207662w
105. Debroy S, van der Hoeven R, Singh KV, et al. Development of a genomic site for gene integration and expression in Enterococcus faecalis. J Microbiol Methods. 2012;90(1):1–8. doi:10.1016/j.mimet.2012.04.011
106. Boccella M, Santella B, Pagliano P, et al. Prevalence and antimicrobial resistance of Enterococcus species: a retrospective cohort study in Italy. Antibiotics. 2021;10(12). doi:10.3390/antibiotics10121552.
107. Alduhaidhawi AHM, AlHuchaimi SN, Al-Mayah TA, et al. Prevalence of CRISPR-Cas Systems and their possible association with antibiotic resistance in Enterococcus faecalis and Enterococcus faecium collected from hospital wastewater. Infect Drug Resist. 2022;15:1143–1154. doi:10.2147/IDR.S358248
108. Hullahalli K, Rodrigues M, Schmidt BD, et al. Comparative analysis of the orphan CRISPR2 Locus in 242 Enterococcus faecalis strains. PLoS One. 2015;10(9):e0138890. doi:10.1371/journal.pone.0138890
109. Hullahalli K, Rodrigues M, Nguyen UT, Palmer K. An attenuated CRISPR-Cas System in Enterococcus faecalis permits DNA acquisition. mBio. 2018;9(3). doi:10.1128/mBio.00414-18
110. Gholizadeh P, Aghazadeh M, Ghotaslou R, et al. Role of CRISPR-Cas system on antibiotic resistance patterns of Enterococcus faecalis. Ann Clin Microbiol Antimicrob. 2021;20(1):49. doi:10.1186/s12941-021-00455-6
111. Burley KM, Sedgley CM. CRISPR-Cas, a prokaryotic adaptive immune system, in endodontic, oral, and multidrug-resistant hospital-acquired Enterococcus faecalis. J Endod. 2012;38(11):1511–1515. doi:10.1016/j.joen.2012.07.004
112. Hullahalli K, Rodrigues M, Palmer KL. Exploiting CRISPR-Cas to manipulate Enterococcus faecalis populations. eLife. 2017;6. doi:10.7554/eLife.26664
113. Gholizadeh P, Aghazadeh M, Ghotaslou R, et al. CRISPR-cas system in the acquisition of virulence genes in dental-root canal and hospital-acquired isolates of Enterococcus faecalis. Virulence. 2020;11(1):1257–1267. doi:10.1080/21505594.2020.1809329
114. Maikova A, Severinov K, Soutourina O. New insights into functions and possible applications of Clostridium difficile CRISPR-Cas system. Front Microbiol. 2018;9:1740. doi:10.3389/fmicb.2018.01740
115. Uruén C, Chopo-Escuin G, Tommassen J, Mainar-Jaime RC, Arenas J. Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics. 2020;10:1. doi:10.3390/antibiotics10010001
116. É R, Woerther PL, Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann Intensive Care. 2015;5(1):61. doi:10.1186/s13613-015-0061-0
117. Sampson TR, Napier BA, Schroeder MR, et al. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc Natl Acad Sci U S A. 2014;111(30):11163–11168. doi:10.1073/pnas.1323025111
118. Cady KC, O’Toole GA. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol. 2011;193(14):3433–3445. doi:10.1128/JB.01411-10
119. Zegans ME, Wagner JC, Cady KC, et al. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191(1):210–219. doi:10.1128/JB.00797-08
120. Southern SJ, Oyston PCF. Genome editing of Francisella tularensis using (CRISPR-Cas9). J Microbiol Methods. 2020;176:106004. doi:10.1016/j.mimet.2020.106004
121. Schunder E, Rydzewski K, Grunow R, Heuner K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol. 2013;303(2):51–60. doi:10.1016/j.ijmm.2012.11.004
122. Uribe RV, Rathmer C, Jahn LJ, et al. Bacterial resistance to CRISPR-Cas antimicrobials. Sci Rep. 2021;11(1):17267. doi:10.1038/s41598-021-96735-4
123. Shabbir MAB, Shabbir MZ, Wu Q, et al. CRISPR-cas system: biological function in microbes and its use to treat antimicrobial resistant pathogens. Ann Clin Microbiol Antimicrob. 2019;18(1):21. doi:10.1186/s12941-019-0317-x
124. Toro M, Cao G, Ju W, et al. Association of clustered regularly interspaced short palindromic repeat (CRISPR) elements with specific serotypes and virulence potential of shiga toxin-producing Escherichia coli. Appl Environ Microbiol. 2014;80(4):1411–1420. doi:10.1128/AEM.03018-13
125. Touchon M, Charpentier S, Pognard D, et al. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology. 2012;158(Pt 12):2997–3004. doi:10.1099/mic.0.060814-0
126. Xu Z, Li M, Li Y, et al. Native CRISPR-cas-mediated genome editing enables dissecting and sensitizing clinical multidrug-resistant P. aeruginosa. Cell Rep. 2019;29(6):1707–1717.e3. doi:10.1016/j.celrep.2019.10.006
127. McDonald ND, Regmi A, Morreale DP, Borowski JD, Boyd EF. CRISPR-Cas systems are present predominantly on mobile genetic elements in Vibrio species. BMC Genomics. 2019;20(1):105. doi:10.1186/s12864-019-5439-1
128. Shabbir MA, Wu Q, Shabbir MZ, et al. The CRISPR-cas system promotes antimicrobial resistance in Campylobacter jejuni. Future Microbiol. 2018;13:1757–1774. doi:10.2217/fmb-2018-0234
129. Liao W, Liu Y, Chen C, et al. Distribution of CRISPR-Cas systems in clinical carbapenem-resistant Klebsiella pneumoniae strains in a Chinese Tertiary hospital and its potential relationship with virulence. Microbial Drug Resist. 2020;26(6):630–636. doi:10.1089/mdr.2019.0276
130. Hille F, Richter H, Wong SP, et al. The biology of CRISPR-Cas: backward and forward. Cell. 2018;172(6):1239–1259. doi:10.1016/j.cell.2017.11.032
131. Borges AL, Davidson AR, Bondy-Denomy J. The discovery, mechanisms, and evolutionary impact of Anti-CRISPRs. Annu Rev Virol. 2017;4(1):37–59. doi:10.1146/annurev-virology-101416-041616
132. Basgall EM, Goetting SC, Goeckel ME, et al. Gene drive inhibition by the anti-CRISPR proteins AcrIIA2 and AcrIIA4 in Saccharomyces cerevisiae. Microbiology. 2018;164(4):464–474. doi:10.1099/mic.0.000635
133. Chowdhury S, Carter J, Rollins MF, et al. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell. 2017;169(1):47–57.e11. doi:10.1016/j.cell.2017.03.012
134. Guo TW, Bartesaghi A, Yang H, et al. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell. 2017;171(2):414–426.e12. doi:10.1016/j.cell.2017.09.006
135. Harrington LB, Doxzen KW, Ma E, et al. A broad-spectrum inhibitor of CRISPR-Cas9. Cell. 2017;170(6):1224–1233.e15. doi:10.1016/j.cell.2017.07.037
136. Pawluk A, Shah M, Mejdani M, et al. Disabling a Type I-E CRISPR-Cas nuclease with a bacteriophage-encoded Anti-CRISPR protein. mBio. 2017;8(6). doi:10.1128/mBio.01751-17.
137. Bondy-Denomy J, Garcia B, Strum S, et al. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature. 2015;526(7571):136–139. doi:10.1038/nature15254
138. Pawluk A, Staals RH, Taylor C, et al. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat Microbiol. 2016;1(8):16085. doi:10.1038/nmicrobiol.2016.85
139. Mahendra C, Christie KA, Osuna BA, et al. Broad-spectrum anti-CRISPR proteins facilitate horizontal gene transfer. Nat Microbiol. 2020;5(4):620–629. doi:10.1038/s41564-020-0692-2
140. Marino ND, Pinilla-Redondo R, Csörgő B, Bondy-Denomy J. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat Methods. 2020;17(5):471–479. doi:10.1038/s41592-020-0771-6
141. Pawluk A, Bondy-Denomy J, Cheung VH, Maxwell KL, Davidson AR. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio. 2014;5(2):e00896. doi:10.1128/mBio.00896-14
142. Meeske AJ, Jia N, Cassel AK, et al. A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity. Science. 2020;369(6499):54–59. doi:10.1126/science.abb6151
143. Shehreen S, Chyou TY, Fineran PC, Brown CM. Genome-wide correlation analysis suggests different roles of CRISPR-Cas systems in the acquisition of antibiotic resistance genes in diverse species. Philos Trans R Soc Lond B Biol Sci. 2019;374(1772):20180384. doi:10.1098/rstb.2018.0384
144. Stanley SY, Maxwell KL. Phage-Encoded Anti-CRISPR Defenses. Annu Rev Genet. 2018;52(1):445–464. doi:10.1146/annurev-genet-120417-031321
145. Rather IA, Kim BC, Bajpai VK, Park YH. Self-medication and antibiotic resistance: crisis, current challenges, and prevention. Saudi J Biol Sci. 2017;24(4):808–812. doi:10.1016/j.sjbs.2017.01.004
146. Shen S, Loh TJ, Shen H, Zheng X, Shen H. CRISPR as a strong gene editing tool. BMB Rep. 2017;50(1):20–24. doi:10.5483/BMBRep.2017.50.1.128
147. Ding Q, Regan SN, Xia Y, et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12(4):393–394. doi:10.1016/j.stem.2013.03.006
148. Malzahn A, Lowder L, Qi Y. Plant genome editing with TALEN and CRISPR. Cell Biosci. 2017;7(1):21. doi:10.1186/s13578-017-0148-4
149. Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi:10.1126/science.aam9321
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.