Back to Journals » Cancer Management and Research » Volume 11
T-lymphoid/myeloid mixed phenotype acute leukemia and early T-cell precursor lymphoblastic leukemia similarities with NOTCH1 mutation as a good prognostic factor
Authors Noronha EP , Marques LVC, Andrade FG , Sardou-Cezar I , dos Santos-Bueno FV, Zampier CDP , Terra-Granado E, Pombo-de-Oliveira MS
Received 1 December 2018
Accepted for publication 6 March 2019
Published 2 May 2019 Volume 2019:11 Pages 3933—3943
DOI https://doi.org/10.2147/CMAR.S196574
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Ahmet Emre Eşkazan
Elda Pereira Noronha, Luísa Vieira Codeço Marques, Francianne Gomes Andrade, Ingrid Sardou-Cezar, Filipe Vicente dos Santos-Bueno, Carolina Da Paz Zampier, Eugênia Terra-Granado, Maria S Pombo-de-Oliveira
Paediatric Haematology-Oncology Program, Research Centre, Instituto Nacional de Câncer, Rio de Janeiro, RJ, Brazil
Purpose: T-lymphoid/Myeloid Mixed phenotype acute leukemia (T/M-MPAL) is ambiguous leukemia which overlaps with early T-cell precursor lymphoblastic leukemia (ETP-ALL). We have revisited the immunophenotyping profile of T/M-MPAL and ETP-ALL to identify differences and/or similarities, as these entities represent a therapeutic challenge in clinical practice.
Patients and methods: A total of 26 ETP-ALL and 10 T/M-MPAL cases were identified among 857 cases of childhood leukemia (T-ALL, n=266 and AML, n=591) before any treatment decisions. The variables analyzed were age strata, sex, clinical features, immunophenotyping, and molecular aberrations. Immunophenotyping was performed in all samples using a panel of cytoplasm and membrane antibodies to identify the lineage and blast differentiation. The mutational status of STIL-TAL1, TLX3, RUNX1, NOTCH1, FBXW7, FLT3, IL7R, RAS, KTM2A, and CDKN2A/B was tested using RT-PCR, FISH, and PCR sequencing methods. The outcomes were assessed in terms of overall survival (OS).
Results: The immunophenotypes were similar in ETP-ALL and T/M-MPAL, regarding the cellular expression of CD34, CD117, CD13/CD33, and CD11b, although CD2 and HLA-DR were more frequent in T/M-MPAL (p<0.01). aMPO positivity and myelomonocyte differentiation were definitive in separating both entities. NOTCH1, FLT3-ITD, and N/KRAS mutations as well as TLX3 and KMT2A rearrangements were found in both ETP-ALL and T/M-MPAL. Thirty-one patients received ALL protocol whereas five had AML therapy. The overall 5-year survival rate (pOS) was 56.4% for patients treated using ALL protocols. No differences were observed between T/M-MPAL (pOS of 57%) and ETP-ALL (pOS of 56%) patients. The prognostic value of NOTCH1mut, was associated with significantly better OS (pOS 90%) than NOTCH1wt, (pOS 37%) (p=0.017).
Conclusion: This research can potentially contribute to NOTCH1 as targeted therapy and prognostic assessment of T-cell mixed phenotype leukemia.
Keywords: T-lymphoid/myeloid mixed phenotype acute leukemia, early T-cell precursor lymphoblastic leukemia, NOTCH1 mutations
Introduction
Acute leukemia (AL) are heterogeneous diseases in which the precise diagnosis characterization of either B-, T-lymphoid or and myeloid lineage is essential to treatment strategy and outcome success. Since the advent of integrative diagnosis of leukemia applying broad cell immunophenotype, karyotype and molecular profiles, unusual AL cases with mixed phenotype have been described.1
In the recent World Health Organization (WHO) classification, T/myeloid mixed phenotype acute leukemia (T/M-MPAL) and B/myeloid mixed phenotype acute leukemia (B/M-MPAL) are distinct entities within the same category of leukemia of ambiguous cell lineage. Even with myeloid markers, the early-T-cell precursor acute lymphoblastic leukemia (ETP-ALL) is not classified by the WHO as leukemia of ambiguous lineage.2 ETP-ALL is classified under T-ALL groups with the strong argument that the behavior of ETP-ALL is like other T-ALL in the most recent children’s oncology group trials and in some adult studies.3,4 Recently, new leukemia of ambiguous cell lineage has been identified. It has been proposed that acute myeloid/T-lymphoblastic leukemia, a specific subset of minimally differentiated myeloid leukemia with T-cell receptor gene rearrangements (TCR-r), should be included in the WHO classification as a diagnostic entity with a distinct molecular profile compared to ETP-ALL and T/M-MPAL.5
In clinical practice, T/M-MPAL and ETP-ALL cases remain a therapeutic challenge. The genomic profiles of ETP-ALL and T/M-MPAL have demonstrated that both diseases might have gene aberrations with potential to target therapy.6,7 It is important to highlight that B/M-MPAL, when associated with BCR-ABL1 and KMT2A/AFF1 genetic abnormalities, determines the treatment strategy.2 The aim of this study was to compare the similarities in immunophenotyping, molecular aberrations, and outcomes in a series of ETP-ALL and T/M-MPAL cases and validate an algorithm of molecular tests at diagnosis to drive prognostic assessment and therapeutic decisions.
Patients and methods
Patients
Diagnostic tests obtained from bone marrow (BM) aspirates and/or peripheral blood samples sent to the Pediatric Haematology–Oncology Program, Research Centre, Instituto Nacional de Cancer, Rio de Janeiro, Brazil, before any treatment decisions, were revisited (2005–2017). Forty-four cases involving AL with an ambiguous phenotype were selected among 857 cases of childhood leukemia (only T-ALL, n=266 and AML, n=591 were included), in which immunophenotyping and molecular tests were performed. The exclusion criteria were B-cell precursor ALL and cases lacking the complete panel of monoclonal antibodies to compare ETP-ALL and T/M-MPAL. Cases of AL with BCR-ABL1 were also excluded.
The European Group for the Immunological Classification of Leukemias criterion8 was used from 2005 to 2010, and all cases with an ambiguous phenotype were reviewed using the WHO classification.2 The definition of ETP-ALL was based on criteria described by Coustan-Smith et al, (2009)9 in all incident T-ALL cases with ambiguous phenotype since the year 2010. The scoring system proposed by Inukai et al, (2012)10 was applied in all cases reviewed in this study. Minimally differentiated AML (AML-M0) with CD7 positivity without TCR rearrangements was included only for comparisons with immunophenotyping. The referring physicians provided an update of the patients’ follow-up.
Mutliparametric flow cytometry
Multiparametric flow cytometry (M-FCM) tests were performed in all samples. The first screening panel was cytoplasmic (cy) CD3, cyCD79/CD22, aMPO, CD7, and CD19 antibodies (MoAb) to identify the leukemia lineage. Then, in samples positive for cyCD3 (T-cell lineage), CD7 and aMPO (Myeloid lineage), a panel of MoAb with T-cell, myeloid and stem cell-associated membrane antigens was applied. The six-color conjugated-MoAbs were performed as described in the supplemental material (Table S1). Samples were acquired in a FACS Canto Flow cytometry (Becton, Dickinson and Company, CA, USA). Data analyses were performed by Infinicyt version 1.8 (Cytognos, Salamanca, Spain) Software.
The analyses were first evaluated in a blast gate identified by CD45low/intermediate expression, then, followed by intracytoplasmatic and membrane analysis of blast gate according to MoAb panel (Figure S1).
Molecular tests
Samples were first subjected to DNA and RNA extraction using commercial kits. Genomic DNA and total RNA were isolated from mononuclear cells from BM samples with the QIAamp® DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany) or the TRIzolTM reagent kit (Invitrogen, Carlsbad, USA), respectively; 2–2.5 μg of RNA were reverse transcribed using First-Strand cDNA Synthesis KitTM (Amersham Pharmacia Biotech Inc., NJ, USA). The integrity of RNA and cDNA was examined by amplifying a fragment of the GAPDH gene.
The STIL-TAL1, RUNX1-RUNX1T1, CBFβ-MYH11, BCR-ABL1, and the presence of the TLX3 transcript were assessed by the reverse transcriptase-polymerase chain reaction technique using primers and conditions as previously described.11,12 KMT2A rearrangements (KMT2A-r) including fusion partners AFF1 (AF4), MLLT3 (AF9), MLLT10 (AF10), MLLT1 (ENL), and MLLT4 (AF6), were identified by RT-PCR according to previous publications.13 Additional fluorescence in situ hybridization with commercial DNA probes (MLL Dual Color Break Apart Rearrangement Probe; Cytocell, UK) was performed to identify KMT2A-r before RT-PCR to identify selected partner genes.
Mutational status of NOTCH1, FBXW7, FLT3, IL7R, RAS, and CDKN2A/B deletions was tested throughout direct sequencing according to methods previously described.14–16
Treatments
Patients were not enrolled in any clinical trials, but received treatments either according to the Brazilian Group for Treatment of Childhood Lymphoblastic Leukemia (GBTLI-ALL 99/09) or to ALL Berlin-Frankfurt- Münster (BFM-ALL 95 and 2002) protocols.17,18 Patients were assigned into the group of a high risk of relapse, according to age strata, leukocyte count (>50×109/L), and T-cell phenotype of both protocols. In summary, all patients received a 4-week induction phase, which included prednisone, vincristine, doxorubicin, L-asparaginase, intrathecal Methotrexate (MTX), Cytarabine (Ara-C), and dexamethasone (MADIT). In the GBTLI-ALL, the consolidation phase, Cyclophosphamide, Ara-C, and 6-mercaptopurine (6-MP) were included. Intensification treatment with MTX 2 g/m2 ×4, 6-MP and MADIT and maintenance of 6-MP, MTX, vincristine, dexamethasone, and MADIT. Patients with more than 5% blast cells in their cerebrospinal fluid after day 14 of induction therapy have received cranial radiotherapy (1200 Gy during the late consolidation phase).18
Statistical analysis
To compare the distribution of categorical variables in leukemia subtypes with the Pearson Chi-square test. When the expected count in at least one cell of the table was less than five, the Fisher’s exact test (two-tailed) was used. Cases were grouped in T/M-MPAL and ETP-ALL, and variables analyzed were age range, sex, clinical features, immunophenotyping, and molecular markers.
Overall survival (OS) was measured from the date of diagnosis to the date of last follow-up or death from any cause. Patients who did not experience an event and/or lost to follow-up were censored. The Kaplan–Meier survival analysis method was used to calculate the 5-year OS. The estimated survival values in ETP-ALL and T/M-MPAL were compared using the log-rank test. p-values <0.05 were considered statistically significant.
All analyses were performed using SPSS 21.0 (SPSS, Chicago, IL, USA, 2004).
Results
The demographic and clinical-biological features of ETP-ALL and T/M-MPAL can be seen in Table 1. Forty-four selected cases were characterized as ETP-ALL (n=26), T/M-MPAL (n=10), and AML-M0 with CD7 positivity (n=8), with equal frequency distribution of variables in each group. Five cases were treated in accordance with the AML BFM-2004 protocol (ETP-ALL, n=2; T/M-MPAL, n=3) and 31 cases were treated using ALL high-risk protocols. The AML-M0 cases were CD34/CD7pos and negative for CD41/CD61, cyCD3, and mCD3, in contrast to the ETP-ALL and T/M-MPAL cases (Table S2). AML-M0 cases positive for CD7 were included in comparisons between ETP-ALL and T/M-MPAL immunophenotype profiles only. AML-M0 positive for CD7 were cCD3 negative and TCR, NOTCH1, FLT3, and RAS wild-type.
 | Table 1 Demographic and main clinical-laboratorial features of early T-cell precursor-ALL, T-lymphoid/myeloid mixed-phenotype acute leukemia, 2005–2017, Brazil |
The summary of the immunophenotyping and molecular analysis results for ETP-ALL and T/M-MPAL cases (n=36) is shown in Figure 1. Detailed M-FCM results for aberrant phenotypes are shown in the supplemental material (Table S2). CD34, CD117, CD13/CD33, and CD11b had similar frequency distributions in both groups, whereas CD2 and HLA-DR were more frequent in the T/M-MPAL group (p<0.01); cyCD3 was present in more than 30% of blast cells, and aMPO was found in more than 20% in T/M-MPAL. Seventeen ETP-ALL cases were previously diagnosed using the criteria initially suggested by Coustan-Smith et al,9 and nine additional cases were identified by applying the scoring system proposed by Inukai et al.10 Four cases (#29, #33, #34, #36), classified as T/M-MPAL, had scores that would classify them as ETP-ALL. In these cases, aMPO expression and/or blast cells with myelomonocyte differentiation markers were used to distinguish between T/M-MPAL and ETP-ALL, an approach that enhances the guidelines provided in the WHO criteria for MPAL.
In the current study, NOTCH1, FLT3-ITD, and N/KRAS mutations as well as TLX3 and KMT2A rearrangements were found in both ETP-ALL and T/M-MPAL cases, whereas FBXW7mut, CDKN2ABdel, and STIL-TAL1 were only found in ETP-ALL cases (Figure 1). KMT2A-r was present in 4/29 cases (13.8%), with two MLLT4 (AF6) and one MLLT3 (AF9) partner genes among the ETP-ALL cases and one MLLT1 (ENL) partner gene in the T/M-MPAL case. In our cohort, NOTCH1mut was the most frequent molecular aberration found in cases with an ambiguous phenotype (ETP-ALL, 38.5%; T/M-MPAL, 20%; p=0.438), following FLT3mut (ETP-ALL, 23.1%; T/M-MPAL, 10%; p=0.645) and RASmut (ETP-ALL, 11.5%; T/M-MPAL, 30%; p=0.317).
There were no differences in patient characteristics such as age strata, sex, white blood cell count (WBC) at diagnosis and molecular alteration, with the exception of the NOTCH1 mutation (p=0.017) as shown in Table 2. Regarding treatment regimen given, 5 out of 36 patients were treated using AML protocols (two ETP-ALL, three T/M-MPAL patients) and they were excluded from the survival analysis by protocol treatment due to small sample size. In four cases (one ETP-ALL and three T/M-MPAL), complete remission was not achieved and the disease progressed until the patients’ death; one ETP-ALL (#2) was alive in the follow-up after undergoing allogeneic hematopoietic stem cell transplantation (alloHSCT). Thirty-one patients were treated initially with ALL protocols (ETP-ALL, n=24; T/M-MPAL, n=7). The overall 5-year survival rate of the 36 patients was 52.3%, mean/months was 37.7±4.54 and confidence intervals of 28.8–46.6 (Table 2). Eight patients underwent alloHSCT, being ETP-ALL (n=6) and T/M-MPAL (n=2) in the first relapse. Death occurred for six patients (ETP-ALL (n=5) and T/M-MPAL (n=1) after alloHSCT due to relapse (n=2) and toxicity (n=4). Data are summarized in Figure 1.
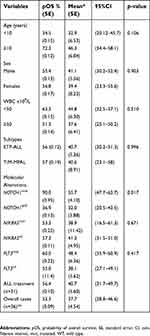 | Table 2 Overall survival of the T-lymphoid/myeloid mixed phenotype acute leukemia and early T-cell precursor lymphoblastic leukemia, 2005–2017, Brazil |
The overall 5-year survival rate was 56.4% for 31 patients treated using ALL protocols. No differences were observed between T/M-MPAL (pOS of 57%) and ETP-ALL (pOS of 56%) patients (Figure 2A). As NOTCH1mut was the most frequent molecular aberration in the cohort and because the treatment administered was approximately the same in most cases, the prognostic value of NOTCH1 status was tested. NOTCH1mut was associated with significantly better OS (pOS of 90%) compared to NOTCH1wt (pOS of 37%) (p=0.017) (Figure 2B).
Discussion
ETP-ALL and T/M-MPAL are closely related entities when it comes to immunophenotyping, genomic alterations, and outcomes.19 Therefore, these entities are a diagnostic and therapeutic challenge owing to their heterogeneity with ambiguous phenotypes and overlapping genomic alterations.
Since childhood ETP-ALL cases can be classified as T/M-MPAL, we revisited the immunophenotype and molecular on a cohort of childhood AL ambiguous phenotypes to test the value of identifying differences or similarities between these diseases in clinical outcomes. Hence, the first step of this study was to determine the methodological criteria to improve the identification of each type. In our cohort, by revisiting the M-FCM, we identified three possible diagnostic categories: ETP-ALL, T/M-MPAL, and AML-M0 with CD7expression. We were able to demonstrate 9.7% of ETP-ALL cases amongst T-ALL, and prevalence in patients older than 10 years of age at diagnosis. The frequency may be lower than previously reported (11% rather than 17%).9,20,21 However, this aspect should take into count that because the complete immunophenotyping was not prospectively performed, especially regarding ETP-ALL identification. The exclusion of cases as probable ETP-ALL (CD1a not available, CD5 weak, CD4-/CD8- with myeloid antigens expression) could explain the lower ETP-ALL case in our T-ALL cohort. Another point is that the high frequency of ETP-ALL reported internationally was observed in children and adults up to 25 years old,4,20 whereas our T-ALL cohort is predominantly of children under 18 years of age. It has been demonstrated that the first identification of ETP-ALL based on Coustan-Smith criteria could significantly underestimate ETP-ALL patients with leukemia with ambiguous phenotype.9,22,23 Applying the scoring system proposed by Inukai et al,10 we were able to characterize nine ETP-ALL cases, adding them to the previous cases identified by Coustan-Smith et al, criteria9 and 10 T/M-MPAL newly diagnosed cases.
Because ETP-ALL and T/M-MPAL share cell antigens which characterize T- and myeloid phenotypes, we wanted to test if molecular aberrations frequently found in T-ALL and AML would help in the distinction of ETP-ALL from T/M-MPAL in our cohort. We observed that STIL-TAL1, FBXW7 mutation and CDKN2A/B deletion were found only in ETP-ALL, while NOTCH1, FLT3, N/KRAS mutations, TLX3-r, and KMT2A-r were present in both T/M-MPAL and ETP-ALL cases.The frequencies of NOTCH1 mutations in ETP-ALL vary worldwide according to series of cases. For instance, Zhang et al, (2012)6 found a lower frequency of NOTCH1 mutations (16%) compared to our series. On the other hand, Zuurbier et al, (2014)23 found very high frequencies of NOTCH1 mutations among ETP-ALL (69%). Alexander et al, (2018)19 found 13 out of 19 ETP-ALL with NOTCH1 mutations (68%). In addition, mutations in NOTCH1 were described frequently in T/M-MPAL.7,19,24 The similarities between T/M-MPAL and ETP-ALL cases regarding KMT2A-r, FLT3, N/KRAS, and NOTCH1 mutations seem to share genomic aberrations with AML.7,24–26 Kotrova et al, have demonstrated in ambiguous lineage leukemia with distinct T and M-blast populations the presence of typical T-ALL mutations that activate the NOTCH1 signaling pathway.27 In mice, aberrations such as KMT2A-ENL and KRASG12D cooperate to the rise of both AML and T-ALL.28 We have found one T/M-MPAL case with KMT2A-ENL, KRAS and NOTCH1 mutation that may be more consistent with Ugale’s animal model.28
The genomic landscape of ETP-ALL has already been described, but only very recently have the genetic basis and differences between T/M-MPAL been deeply explored with the distinct profiles of ambiguous lineage leukemia.6,19,29 Authors showed that ETP-ALL and T/M-MPAL are genomically similar, characterized by mutations in regulators of hematopoietic stem cell, although, variation in mutations within the transcription factor gene have been observed. For example, the aberrations of core transcription factors that drive T-cell (eg, TAL1, TLX1,) were less frequent in T/M-MPAL.19 Our findings validate this previous study, regarding NOTCH1 and RAS mutations found in T/M-MPAL.19 FLT3 mutated were not found at similar frequencies in T/M MPAL and ETP-ALL.
Recently, in an international study of leukemia with B/M-MPAL, the event-free survival of patients treated with ALL–type therapy was demonstrated to be superior to those who received AML–type, or with combined-type treatment.30,31 Our data regarding T/M-MPAL are aligned with the findings of Hrusak et al31.
In our cohort, five patients were treated as AML with a dismal outcome as opposed to cases that were treated based on ALL-therapy [(56.4% (31.7–49.7)]. The overall survival of patients with ETP-ALL and T/M-MPAL in our cohort was similar to the results obtained in ETP-ALL patients treated according to AIEOP-BFM 2009 [(55.6% (28.6–75.9)].21 In the AIEOP centers with AIEOP-BFM protocols, the accuracy of the ETP-ALL diagnosis was confirmed by the score proposed by Inukai et al10. Conversely, the ETP-ALL enrolled into the MRC UK-ALL 2003 trial had an OS [82.4% (69.7–95.1%0] in a 5-year followup.20
A noteworthy limitation of our study is the sample size, which is to be expected with a rare disease, but which limits the interpretation of the data, particularly the outcome between ETP-ALL and T/M-MPAL compared with other determinants, such as age, sex, WBC, minimal residual disease and NOTCH1, RAS, FLT3, and KMT2A-r. The outcome in patients treated either with chemotherapy alone or with both chemotherapy and alloHSCT was not performed due to the sample size.
One strong finding in our study was to demonstrate that ETP-ALL and T/M-MPAL, irrespective of similarities or differences, have NOTCH1 mutations as a good predictive responsive treatment in both ambiguous leukemia subtypes. The presence of NOTCH1 mutations was associated with significantly better OS.
Conclusion
In conclusion, we demonstrated that ETP-ALL and T/M-MPAL are closely related in terms of immunophenotyping, genetic aberrations, and outcomes. In both diseases, genetic mutations were in NOTCH1, FLT3, RAS, and KMT2A-r, which should be taken into account for potential innovative therapies in front-line treatments. This is especially true regarding NOTCH1 mutations for prognostic assessment and risk-stratification of these cases.
Ethics approval and informed consent
Patients were treated in accordance with the Declaration of Helsinki medical ethical standards. The Research Ethical Committee of the Instituto Nacional de Cancer, Rio de Janeiro, Brazil approved the study (CEP/INCA#117/12; CEP-CONEP: PB #888.277). A parent or legal guardian of all children and adolescent have provided the written informed consent for samples tests.
Acknowledgments
Brazilian Study Group for Childhood Leukemia (BSGCL) members: Rebeca Ferreira Marques, Anna Carolina Silva Dias, Juliana Teixeira Costa, Claudia Teresa de Oliveira, Cesar Casagranda, Geni Ramos Vera, Gustavo Ribeiro Neves, Isis Maria Quezado Magalhães, José Carlos Cordoba, Ricardo Camargo, Patrícia Carneiro de Brito, Geraldo Pedral Sampaio, Raimundo Antônio Gomes Oliveira, Renata Pereira and Sidnei Epelman provided samples, clinical characteristics and patient follow-up data.
Hospital de Clínicas de Porto Alegre, Porto Alegre, RS, Brazil; Hospital Araújo Jorge, Goiania, GO, Brazil; Hospital Martagão Gesteira, Salvador, BA, Brazil; Hospital Amaral Carvalho, Jaú, SP, Brazil; Hospital Aldenora Bello, São Luís, MA, Brazil; Hospital Sarina Rolin Caracante, Sorocaba, SP, Brazil; Hospital da Criança de Brasília Jose Alencar, Brasília, DF, Brazil; Centro de Pesquisa Clínica - HUUFMA, São Luís, MA, Brazil; Hospital dos Servidores do Estado, Rio de Janeiro, RJ, Brazil; Hospital Santa Marcelina, São Paulo, SP, Brazil. EPN is supported by the Brazilian Ministry of Health (INCA-Brazil). MSPO is a scholar of Conselho Nacional de Pesquisas -CNPq (#301594/2015-5). This Project has received support of Childhood and Adolescent Cancer Association-TUCCA, São Paulo, Brazil.
Author contributions
All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bene MC. Biphenotypic, bilineal, ambiguous or mixed lineage: strange leukemias! Haematologica. 2009;94(7):891–893. doi:10.3324/haematol.2009.007799
2. Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):
3. Wood BL, Winter SS, Dunsmore KP, Devidas M. T-lymphoblastic leukemia (T-ALL) shows excellent outcome, lack of signifi cance of the early thymic precursor (ETP) immunophenotype, and validation of the prognostic value of end-induction minimal residual disease (MRD) in children’s oncology group (COG). Blood. 2014;124(21):1.
4. Bond J, Graux C, Lhermitte L, et al. Early response–based therapy stratification improves survival in adult early thymic precursor acute lymphoblastic leukemia: a group for research on adult acute lymphoblastic leukemia study. J Clin Oncol. 2017;35(23):2683–2691. doi:10.1200/JCO.2016.71.8585
5. Gutierrez A, Kentsis A. Acute myeloid/T-lymphoblastic leukaemia (AMTL): a distinct category of acute leukaemias with common pathogenesis in need of improved therapy. Br J Haematol. 2018;180(6):919–924. doi:10.1111/bjh.15129
6. Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi:10.1038/nature10725
7. Eckstein OS, Wang L, Punia JN, et al. Mixed-phenotype acute leukemia (MPAL) exhibits frequent mutations in DNMT3A and activated signaling genes. Exp Hematol. 2016;44(8):740–744. doi:10.1016/j.exphem.2016.05.003
8. Bene MC, Castoldi G, Knapp W, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leuk Off J Leuk Soc Am Leuk Res Fund, UK. 1995;9(10):1783–1786. doi:10.1002/cpp.1797
9. Coustan-Smith E, Mullighan CG, Onciu M, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10(2):147–156. doi:10.1016/S1470-2045(08)70314-0
10. Inukai T, Kiyokawa N, Campana D, et al. Clinical significance of early T-cell precursor acute lymphoblastic leukaemia: results of the Tokyo Children’s Cancer Study Group Study L99-15. Br J Haematol. 2012;156(3):358–365. doi:10.1111/j.1365-2141.2011.08955.x
11. van Dongen JJM, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13:1901–1928. doi:10.1038/sj.leu.2401592
12. Mansur MB, Emerenciano M, Brewer L, et al. SIL-TAL1 fusion gene negative impact in T-cell acute lymphoblastic leukemia outcome. Leuk Lymphoma. 2009;50(8):1318–1325. doi:10.1080/10428190903040014
13. Burmeister T, Meyer C, Gröger D, Hofmann J, Marschalek R. Evidence-based RT-PCR methods for the detection of the 8 most common MLL aberrations in acute leukemias. Leuk Res. 2015;39(2):242–247. doi:10.1016/j.leukres.2014.11.017
14. Zenatti PP, Ribeiro D, Li W, et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet. 2011;43(10):932–941. doi:10.1038/ng.924
15. Mansur MB, Hassan R, Barbosa TC, et al. Impact of complex NOTCH1 mutations on survival in paediatric T-cell leukaemia. BMC Cancer. 2012;12(1):9. doi:10.1186/1471-2407-12-9
16. Andrade FG, Noronha EP, Brisson GD, et al. Molecular characterization of pediatric acute myeloid leukemia: results of a multicentric study in Brazil. Arch Med Res. 2016;47(8):656–667. doi:10.1016/j.arcmed.2016.11.015
17. Möricke A, Reiter A, Zimmermann M, et al. Risk-adjusted therapy of acute lymphoblastic leukemia can decrease treatment burden and improve survival: treatment results of 2169 unselected pediatric and adolescent patients enrolled in the trial ALL-BFM 95. Blood. 2008. doi:10.1182/blood-2007-09-112920
18. Scrideli CA, Assumpção JG, Ganazza MA, et al. A simplified minimal residual disease polymerase chain reaction method at early treatment points can stratify children with acute lymphoblastic leukemia into good and poor outcome groups. Haematologica. 2009. doi:10.3324/haematol.2008.003137
19. Alexander TB, Gu Z, Iacobucci I, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature. 2018. doi:10.1038/s41586-018-0436-0
20. Patrick K, Wade R, Goulden N, et al. Outcome for children and young people with Early T-cell precursor acute lymphoblastic leukaemia treated on a contemporary protocol, UKALL 2003. Br J Haematol. 2014;166:421–424. doi:10.1111/bjh.12882
21. Conter V, Valsecchi MG, Buldini B, et al. Early T-cell precursor acute lymphoblastic leukaemia in children treated in AIEOP centres with AIEOP-BFM protocols: a retrospective analysis. Lancet Haematol. 2016;3:e80–e86. doi:10.1016/S2352-3026(15)00254-9
22. Gutierrez A, Dahlberg SE, Neuberg DS, et al. Absence of biallelic TCRgamma deletion predicts early treatment failure in pediatric T-cell acute lymphoblastic leukemia. J Clin Oncol. 2010;28(24):3816–3823. doi:10.1200/JCO.2010.28.3390
23. Zuurbier L, Gutierrez A, Mullighan CG, et al. Immature MEF2C-dysregulated T-cell leukemia patients have an early T-cell precursor acute lymphoblastic leukemia gene signature and typically have non-rearranged t-cell receptors. Haematologica. 2014;99(1):94–102. doi:10.3324/haematol.2013.090233
24. Wouters BJ, Jorda MA, Keeshan K, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110(10):3706–3714. doi:10.1182/blood-2007-02-073486
25. Oliveira JL, Kumar R, Khan SP, et al. Successful treatment of a child with T/myeloid acute bilineal leukemia associated with TLX3/BCL11B fusion and 9q deletion. Pediatr Blood Cancer. 2011;56(3):467–469. doi:10.1002/pbc.22850
26. Matlawska-Wasowska K, Kang H, Devidas M, et al. MLL rearrangements impact outcome in HOXA-deregulated T-lineage acute lymphoblastic leukemia: a Children’s Oncology Group Study. Leukemia. 2016;30(9):1909–1912. doi:10.1038/leu.2016.60
27. Kotrova M, Musilova A, Stuchly J, et al. Distinct bilineal leukemia immunophenotypes are not genetically determined. Blood. 2016;128:2263–2266. doi:10.1182/blood-2016-07-725861
28. Ugale A, Säwén P, Dudenhöffer-Pfeifer M, Wahlestedt M, Norddahl GL, Bryder D. MLL-ENL-mediated leukemia initiation at the interface of lymphoid commitment. Oncogene. 2017;36(22):3207–3212. doi:10.1038/onc.2016.470
29. Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017. doi:10.1038/ng.3909
30. Mejstrikova E, Volejnikova J, Fronkova E, et al. Prognosis of children with mixed phenotype acute leukemia treated on the basis of consistent immunophenotypic criteria. Haematologica. 2010;95(6):928–935. doi:10.3324/haematol.2009.014506
31. Hrusak O, De Haas V, Stancikova J, et al. International cooperative study identifies treatment strategy in childhood ambiguous lineage leukemia. Blood. 2018:1–12. doi:10.1182/blood-2017-12-821363
Supplementary Materials
 | Table S1 Panel of monoclonal antibodies and fluorochromes used and revisited in the diagnostic panel |
 | Table S2 Summary of the immunophenotypic profile of acute leukemia with aberrant phenotypes, Brazil, 2005–2017 |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



