Back to Journals » Cancer Management and Research » Volume 11
TCDD-Inducible Poly-ADP-Ribose Polymerase (TIPARP), A Novel Therapeutic Target Of Breast Cancer
Authors Cheng L, Li Z, Huang YZ, Zhang X, Dai XY, Shi L, Xi PW, Wei JF, Ding Q
Received 12 June 2019
Accepted for publication 17 September 2019
Published 18 October 2019 Volume 2019:11 Pages 8991—9004
DOI https://doi.org/10.2147/CMAR.S219289
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Bilikere Dwarakanath
Lin Cheng,1,2,* Zhi Li,1,* Yu-Zhou Huang,1,* Xu Zhang,1 Xin-Yuan Dai,1 Liang Shi,1 Pei-Wen Xi,1 Ji-Fu Wei,3 Qiang Ding1
1Jiangsu Breast Disease Center, The First Affiliated Hospital with Nanjing Medical University, Nanjing 210029, People’s Republic of China; 2Department of Breast Surgery, The Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University, Changzhou 213003, People’s Republic of China; 3Research Division of Clinical Pharmacology, The First Affiliated Hospital with Nanjing Medical University, Nanjing 210029, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qiang Ding
Jiangsu Breast Disease Center, The First Affiliated Hospital with Nanjing Medical University, 300 Guangzhou Road, Nanjing 210029, People’s Republic of China
Tel +86-25-83673123
Email [email protected]
Ji-Fu Wei
Research Division of Clinical Pharmacology, The First Affiliated Hospital with Nanjing Medical University, 300 Guangzhou Road, Nanjing 210029, People’s Republic of China,
Tel +86-25-68306984
Email [email protected]
Background: TIPARP (TCDD-inducible poly-ADP-ribose polymerase), a mono-ADP-ribosyltransferase and a transcriptional repressor of aryl hydrocarbon receptor (AHR), was one of the potential therapeutic targets for human cancers identified by CRISPR–Cas9 screens recently. Studies about TIPARP on cancers are scarce till now, most of which just focus on expressions, while the functions have not been widely reported yet. Moreover, the TIPARP prognostic significance and therapeutic value of breast cancer is also uncertain.
Methods: The present study was performed to comprehensively analyze the expression pattern, prognostic effect, potential therapeutic function of TIPARP in breast cancer by pooling all currently available databases online including Oncomine, UALCAN, bc-GenExMiner, Kaplan–Meier Plotter, COSMIC, UCSC Xena, STRING, DAVID and Comparative Toxicogenomics Database. Further, we also performed several cell biology experiments including RT-qPCR, Western blot and CCK-8 in cellular and clinical sample levels to confirm the conclusions from bioinformatics analysis.
Results: TIPARP was expressed lower in tumor tissues comparing with normal tissues. Meanwhile, several clinical parameters of breast cancer patients were correlated with TIPARP expression. Further, higher TIPARP expression was related to preferable survival. Moreover, the mutations and DNA methylation of TIPARP might contribute to TIPARP dysregulation in breast cancer. Interactors with TIPARP were significantly enriched in telomere maintenance, telomere organization and mainly participated in pathways in cancer. Finally, several common drugs including metformin were observed to up-regulate the expression of TIPARP.
Conclusion: TIPARP might act as a preferable prognostic marker of breast cancer through multiple biological processes such as DNA methylation, mutation as well as pathway related to telomere and so on. TIPARP could be considered as a potential therapeutic target for breast cancer. However, large-scale and comprehensive research is needed to clarify our results.
Keywords: TIPARP, breast cancer, therapeutic target
Introduction
Breast cancer is the most common malignant tumor and remains a major cause of death in women.1 With the development of treatment, including surgery, chemotherapy, radiotherapy, endocrine therapy and target therapy, both disease-free survival and overall survival of breast cancer have been significantly improved. However, after systemic therapies, there are still some patients dying of breast cancer, especially for advanced breast cancer. These patients do not benefit from targeted therapies, partly owing to a limited knowledge of candidate therapeutic targets.2 Therefore, identifying novel and specific therapeutic targets for breast cancer treatment has attracted much attention from worldwide.
Recently, Behan identified 628 new cancer therapeutic targets by performing genome-scale CRISPR-Cas9 screens in 324 human cancer cell lines from 30 cancer types. The framework informed a data-driven list of prioritized therapeutic targets that would be the strong candidates for the development of cancer drugs. It can also inform the initial stages of drug development by contributing to a new, diverse and more effective portfolio of cancer drug targets.3 However, most priority targets need more evidence to support their role as drug targets in cancer treatment.
To further evaluate the priority targets above, we chose TIPARP (TCDD-inducible poly-ADP-ribose polymerase) as a promising drug target in breast cancer and performed confirmatory studies for it. TIPARP is a mono-ADP-ribosyltransferase and a transcriptional repressor of aryl hydrocarbon receptor (AHR), revealing a novel negative feedback loop in AHR signaling.4 TIPARP and AHR have been reported to play an important role in regulating the immune functions, inflammation and gastrointestinal health.5–8 However, the studies about the role of TIPARP in cancers are scarce till now. Most studies just focused on the expression in tumors. For example, AHR signaling pathway genes including TIPARP have been reported to be upregulated in human meningioma.9 TIPARP also showed significantly reduced expression in ovarian cancer cells, compared to normal primary human ovarian surface epithelial cells.10 To our best knowledge, there are extremely few studies about TIPARP on breast cancer. So, the role of TIPARP in breast cancer and the underlying molecular mechanisms responsible for breast cancer development are still unclear. Numerous studies should be performed to investigate the role of TIPARP in breast cancer.
In the present study, we performed a deep bioinformatics analysis of the clinical parameters and survival data as well as biological functions and molecular mechanisms related to TIPARP in breast cancer patients using several online databases. Moreover, we also performed several experiments to confirm the conclusions from bioinformatics analysis in cellular and clinical sample levels.
Materials And Methods
ONCOMINE Data-Mining Analysis
ONCOMINE (www.oncomine.org), an online web-based cancer database for RNA and DNA sequences, is used to facilitate data-mining the transcriptional expressions of genes in 20 types of cancer.11 Transcriptional expression of TIPARP in cancer samples was compared with that in normal individuals using Student’s t-test. Statistically significant values and fold change were demarcated as P-value ≤1E-4 and 2, respectively.
UALCAN
UALCAN (http://ualcan.path.uab.edu/) is a user-friendly, interactive web resource for analyzing transcriptome data of cancers from The Cancer Genome Atlas (TCGA).12 The mRNA expression of TIPARP in breast cancer and normal tissues as well as different stages of breast cancer was detected using the UALCAN web portal. Moreover, the promoter methylation level of TIPARP in breast cancer and normal tissues was obtained from UALCAN.
bc-GenExMiner v4.2
Breast Cancer Gene-Expression Miner v4.2 (bc-GenExMiner v4.2) is a statistical mining tool of incorporates' 264 independent datasets and three classical mining functions: expression, prognosis and correlation.13,14 Expression of TIPARP was analyzed with clinical parameters as well as the relevance with metastatic relapse events.
Kaplan–Meier Plotter
The Kaplan-Meier Plotter (http://kmplot.com/analysis/), a platform containing gene expression information and survival data of 5143 clinical breast cancer patients, was applied to verify the prognostic value of TIPARP gene in overall survival (OS), relapse-free survival (RFS), distant metastasis-free survival (DMFS) and post-progression survival (PPS).15
COSMIC Analysis
The Catalog of Somatic Mutations in Cancer (COSMIC) database (http://www.sange r.ac.uk/cosmic/), the world’s largest and most comprehensive resource for exploring the impact of somatic mutations in human cancer,16 was performed to analyse the mutations of TIPARP. Pie charts were generated for a distribution overview and substitutions on the coding strand in cancer.
UCSC Cancer Genomics Browser Analysis
The heatmap of TIPARP expression and DNA methylation status was identified using data in TCGA breast cancer cohort (TCGA-BRCA) by performing the UCSC (University of California at Santa Cruz) Cancer Genomics Browser (http://xena.ucsc.edu/).17
String
We analyzed the functional protein association network by STRING database (https://string-db. org/).18 Protein name: TIPARP and organism: homo sapiens were analyzed.
David
The Database for Annotation, Visualization and Integrated Discovery (DAVID)(https://david.ncifcrf.gov/home.jsp) provides a set of complete annotation tools to help us understand the biological significance of a large number of genes and classifies gene function and analyses the functional annotation clustering and functional annotation table for a list of given genes.19 TIPARP-related genes were submitted to DAVID for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Visualization of the results of GO and KEGG analysis was done using ggplot2 package of R software.
Comparative Toxicogenomics Database
The Comparative Toxicogenomics Database (CTD, http://ctdbase.org/) is a robust, publicly available database for toxicogenomic information. It provides manually curated key information about chemical-gene/protein interactions, chemical-disease and gene-disease relationships, from peer-reviewed scientific literatures.20 Given a target gene, CTD can provide the corresponding chemicals sorted by the interactions between them. Candidate treatments for regulating the expression of TIPARP were predicted using CTD with default parameters.
Breast Tissue Samples
30 pairs of breast tissue samples used in real-time quantitative PCR (RT-qPCR) were obtained from the First Affiliated Hospital of Nanjing Medical University, China, between 2014 and 2016. The collection and use of the samples was reviewed and approved by the Institutional Ethics Committee of the First Affiliated Hospital of Nanjing Medical University. All patients provided written informed consent, and that this was conducted in accordance with the Declaration of Helsinki.
RNA Isolation And RT-qPCR Analysis
Total RNA was isolated using Trizol reagent (TaKaRa, Japan), and 1000 ng RNA was reverse-transcribed into cDNA using Primescript RT Reagent (TaKaRa, Japan). The RT-PCR was performed using FastStart Universal SYBR Green Master (Roche, Switzerland) in a RT-PCR instrument (Applied Biosystems, USA), and β-Actin was used as endogenous control. The following PCR primers were used:
TIPARP forward, 5′-TGCACCCAGTTTCAAGTGAT3-′
TIPARP reverse, 5′- GTTCACCAGCTCAAACACGA3-′
β-Actin forward, 5′-GCTGTGCT ATCCCTGTACGC3-′
β-Actin reverse, 5′-TGCCTCAGGGCAGCGGAACC3-′
Cell Culture
The human breast cancer cell lines BT474, MDA-MB-231, T47D, MDA-MB-453, SUM1315, MCF-7 and normal mammary epithelial cell line MCF-10A were obtained from American Type Culture Collection (Manassas, USA) and incubated in a humidified atmosphere of 5% CO2 in air at 37°C and fed with the culture medium of High glucose Dulbecco’s Modified Eagle Medium, supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin solution. For routine passages, cultures were split 1:3 when they reached 80–90% confluence generally every 2–3 days. All experiments were performed on exponentially growing cells.
CCK-8 Assay
We assessed cell viability through cell counting kit-8 assay (CCK-8) according to the manufacturer’s protocol (Dojindo, Japan). Briefly, 5 × 103 cells were seeded into a 96-well plate in triplicate and 8 hrs later, Metformin (Sigma, USA) was added into the wells at the indicated final concentrations (0, 1, 5, 10, 20 and 40 μM). After incubation with metformin for 24 hrs, 48 hrs and 72 hrs, we added the CCK-8 solution (10 μL) into each well, and the plates were incubated for 2 hrs at 37°C. The absorbance of cells at 450 nm (OD450) was measured in a microplate reader (Bio-Rad, USA).
Western Blot
Tissues and cells treated with/without metformin were lysed by RIPA buffer containing protease inhibitors (Sigma, USA). Protein extractions were separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, USA). After incubated with the anti-TIPARP antibody (1:1000, Abcam, USA) and anti-β-Actin antibody (1:1000, Cell Signaling Technology, USA), the membranes were then incubated with peroxidase (HRP)-conjugated secondary antibody (1:1000, Cell Signaling Technology, USA). After washes, signals were detected using a chemiluminescence system (Bio-Rad, USA) and analyzed using Image Lab Software.
Statistical Analysis
Experiments were repeated in triplicate, unless otherwise specified. The data were analyzed using the SPSS 20.0 software (Chicago, USA). We analyzed the statistical significance of the differences between groups using Students t-test and oneway ANOVA followed by Bonferroni test, and a statistically significant difference was considered at the level of P < 0.05.
Results
Reduced Expression Of TIPARP In Breast Cancer Tissues
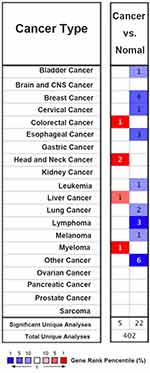
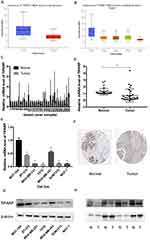
Firstly, the expression of TIPARP in 20 types of cancer was measured and compared to normal tissues using the Oncomine online database (Figure 1). We found that decreased TIPARP (blue) was observed in bladder cancer, cervical cancer, esophageal cancer, leukemia, lung cancer, lymphoma, melanoma and especially breast cancer. Whereas, increased level of TIPARP (red) was observed in colorectal cancer, head and neck cancer, liver cancer and myeloma. Consistently, using UALCAN website, we also found that lower TIPARP was expressed in breast cancer tissues than in normal tissues (Figure 2A, P<0.05). Next, we focused on whether mRNA expression of TIPARP was related to cancer stage in individual patients. As shown in Figure 2B, the results indicated that patients with a more advanced stage of breast cancer tended to express lower levels of TIPARP.
To verify the results above, we further compared the TIPARP expression in breast cancer tissues and adjacent normal tissues of patients in our hospital and found that TIPARP was expressed lower in breast cancer tissues both in mRNA level (Figure 2C, D) and protein level (Figure 2H), consistent with the results from databases (Figure 2C, D, P<0.05). The mRNA expression of TIPARP was also lower in breast cancer cells compared to normal breast cell line MCF-10A both in mRNA level (Figure 2E) and protein level (Figure 2G). Immunohistochemistry from the Human Protein Atlas database confirmed the lower expression of TIPARP in breast cancer in protein level (Figure 2F).
TIPARP Expression And Clinical Parameters Of Breast Cancer Patients
Using bc-GenExMiner v4.2 software, we implemented Welch’s test to compare the abnormal expression of TIPARP among different groups of patients according to clinical pathological features. For age criteria, TIPARP was significantly elevated in ≤51-year group with respect to >51-year group (Figure 3A). As we know, the Scarff, Bloom & Richardson (SBR) is a histological grade that evaluates tubule formation, nuclear characteristics of pleomorphism and mitotic index.21 Based on tumor size, lymph node stage and tumor grade, the Nottingham Prognostic Index (NPI) is used to stratify patients into additional prognostic groups. The SBR grade and NPI index are two wonderful prognostic model for breast cancer.22 More advanced SBR grade and NPI index were associated with lower TIPARP level (Figure 3B and C). ER-positive or PR-positive breast cancer patients tended to express higher TIPARP gene compared with ER-negative or PR-negative patients (Figure 3D and E). Regarding nodal status and HER-2 status, there was no significant difference between positive and negative group (Figure 3F and G). Moreover, TIPARP was significantly increased in non-triple-negative and non-basal-like breast cancer patients compared to triple-negative and basal-like breast cancer patients (Figure 3H and I).
TIPARP Expression And Survival Data Of Breast Cancer Patients
We then investigated the prognostic value of TIPARP. The Kaplan–Meier curves indicated that higher level of TIPARP correlated with preferable RFS, OS, DMFS and PPS (Figure 4A–D). To confirm the prognostic value, we also obtained data about metastatic relapse-free survival of TIPARP from bc-GenExMiner and found that patients with decreased TIPARP expression presented worse survival (Figure 4E). The pooled results revealed that low TIPARP expression was correlated with longer survival.
TIPARP Mutation In Human Cancer
The pie chart including the information of mutations of nonsense substitution, missense substitution, inframe insertion, synonymous substitution, frameshift insertion, frameshift deletion and inframe deletion was generated by COSMIC. Missense substitution rate was 73% while synonymous substitution rate was 18% of mutant samples of human cancer (Figure 5A). In the study, the profile of mutations was displayed using the six substitution subtypes: C>A, C>G, C>T, T>A, T>C and T>G. Respectively, human cancer had 44% C > T, 22% T > C and 15% C > A mutation in TIPARP coding strand (Figure 5B). Moreover, the pie charts of mutation types for mutational signatures of human cancers were displayed according to the data from COSMIC. We also drew the pie charts of mutation types identified from 53 treatments according to data from an excellent research published in Cell recently.23 Interestingly, comparing with other treatments, semustine resulted in 52% C > T, 13% T > C and 13% C > A mutation, which was also similar to the mutation of TIPARP (Figure 5C, the data for other treatments are not shown).
TIPARP DNA Methylation Status In Human Breast Cancer
Heatmap of TIPARP expression and DNA methylation status revealed that its expression might be negatively related with DNA methylation in breast cancer (Figure 6A). Consistently, using UALCAN website, we also found that higher promoter methylation level of TIPARP was expressed in breast cancer tissues than in normal tissues (Figure 6B, P<0.05).
TIPARP Related Protein–Protein Interaction Analysis
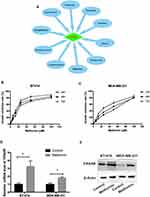
STRING is a database known to be used to predict protein–protein interactions. The co-expression analysis revealed that TIPARP was directly co-expressed with 10 interactors including poly(ADP-ribose) polymerase family member 16 (PARP 16), PIN2/TERF1-interacting telomerase inhibitor 1 (PINX1), intracellular hyaluronan-binding protein 4 (HABP4), cytochrome P450 1A1 (CYP1A1), poly [ADP-ribose] polymerase 3 (PARP3), cytochrome P450 1B1 (CYP1B1), immunoglobulin superfamily DCC subclass member 3 (IGDCC3), aryl hydrocarbon receptor (AHR), poly(ADP-ribose) polymerase family member 6 (PARP 6) and indolethylamine N-methyltransferase (INMT). Among the directly co-expressed 10 interactors, some have been reported to play an important role in breast cancer, such as PINX124 and PARP3.25 Meanwhile, TIPARP interacted indirectly with other 11 proteins (Figure 7A).
 |
Figure 7 TIPARP-related protein–protein interaction analysis. (A) PPI network of TIPARP. (B) GO analysis of interactors of TIPARP. (C) KEGG analysis of interactors of TIPARP. |
GO Functional And Pathway Enrichment Analysis
To determine the function and mechanisms of these 21 interactors mentioned above, all of them were submitted to the DAVID for GO analysis and KEGG pathway analysis. We found that the interactors were significantly enriched in response to xenobiotic stimulus, telomere maintenance, telomere organization and response to organic substance and so on (Figure 7B). According to the KEGG pathway enrichment analysis, the 21 related genes mainly participated in pathways in cancer, tryptophan metabolism and steroid hormone biosynthesis and metabolism of xenobiotics by cytochrome P450 (Figure 7C).
TIPARP Acting As A Target Of Several Drugs
Using CTD, we found a list of chemicals that interacted with TIPARP. From the 151 chemicals, we picked 10 “old” drugs used commonly for treatment of several diseases, including breast cancer, diabetes, bacterial infection and so on. As shown in Figure 8A, the 10 drugs including tamoxifen, raloxifene, estradiol, ethanol, gentamicins, metformin, acetaminophen, zoledronic acid, rosiglitazone and testosterone have been demonstrated to improve the mRNA expression of TIPARP. To substantiate the results, we investigated the effect of metformin on TIPARP. Metformin showed growth inhibition in BT-474 and MDA-MB-231 breast cancer cell lines in both dose-dependent and time-dependent manners (Figure 8B and C). After exposure to 10 μM metformin for 48 hrs, metformin increased the mRNA and protein expression of TIPARP (Figure 8D and E), consist with the result from CTD database.
Discussion
Despite the development in early clinical diagnosis and treatment strategies, breast cancer remains a significant global burden.26 Therefore, the development of more effective drugs for breast cancer patients is urgently needed. As is well known, identifying novel drug targets according to the molecular features of tumors could expand the range of drugs and then improve the success rates of new cancer therapies for breast cancer.27 In the present study, we performed a comprehensive bioinformatics analysis as well as several experiments to investigate the potential therapeutic role of TIPARP in breast cancer.
TIPARP expression decreased in breast cancer significantly and a more advanced stage of breast cancer tended to express lower levels of TIPARP. Consistently, TIPARP also expressed lower in breast cancer tissues, compared to adjacent normal tissues of patients from our hospital, confirming the results from databases online. Meanwhile, ER and PR status were found to be positively correlated with TIPARP expression. Conversely, basal-like status, TNBC status, SBR grade and NPI index were negatively correlated with TIPARP expression. It is generally known that breast cancer patients with ER- or PR-positive, non-basal-like or non-triple-negative status have a preferable outcome.28 We further investigated the prognostic value of TIPARP in breast cancer using Kaplan–Meier plotter and bc-GenExMiner tool, and the pooled results revealed that low TIPARP expression was correlated with longer OS, RFS, DMFS and PPS. Together, as there is no research about the role of TIPARP in breast cancer, it is the first time to report that TIPARP might act as a preferable prognostic marker of breast cancer.
Then, we tried to investigate the mechanisms of TIPARP dysregulation in breast cancer. Through examining its DNA methylation status in TCGA, we observed a negative correlation between methylation status and TIPARP expression. These findings suggested that DNA methylation might be an important mechanism of TIPARP dysregulation in breast cancer, as the same as many other biomarkers, for instance, MEG3 expression gradually decreased with increasing DNA methylation in breast cancer.29
To explore the mechanisms of TIPARP acting as a preferable prognostic marker in breast cancer, we further analyzed the mutation of TIPARP using COSMIC and performed PPI analysis. In TIPARP coding strand, human cancer had 44% C > T, 22% T > C and 15% C > A mutation, respectively. This shared a similar pattern with mutation types identified from semustine according to Kucab.23 Notably, the mutation types of TIPARP were quite different from those identified from common environmental agents such as radiation, aldehydes and so on, suggesting that the mutation of TIPARP was not caused by single environmental agents. Semustine was known as a drug for treatment of brain, gastrointestinal cancers and melanomas.30 The results above suggested that exposure to semustine for a long time might induce the mutation of TIPARP, therefore promote the development of breast cancer. Next, PPI as well as GO and KEGG analysis demonstrated that interactors with TIPARP significantly enriched in telomere maintenance, telomere organization and mainly participated in pathways in cancer. As we know, telomere plays an important role in regulating cell growth and division. And telomerase is overexpressed in 80–95% of cancers and likely to participate in cell malignant transformation.31 TIPARP dysregulation may play an important role in breast cancer through the pathway related to telomere.
Finally, we investigated the role of TIPARP acting as a potential therapeutic target of breast cancer. Several common drugs could up-regulate the expression of TIPARP, one of which was metformin. Metformin is the first-line therapy for type 2 diabetes mellitus and one of the most widely prescribed oral antidiabetic medications, having been widely used for over 40 years. In recent years, several studies suggest a potential role for metformin in anti-cancer therapy including breast cancer.32,33 Our observations might provide a new potential mechanism for metformin treating breast cancer.
In conclusion, over the past several decades, much research has focused on identifying new prognostic markers and therapeutic targets in order to make better clinical decisions and improve therapy and outcomes of breast cancer. The present study was performed to comprehensively analyze the expression pattern, prognostic effect and potential therapeutic function of TIPARP in breast cancer by pooling all currently available data online as well as performing experiments. TIPARP might act as a preferable prognostic marker of breast cancer through multiple biological processes such as DNA methylation, mutation as well as pathway related to telomere and so on. More in-depth experiments are needed to validate the value of TIPARP acting as a therapeutic target of breast cancer.
Abbreviations
TIPARP, TCDD-Inducible Poly-ADP-Ribose Polymerase; AHR, Aryl hydrocarbon receptor; ER, Estrogen receptor; PR, Progesterone receptor; AR, Androgen receptor; HER-2, Epidermal growth factor receptor-2; SBR, Scarff-Bloom-Richardson; NPI, Nottingham prognostic index; RFS, Relapse-free survival; OS, Overall survival; DMFS, Distant metastasis-free survival; PPS, Post-progression survival; TCGA, The Cancer Genome Atlas; PPI, Protein–Protein interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Acknowledgments
This work was financially supported by Natural Science Foundation of China (81502294,81572595) and Changzhou Science and Technology Program (CJ20159044).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Fan L, Strasser-Weippl K, Li -J-J, et al. Breast cancer in China. Lancet Oncol. 2014;15(7):e279–e289. doi:10.1016/S1470-2045(13)70567-9
2. Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi:10.1038/nm.4333
3. Behan FM, Iorio F, Picco G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568(7753):511–516. doi:10.1038/s41586-019-1103-9
4. MacPherson L, Tamblyn L, Rajendra S, Bralha F, McPherson JP, Matthews J. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 2013;41(3):1604–1621. doi:10.1093/nar/gks1337
5. Marshall NB, Kerkvliet NI. Dioxin and immune regulation: emerging role of aryl hydrocarbon receptor in the generation of regulatory T cells. Ann N Y Acad Sci. 2010;1183:25–37. doi:10.1111/j.1749-6632.2009.05125.x
6. Stevens EA, Mezrich JD, Bradfield CA. The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology. 2009;127(3):299–311. doi:10.1111/j.1365-2567.2009.03054.x
7. Busbee PB, Rouse M, Nagarkatti M, Nagarkatti PS. Use of natural AhR ligands as potential therapeutic modalities against inflammatory disorders. Nutr Rev. 2013;71(6):353–369. doi:10.1111/nure.12024
8. Esser C. Biology and function of the aryl hydrocarbon receptor: report of an international and interdisciplinary conference. Arch Toxicol. 2012;86(8):1323–1329. doi:10.1007/s00204-012-0818-2
9. Talari NK, Panigrahi MK, Madigubba S, Phanithi PB. Overexpression of aryl hydrocarbon receptor (AHR) signalling pathway in human meningioma. J Neurooncol. 2018;137(2):241–248. doi:10.1007/s11060-017-2730-3
10. Goode EL, Chenevix-Trench G, Song H, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42(10):874–879. doi:10.1038/ng.668
11. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia (New York, NY). 2004;6(1):1–6. doi:10.1016/S1476-5586(04)80047-2
12. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
13. Jezequel P, Campone M, Gouraud W, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131(3):765–775. doi:10.1007/s10549-011-1457-7
14. Jezequel P, Frenel JS, Campion L, et al. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database (Oxford). 2013;2013:bas060. doi:10.1093/database/bat081
15. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi:10.1007/s10549-009-0674-9
16. Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. doi:10.1093/nar/gku1075
17. Haeussler M, Zweig AS, Tyner C, et al. The UCSC genome browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):D853–d858. doi:10.1093/nar/gky1095
18. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi:10.1093/nar/gku1003
19. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
20. Davis AP, Grondin CJ, Johnson RJ, et al. The comparative toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47(D1):D948–d954. doi:10.1093/nar/gky868
21. Bansal C, Pujani M, Sharma KL, Srivastava AN, Singh US. Grading systems in the cytological diagnosis of breast cancer: a review. J Cancer Res Ther. 2014;10(4):839–845. doi:10.4103/0973-1482.140979
22. Lee AH, Ellis IO. The Nottingham prognostic index for invasive carcinoma of the breast. Pathol Oncol Res. 2008;14(2):113–115. doi:10.1007/s12253-008-9067-3
23. Kucab JE, Zou X, Morganella S, et al. A compendium of mutational signatures of environmental agents. Cell. 2019;177(4):821–836.e816. doi:10.1016/j.cell.2019.03.001
24. Shi M, Cao M, Song J, et al. PinX1 inhibits the invasion and metastasis of human breast cancer via suppressing NF-kappaB/MMP-9 signaling pathway. Mol Cancer. 2015;14:66. doi:10.1186/s12943-014-0278-9
25. Beck C, Rodriguez-Vargas JM, Boehler C, et al. PARP3, a new therapeutic target to alter Rictor/mTORC2 signaling and tumor progression in BRCA1-associated cancers. Cell Death Differ. 2019;26(9):1615–1630. doi:10.1038/s41418-018-0233-1
26. Duffy MJ, Walsh S, McDermott EW, Crown J. Biomarkers in breast cancer: where are we and where are we going? Adv Clin Chem. 2015;71:1–23. doi:10.1016/bs.acc.2015.05.001
27. Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31(15):1806–1814. doi:10.1200/JCO.2012.46.8934
28. Bagaria SP, Ray PS, Sim MS, et al. Personalizing breast cancer staging by the inclusion of ER, PR, and HER2. JAMA Surg. 2014;149(2):125–129. doi:10.1001/jamasurg.2013.3181
29. Cui X, Yi Q, Jing X, et al. Mining prognostic significance of MEG3 in human breast cancer using bioinformatics analysis. Cell Physiol Biochem. 2018;50(1):41–51. doi:10.1159/000493956
30. Agarwal S, Chadha D, Mehrotra R. Molecular modeling and spectroscopic studies of semustine binding with DNA and its comparison with lomustine-DNA adduct formation. J Biomol Struct Dyn. 2015;33(8):1653–1668. doi:10.1080/07391102.2014.968874
31. Ruden M, Puri N. Novel anticancer therapeutics targeting telomerase. Cancer Treat Rev. 2013;39(5):444–456. doi:10.1016/j.ctrv.2012.06.007
32. Chae YK, Arya A, Malecek MK, et al. Repurposing metformin for cancer treatment: current clinical studies. Oncotarget. 2016;7(26):40767–40780. doi:10.18632/oncotarget.8194
33. Jara JA, Lopez-Munoz R. Metformin and cancer: between the bioenergetic disturbances and the antifolate activity. Pharmacol Res. 2015;101:102–108. doi:10.1016/j.phrs.2015.06.014
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.