Back to Journals » International Journal of Nanomedicine » Volume 17
Systematic Identification of Genomic Markers for Guiding Iron Oxide Nanoparticles in Cervical Cancer Based on Translational Bioinformatics
Authors Zhou H, Tian J, Sun H, Fu J, Lin N , Yuan D, Zhou L, Xia M, Sun L
Received 24 February 2022
Accepted for publication 7 June 2022
Published 29 June 2022 Volume 2022:17 Pages 2823—2841
DOI https://doi.org/10.2147/IJN.S361483
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Haohan Zhou,1,2,* Jiayi Tian,3,* Hongyu Sun,1 Jiaying Fu,1 Nan Lin,1 Danni Yuan,1 Li Zhou,3 Meihui Xia,3 Liankun Sun1
1Key Laboratory of Pathobiology, Ministry of Education, Department of Pathophysiology, College of Basic Medical Sciences, Jilin University, Changchun, 130021, People’s Republic of China; 2Department of Orthopaedic Oncology, Changzheng Hospital, Second Military Medical University, Shanghai, 200000, People’s Republic of China; 3First Hospital, Jilin University, Changchun, 130021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liankun Sun, Key Laboratory of Pathobiology, Ministry of Education, Department of Pathophysiology, College of Basic Medical Sciences, Jilin University, Changchun, 130021, People’s Republic of China, Email [email protected] Meihui Xia, First Hospital, Jilin University, Changchun, 130021, People’s Republic of China, Email [email protected]
Purpose: Magnetic iron oxide nanoparticle (MNP) drug delivery system is a novel promising therapeutic option for cancer treatment. Material issues such as fabrication and functionalized modification have been investigated; however, pharmacologic mechanisms of bare MNPs inside cancer cells remain obscure. This study aimed to explore a systems pharmacology approach to understand the reaction of the whole cell to MNPs and suggest drug selection in MNP delivery systems to exert synergetic or additive anti-cancer effects.
Methods: HeLa and SiHa cell lines were used to estimate the properties of bare MNPs in cervical cancer through 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) and enzyme activity assays and cellular fluorescence imaging. A systems pharmacology approach was utilized by combining bioinformatics data mining with clinical data analysis and without a predefined hypothesis. Key genes of the MNP onco-pharmacologic mechanism in cervical cancer were identified and further validated through transcriptome analysis with quantitative reverse transcription PCR (qRT-PCR).
Results: Low cytotoxic activity and cell internalization of MNP in HeLa and SiHa cells were observed. Lysosomal function was found to be impaired after MNP treatment. Protein tyrosine kinase 2 beta (PTK2B), liprin-alpha-4 (PPFIA4), mothers against decapentaplegic homolog 7 (SMAD7), and interleukin (IL) 1B were identified as key genes relevant for MNP pharmacology, clinical features, somatic mutation, and immune infiltration. The four key genes also exhibited significant correlations with the lysosome gene set. The qRT-PCR results showed significant alterations in the expression of the four key genes after MNP treatment in HeLa and SiHa cells.
Conclusion: Our research suggests that treatment of bare MNPs in HeLa and SiHa cells induced significant expression changes in PTK2B, PPFIA4, SMAD7, and IL1B, which play crucial roles in cervical cancer development and progression. Interactions of the key genes with specific anti-cancer drugs must be considered in the rational design of MNP drug delivery systems.
Keywords: drug delivery, anti-cancer drugs, PTK2B, PPFIA4, SMAD7, IL1B
Introduction
Conventional drug administration in cancer treatment is mainly nonselective, and many anti- cancer drugs are expelled out of cancer cells or cleared rapidly from the body, reducing chemotherapeutics efficiency and resulting in off-target side effects.1,2 A promising strategy to deal with this challenge is the nano-drug delivery platform, which carries anti-cancer drugs and enhances their local concentration at the tumor site.3–7 Magnetic iron oxide nanoparticles (MNPs) occupy a privileged position among other nano-drug delivery platforms,8,9 given their properties in imaging contrast and magnetic drug targeting.10–12 Although the MNP is designed as a carrier of the drugs and exhibits excellent biocompatibilities and low toxicities,13–15 the effects of the MNP itself should not be ignored. However, currently, much effort is devoted to addressing several important issues such as MNP fabrication, functionalized modification, drug conjugation, and delivery selectivity,16–19 and the focus on elucidating the molecular pathogenesis of MNP in cancer cells is lacking.
Magnetic iron oxide nanoparticles have been successfully applied in the in vivo delivery of doxorubicin to high-grade intramedullary spinal cord tumors in rats.20 In in vitro models of hepatocellular carcinoma and prostate cancer, MNP delivery systems loaded with quercetin or docetaxel also exhibited potent anti-cancer efficacy.21,22 However, it should be noted that a series of cascade reactions are supposed to be triggered by the MNP itself, which might impact the effects of anti-cancer drugs at the molecular level. However, despite the growing evidence for interaction among MNPs and bio-macromolecules,23–25 information on the molecular mechanism of MNPs inside cancer cells and their interaction with anti-cancer drugs is very limited, which might be extremely important in clinic applications.
The systems pharmacology approach has been widely used in monitoring the effects of drugs and screening potential drug candidates.26 This approach develops a global understanding of the drug’s pathophysiology and does not focus on the single interaction between a drug and its target or pathway. The Food and Drug Administration has recognized that gene expression profiling is a promising approach for advancing the development of medical products.27 Owing to the unique property of MNPs, their molecular effects cannot be predicted, neither from the known chemical agents nor from iron metabolism. Therefore, this study rationalized that a non-hypothesis-driven systems pharmacology approach using whole genome expression profiling is likely to provide a broad insight into the biological effects of MNPs.
In the last few years, the progress in biological computational methods has made it possible to analyze genetic aberrations and molecular mechanisms and provide promising tools in medical oncology with great clinical applications.28–30 However, these approaches have not been fully used in the field of nano-chemotherapy. In this study, bioinformatics data mining coupled with clinical data analysis were employed to achieve the systems pharmacology approach. The properties of MNPs were evaluated in cervical cancer cell lines, and four key genes relevant for MNP pharmacology, clinical features, somatic mutation, and immune infiltration in cervical cancer were identified. This study also showed that the four key genes were significantly correlated with the lysosome gene set. Potential targeting drugs of key genes were also presented.
This study validated the properties of MNPs in two cervical cancer cell lines, employed biological computational methods and identified protein tyrosine kinase 2 beta (PTK2B), liprin-alpha-4 (PPFIA4), mothers against decapentaplegic homolog 7 (SMAD7), and interleukin (IL) 1B as key genes of MNP onco-pharmacology in cervical cancer. Moreover, potential target drugs were also proposed for further investigation. This study can increase our understanding of the pharmacologic mechanism of MNP treatment in tumors, which is necessary when designing an effective MNP-based drug delivery system.
Materials and Methods
Chemicals
Chemicals, including iron (II) chloride tetrahydrate (FeCl2.4H2O), iron (III) chloride hexahydrate (FeCl3.6H2O), ethanol (EtOH), and ammonium hydroxide solution (NH4OH), were obtained from Merck (Darmstadt, Germany).
The bare Fe3O4 nanoparticles (Fe3O4 NPs) were prepared via the co-precipitation method.31 Briefly, FeCl3·6H2O (0.02 mol/5.38 g) was mixed with FeCl2·4H2O (0.01 mol/1.98 g) in deionized water (100 mL) under a flow of nitrogen. After 10 min, NH4OH (25%, 10 mL) was added dropwise into the above reaction with vigorous stirring at 80 °C for 30 min. The resulting black solution was filtered and washed with deionized water (100 mL) three times. The Fe3O4 NPs loaded with coumarin 6 (C6) used in this study were synthesized according to a previously published technique.32 For the cellular study, Fe3O4 NPs were sterilized with ethanol, and a bath sonicator was used to disperse the products into a colloid which was stored at 4 °C for further experiments.
Cell Culture and Treatment
Human cervical cancer HeLa and SiHa cells were obtained from the China Academy of Chinese Medical Sciences (Beijing, China) and were cultured under ideal growth conditions (37 °C, 5% CO2, and 95% air atmosphere) using RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., USA) containing 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc., USA), 100 U/mL penicillin, and 100 µg/mL streptomycin. HeLa and SiHa cells were incubated in the presence of 0, 50, 100, 200, and 400 μg/mL MNP. Primarily, events under 100 μg/mL MNP exposure over 24 h were chosen for the following experiments.
MTT Assays
Cytotoxicity assessment was performed using the 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) assay. HeLa and SiHa cells were plated in 96-well plates at a density of 1×104 cells/well and cultured with RPMI-1640 medium. After 24 h of incubation, the cells were supplemented with MNP (10 μg) at a final concentration of 100 μg/mL and incubated for a further 24 h. Control cells were incubated in RPMI-1640 medium with 10% FBS, and H2O2 (0.5 mM) was employed as the positive control. After incubation, 10 μL of 5 mg/mL MTT in 90 μL of RPMI-1640 medium was added to each well. After 4 h, the formazan crystals were dissolved by adding 150 μL of dimethylsulfoxide. The absorption of the formazan solution was measured using a Fluostar Omega microplate reader (BMG Labtech, Germany) at a wavelength of 570 nm.
Cell Fluorescence Imaging
Magnetic iron oxide nanoparticles were labeled with C6 for cell imaging, and LysoTracker Red (LTR) was utilized to indicate lysosomes. Bright yellow fluorescence indicated the colocalization of the MNP and lysosome in HeLa cells. Quantitative colocalization analysis was performed using ImageJ software 1.52a (http://rsb.info.nih.gov/ij/). The LTR (Beyotime, China) probes are fluorescent acidotropic probes used for labeling and tracking acidic organelles in live cells; the variance in fluorescence intensity may also imply lysosomal pH changes. Acridine orange (AO; Merck, Germany), a lysosomotropic weak base that accumulates at high concentrations in intact lysosomes because of proton trapping was also used as the shift of granular red fluorescence to diffused green fluorescence may monitor either lysosomal leakage or change in lysosomal pH. Briefly, HeLa and SiHa cells seeded (5 × 104 cells/well) on coverslips placed in 24-well plates were treated with or without 100 μg/mL MNP (24 h), followed by loading with 5 μg/mL AO for 30 min and Hoechst 33,342 for 5 min, or 100 nM LTR for 30 min under specific growth conditions (37 °C, 5% CO2). Then, the slides were rinsed twice with warm (37 °C) PBS (1 mL) and observed under a Zeiss Axio Imager Z2 (Carl Zeiss, Germany).33
Cathepsin-B (CTSB) Activity Assay
The lysosomal function was evaluated by detecting CTSB-specific activity using Z-FR-AMC (R&D Systems, USA), a synthetic fluorogenic peptide substrate for CTSB. The cathepsin-B-specific activities of the HeLa and SiHa cells treated with MNPs were measured in an assay buffer containing 200 mM NaOAc, 0.1% Brij 35, 0.1% Triton X-100, and 1 mM EDTA at pH 6.7. Briefly, 9 μL of cellular extraction and 1 μL of the CTSB substrate Z-FR-AMC (100 mg/mL) were added to 90 μL of the assay buffer. Then, the samples were incubated in each well of a black 96-well plate at 37 °C for 1 h. The relative fluorescence units, corresponding to the CTSB cleavage of the substrates, were measured at an excitation wavelength of 355 nm and emission wavelength of 460 nm in a Fluostar Omega microplate reader (BMG Labtech, Germany).
Apoptosis Analysis by Flow Cytometry
Apoptosis was determined by Annexin-V and propidium iodide (PI) staining, according to the manufacturer’s protocol (556,547; BD Biosciences). Briefly, HeLa and SiHa cells were seeded (2 × 105 cells/well) into six-well plates. After treatments, the cells were collected and washed and then stained with Annexin-V and PI. Cells were quantified using a guava easyCyte flow cytometer (Millipore).
Quantitative PCR
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA). Then, 2 μg of total RNA was used to synthesize cDNA using the Omniscript Reverse Transcription kit (TaqMan, Applied Biosystems, USA). Quantitative reverse transcription PCR (qRT-PCR) was performed to estimate the mRNA levels of PPFIA4, PTK2B, IL1B, and SMAD7 using SYBR SuperMix (TransGen Biotech, CA, USA). All reactions were performed in duplicate on a Bio-Rad CFX96 Touch system (Bio-Rad, Hercules, CA, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control, and the relative expression level for each gene was calculated using the 2−ΔΔCt relative quantification method and normalized to that of endogenous housekeeping gene. All reactions were performed using the following cycling parameters: initial denaturation at 94 °C for 30s; followed by 45 cycles of 94 °C for 5s, 60 °C for 15s, and 72 °C for 10s. The primers used were as follows: PTK2B gene, 5′-GAAGTCCGATGAGATCCACTGG-3′ (sense) and 5′-CTTCTGGCAAGTAGCGGATTT-3′ (anti-sense); PPFIA4 gene, 5′-CTCTGCGGATGTTGTCTCCC-3′ (sense) and 5’-ATGCTGCCACTGGTTACACG-3′ (anti-sense); IL1B gene, 5′-ATGATGGCTTATTACAGTGGCAA-3′ (sense) and 5’-GTCGGAGATTCGTAGCTGGA-3′ (anti-sense); SMAD7 gene, 5′-GGACGCTGTTGGTACACAAG-3′ (sense) and 5′-GCTGCATAAACTCGTGGTCATTG-3′ (anti-sense); GAPDH gene, 5′-ACAACTTTGGTATCGTGGAAGG-3’ (sense) and 5′-GCCATCACGCCACAGTTTC-3′ (anti-sense); and PIK3R2 gene, 5′-AATGCAGCAAGGAATACCTGG-3′ (sense) and 5′-GCTCTCATGGATCTCGGCAA-3′ (anti-sense).
Microarray Data of HeLa Cells Treated with MNPs
The microarray datasets of GSE15248 were downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/). These RNA profiles were based on the GPL6102 platforms (Illumina human-6 v2.0 expression beadchip) containing a total of three MNP-treated HeLa cell samples and three control samples.
Identification of Differentially Expressed Genes (DEGs) and Hub Genes
The downloaded matrix files were converted and processed in R (version 3.5.3) software with the “Affy” package to read the expression data for the CEL files. Gene differential expression analysis was performed using the “limma” package, after which the genes were sorted according to the log fold-change values. Finally, the genes with P < 0.05 and |logFC| > 0.5 were considered as DEGs. The “pheatmap” and “ggplot2” packages were used to draw the heatmap and volcano plot of the top ranked genes and DEGs, respectively.
The protein–protein interaction (PPI) network was also constructed by uploading all DEGs to the STRING database (https://string-db.org/). Then, the PPI network was further analyzed in Cytoscape software (www.cytoscape.org/). CytoHubba, a Cytoscape plug-in, sorts the genes by analyzing 12 parameters, including MCC, DMNC, MNC, DEGREE, EPC, BOTTLENECK, EcCentricity, CLOSENESS, RADIALITY, BETWEENNESS, STRESS, and ClusteringCoefficient. The top 50 genes ranked by each parameter were recorded. The genes sorted by eight or more parameters were selected as the hub genes being highly essential in the functional network.
Survival Analysis and Efficacy Evaluation
Expression data and survival information of 310 patients with cervical cancer from The Cancer Genome Atlas (TCGA) were obtained using the online software SangerBox (http://sangerbox.com). A Kaplan–Meier survival plot was constructed using the “survival”, “survminer”, and “ggplot2” packages in R software. Differential expression of the key genes among the clinical features was also plotted as a dot-box plot graph with the Kruskal–Wallis test.
Somatic Mutation of Key Genes
Genetic alterations of the key genes across the patients with cervical cancer were explored in the CBio Cancer Genomics Portal (http://www.cbioportal.org/). Oncoplots for the mutation spectrum visualization and Lollipop plots for the amino acid changes were processed with the “maftools” package in R software. Mutually exclusive or co-occurring sets of genes were also detected using somatic interaction function in the “maftools” package, which performs the pair-wise Fisher’s exact test to detect significant pairs of genes.
Correlation of Key Genes with Immune Infiltration
Systematic analysis of the immune infiltrates was conducted through Tumor Immune Estimation Resource (TIMER), which is a comprehensive resource with diverse cancer types (https://cistrome.shinyapps.io/timer/). The TIMER applies a previously published statistical algorithm, that is, deconvolution, to estimate the abundance of immune infiltrating levels from gene expression profiles. Based on 310 patients with cervical cancer from TCGA, the correlation of the expression of four key genes with the abundance of immune infiltrates was analyzed, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, dendritic cells, and tumor purity via gene modules. Finally, Spearman correlation and the estimated statistical significance were displayed as scatter-plots. The gene expression level was displayed with log2 RSEM.
Correlation of Key Genes with Lysosomes
Patients with cervical cancer were divided into two groups according to the median expression values of each key gene. First, Gene Set Enrichment Analysis (GSEA) was applied using the Java GSEA implementation to determine whether the defined set of lysosomal genes shows statistically significant concordant differences. The number of permutations was set as 1000, and c2.cp.kegg.v6.2.symbols.gmt was downloaded as the reference gene set. A false discovery rate q value < 0.05 and nominal P < 0.25 were set as the cut-off threshold. Second, correlations among key genes and individual lysosomal genes were calculated as Pearson’s correlation coefficients and processed using the “ggstatsplot” package in R software.
Identification of Potential Anti-Cancer Drugs Suitable for MNP Delivery
CBioPortal was used to explore the correlation among the four key genes, their most frequently altered neighbor genes, and anti-cancer drugs to screen new potential anti-cancer drugs suitable for MNP delivery.
Chemotherapy Resistance Gene Informatics Analysis
The Cancer Genome Atlas gene expression data were used for chemotherapy resistance analysis. As shown in Table 1, most of the patients received cisplatin, whereas the sample size of patients that received other drug regimens was too small to perform gene expression analysis. Finally, 106 patients that received cisplatin were selected for further analysis. Neoplasm cancer status was selected as the indicator of drug resistance. Expression data were transformed into transcripts per kilobase million before analysis in SangerBox software (Mugu Tech, Hangzhou, China). Finally, the genes with P < 0.05 and |logFC| > 0.5 were considered as DEGs. Hub genes were also explored with the PPI network and CytoHubba.
 |
Table 1 Correlation Between the Clinicopathologic Variables and the Four Key Genes mRNA Expression in Cervical Cancer |
Statistical Analyses
GraphPad Prism 8 statistical software (GraphPAD Software Inc., San Diego, CA, USA) was used for experimental data statistical analyses and image processing. Differences between groups were determined by the unpaired Student’s t-test.
Results
Cytotoxic Effect of MNP
The transmission electron microscopy images of MNPs in Figure 1A–C show a spherical morphology with a size of 20−100 nm. Magnetic iron oxide nanoparticles had a good magnetic response in the external magnetic field (Figure 1D). No cytotoxicity was observed even at the highest particle loading level (400 μg/mL) in the culture medium (Figure 1E, F, H, I). The effects of MNPs on apoptosis of cervical cancer cells were further investigated. HeLa (Figure 1G) and SiHa (Figure 1J) cells treated with MNP for 24 h and 72 h were stained with fluorescein isothiocyanate-labeled Annexin-V/PI and subsequently subjected to flow cytometry analysis. The results revealed that MNP (100 μg/mL) did not significantly increase the number of apoptotic cervical cancer cells.
MNP Induced Inhibition of Lysosomal Function
To investigate the successful cell internalization of MNPs, colocalization experiments were conducted using fluorescence microscopy. The colocalization correlation coefficient was determined to be 0.95 (Figure 2A), indicating the internalization of MNP in the lysosome. A decrease in relative fluorescence units of Z-FR-AMC was observed, as a sign of decline in CTSB enzymatic activity (Figure 1B and C), indicating decreased lysosomal function. Lysosomotropic probes were applied to evaluate the effect of MNP on lysosomal pH in the HeLa and SiHa cells. LysoTracker Red exhibits red fluorescence in the lysosome in a pH-dependent manner, and a decrease in red fluorescence indicates an increase in lysosomal pH. The 24-h treatment with 100 μg/mL MNP neutralized the lysosomal pH, which was displayed as decreased red fluorescence of LTR compared with that of the control cells (Figure 1B and C). In addition, AO staining presents red fluorescence when it is accumulated within lysosomes owing to it being highly protonated but shows green fluorescence after diffusion into the cytosol. Thus, an increase in diffuse green fluorescence with a decrease in punctate red fluorescence suggests an elevated lysosomal pH. The 24-h treatment of HeLa and SiHa cells with 100 μg/mL MNP led to a decrease in the red fluorescence and increase in the green fluorescence with AO staining (Figure 1F–G).
Identification of DEGs After MNP Treatment
Microarray datasets GSE15248 obtained from the Gene Expression Omnibus database were based on the GPL6102 platforms, which included three MNP-treated HeLa cell samples and three control samples. The results of the GSE15248 provided insights into the potential mechanisms underlying the effects of MNP on cervical cancer cell function. After normalization, 179 DEGs were identified, of which 67 were significantly upregulated genes and 112 were downregulated genes. The results are displayed in the heatmap and volcano plots for DEG expression (Supplementary Figure S1).
Identification of Key Genes Associated with the Survival Time and Clinical Features in Patients with Cervical Cancer
To evaluate the involvement of the above-identified hub genes in cervical cancer, the survival data of 310 patients from the TCGA were used for Kaplan–Meier survival analysis using the Log rank test. Four out of 44 hub genes demonstrated significant associations with overall survival (Figure 3). Patients with high expression of receptor-like protein tyrosine phosphatases-interacting protein (liprin) alpha 4 (PPFIA4) (P = 0.0031; Figure 3A), SMAD7 (P = 0.044; Figure 3B), and IL1B (P = 0.0085; Figure 3C) had a poor overall survival, whereas patients with high expression of PTK2B exhibited a better performance in survival time (P = 0.017; Figure 3D). The four genes were identified as key genes for further analysis. The relationship between the clinical features and the expression of the four key genes was also analyzed (Table 1).
 |
Figure 3 Four key genes were associated with overall survival time and clinical features in cervical cancer patients. Survival analysis of PPFIA4 (A), SMAD7 (B), IL1B (C), and PTK2B (D) expression levels associating with overall survival time in 310 TCGA cervical cancer patients using Kaplan–Meier curves. The patients were divided into 2 groups, based on median gene expression value as a cut-off point. (E–G) The expression of PPFIA4 was highly associated with clinical stage with a p value of 2e-7 (E), and tumor (T) classification with a p value of 7.7e-06 (F). The expression of IL1B was significantly associated with tumor (T) classification with a p value of 0.0072 (G). The association between genes expression level and clinical features was estimated by Kruskal–Wallis test. (see also Figure S2). |
Further subgroup analysis was conducted among patients with different clinical stage, histologic grade, and tumor (T) and lymph node (N) classification. High PPFIA4 expression correlated with a poor overall survival in patients with clinical stages I/II (P = 0.029), T1/T2 (P = 0.021), and N0 (P = 0.002). High SMAD7 expression correlated with a poor overall survival in patients with clinical stages I/II (P = 0.048), histologic grade G3/G4 (P = 0.034), and T1/T2 (P = 0.031). High IL1B expression correlated with a poor overall survival in patients with clinical stage I/II (P = 0.036), histologic grade G1/G2 (P = 0.027), T1/T2 (P = 0.028), and N0 (P = 0.033). High PTK2B expression correlated with a better overall survival in patients with histologic grade G3/G4 (P = 0.016) and T1/T2 (P = 0.013) (Supplementary Figure S2).
Somatic Mutations of Key Genes with Various Mutation Patterns in Patients with Cervical Cancer
Genetic alteration of the oncoplot in CBioPortal was used to show the alteration status of the four key genes in patients with cervical cancer. As shown in Figure 4, the four key genes were altered in a total of 71 (22.9%) out of 310 patients (Figure 4A), among which PPFIA4 was altered the most, with an alteration frequency of 11%. Mutational hotspots of PPFIA4 and PTK2B showed that PPFIA4 has two mutation types (S743F/R1007M/G1134D-Missense, X1087-Splice) (Figure 4B) and PTK2B has three mutation types (R159Q/E494K/R586H/R936L-Missense, X369-Splice, I597Hfs*2-Frameshift) (Figure 4C). The mutation spots of PPFIA4 were highly clustered, which is the feature of a proto-oncogene. The mutation spots were discrete in PTK2B, as expected in a tumor suppressor gene. Mutation interaction of the key genes with the top mutated genes was also explored (Figure 4D) in patients with cervical cancer. PPFIA4 and PTK2B had a significant co-occurrence of mutation, with seven of the top mutated genes (Figure 4E).
Key Genes Play a Role in Immune Infiltration in Cervical Cancer
Immune infiltration was investigated using the TIMER deconvolution algorithm, which is more reliable than other methods. Analysis showed that PTK2B expression level had significant positive correlations with infiltrating levels of neutrophils (r = 0.38, P = 6.04e-11), dendritic cells (r = 0.317, P = 7.2e-08), CD8+ T cells (r = 0.217, P = 3.01e-04), and CD4+ T cells (r = 0.18, P = 2.7e-03) in patients with cervical cancer (Figure 5A), whereas there were significant negative correlations of PPFIA4 expression with infiltrating levels of CD8+ T cells (r = –0.201, P = 7.96E-04) and neutrophils (r = –0.172, P = 4.09e-03) (Figure 5B). In addition, expression levels of PTK2B and PPFIA4 had significant negative (r = –0.283, P = 1.59E-06) and positive correlations (r = –0.20, P = 8.01E-04), respectively, with tumor purity in patients with cervical cancer (Figure 5A and B). Significant correlations between immune infiltration and the expression levels of SMAD7 and IL1B were also observed (Figure 5C and D). These findings demonstrated that the four key genes, especially PTK2B and PPFIA4, play a specific role in immune infiltration in cervical cancers.
Key Genes Significantly Correlated with the Lysosome
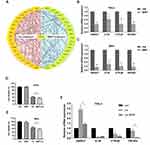
To determine whether lysosomes are associated with key genes in patients with cervical cancer, GSEA associated with the expression of key genes was conducted. The GSEA revealed significant differences (nominal P-value < 0.05, false discovery rate q value < 0.25) in the enrichment of the lysosome gene set for the expression of PTK2B and PPFIA4 (c2.cp.kegg.v6.2.symbols.gmt) (Figure 6A and B). The correlations among PTK2B, PPFIA4, and lysosomal genes were then evaluated by two methods. First, mutation interactions of these genes in patients with cervical cancer were explored, which showed significant co-occurrence of mutations in 26 lysosomal genes and in PPFIA4 and PTK2B (Figure 6C). Second, correlation analyses of gene expression levels were also performed, and strong correlations between key genes and certain lysosomal genes were observed. The results are displayed as Pearson’s correlation coefficient bubble-plots (Figure 6D), heatmap (Figure 6E), and scatter-plots (Figure 6F).
Network of Key Genes and Anti-Cancer Drugs
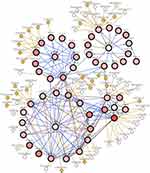
Figure 7 shows the relationship between the key genes and the other 46 most frequently altered neighbor genes. The correlations between anti-cancer drugs and the key genes were also illustrated. PTK2B and IL1B were the direct targets of anti-cancer drugs (genistein, leflunomide, and gallium nitrate). Although there was no anti-cancer drug directly targeting PPFIA4 and SMAD7, many of their most frequently altered neighbor genes have been identified as the targets of anti-cancer drug amounts.
Relationship Between Clinical Features and Key Gene Expression
The relationships between the expression of the four key genes and clinical features were also explored. The expression of PPFIA4 was highly associated with clinical stage (P = 2e-7; Figure 3E) and tumor (T) classification (P = 7.7e-06; Figure 3F). The expression of IL1B was significantly associated with tumor (T) classification (P = 0.0072; Figure 3G). In addition, the expression of PPFIA4 was associated with age and radiation treatment and was thus unsuitable for further analysis (Table 1).
Chemotherapy Resistance Analysis
A total of 1288 DEGs were identified, of which 319 were significantly upregulated genes, and 969 were downregulated genes. The top genes sorted by six or more parameters in the CytoHubba plug-in were identified as hub genes. PIK3R2 was identified as a common hub gene in both MNP treatment and chemotherapy resistance (Figure 8A).
The validation of the key genes was performed at the mRNA level in both the HeLa and SiHa cells by qRT-PCR. The MNP treatment significantly downregulated the mRNA expression levels of PTK2B, IL1B, SMAD7, and PIK3R2 (Figure 8B and C).
The MTT assay was performed in HeLa and SiHa cells to explore whether MNP enhanced the killing effects of cisplatin in cervical cancer. The results show that MNP increased the sensitivity of HeLa and SiHa cells to cisplatin after 24 h of treatment (Figure 8D and E). The qRT-PCR demonstrated that a combination of MNP with cisplatin significantly reduced the expression levels of SMAD7 and PIK3R2 in HeLa cells (Figure 8F).
Discussion
In the present study, we showed that MNP has no cytotoxicity in cervical cancer cells; however, changes in the ratio of 650 to 540 nm fluorescence of AO were observed. The LTR red fluorescence decreased, indicating a decline in the lysosomal proton gradient. The altered fluorescence could also reflect lysosomal membrane permeabilization, besides an elevated lysosomal pH indicator.33 However, the loss of the ability to retain lysosomotropic dyes can be attributed to either lysosomal membrane permeabilization or proton gradient dissipation. Nonetheless, it indicates lysosomal destabilization.34 Overall, these results demonstrated that MNP interferes with lysosomes but has no toxicity in cancer cells, which are the two most important properties of an MNP as a carrier in drug delivery systems. A previous study demonstrated that MNP was transported into lysosomes, where it interfered with the lysosomal hydrolases and caused lysosome instability,35,36 which is consistent with our findings. Lysosomes are thought to play a crucial role in regulating the biological activity of MNPs. Accumulating data shows that malignant transformation is associated with dramatic alterations in lysosomal structure and functions to meet increased metabolic demands. Furthermore, lysosomal cathepsins are involved in tumor progression and invasion. Overall, these changes greatly sensitize cancer cells to lysosomal destabilization.37,38 Thus, the intrinsic properties of the MNP involved in lysosomes make it a promising material as a chemotherapeutic carrier. The kinetics of nanoparticle dissolution were found to be strongly dependent on the lysosomal pH and lysosomal membrane permeabilization, only in acidic conditions the MNP significantly released iron ions in solution.39–41 The effects of lysosomal destabilization on the dissolution of iron oxide nanoparticles should be emphasized.42
A systems approach to address the issues of onco-pharmacologic mechanism of drugs is recognized to have advantages over the conventional hypothesis-driven methodology, which dominantly identifies a single target or its associated pathway.23,24,43–45 Here, we followed a data-driven approach, which provides an unbiased view of alterations in cancer cells in response to MNP exposure by statistically analyzing whole-genome expression profiling without a previously defined hypothesis. Jameel et al studied transcriptional alterations following MNP exposure in cervical cancer cells and showed that MNP interferes with transforming growth factor-β (TGF-β) signaling by inhibiting the expression of ID and SMAD genes.46 The identified genes were partially different from our results, and notably, the authors selected the genes for further investigations by ranking the false discovery rate, which would lead to the loss of functional genes. In the present study, we employed CytoHubba to explore important nodes or hubs by ranking 12 parameters and excluded genes without clinical relevance in cervical cancer. Navin et al proposed a systems pharmacology approach to identify genes possibly relevant for cisplatin resistance,47 which provided a broad insight into the molecular mechanisms. We made improvements by analyzing multi-omics data and associating key genes with clinical phenotypes.
This work identified PPFIA4, PTK2B, SMAD7, and IL1B as key players in MNP onco-pharmacology. PPFIA4, also known as liprin-α4, is a member of the liprin gene family.48 PPFIA4 was found to contribute to the proliferation of SBC-5 small-cell lung cancer cells both under normoxia and hypoxia.49,50 The protein tyrosine kinase PTK2B, also known as PYK2, is a non-receptor tyrosine kinase acting as a signaling mediator involved in tumorigenesis, invasion, and metastasis.51–53 A completely opposite role of PTK2B in cancer was also reported in ovarian carcinoma cells and prostate cancer cells as a tumor suppressor,54,55 which is in agreement with our observation that high expression levels of PTK2B are correlated with improved survival in patients with cervical cancer. Interleukin-1β genes encode an inflammatory cytokine IL-1β, which favors the maintenance of inflammatory tumor microenvironment that contributes to the acquisition of the aggressive phenotype in cancer cells.56 SMAD7 is an intracellular protein that allows transmembrane delivery of TGF-β. Similar to PTK2B, SMAD7 also possesses dual roles as a tumor suppressor and activator depending on tumor types.57,58
The findings of drug interaction analysis showed that PTK2B was the target of genistein and leflunomide, and IL1B was the target of gallium nitrate. Considering the upregulation of these two genes upon MNP treatment in cervical cancer cells, genistein, leflunomide, and gallium nitrate were recommended as potential partners of MNP in drug delivery systems for further investigation. No drug targeting PPFIA4 and SMAD7, which might be the promising targets of new anti-cancer drugs, was identified. Genistein as an active soy isoflavone has been found to mediate anti-cancer effects. Hua-Yan et al showed that compared to free genistein, the MNP-genistein delivery system exhibits a significantly enhanced inhibition effect on the gastric cancer cell line SGC-7901.59,60 Leflunomide, as an inhibitor of dihydroorotase dehydrogenase, was initially approved for the treatment of rheumatoid arthritis. Recently, leflunomide was also identified as a potent anti-cancer drug.61 Pund et al explored the delivery of leflunomide by nanoemulgel in melanoma and observed a significantly enhanced therapeutic response.62
The four key genes identified have been reported to affect the sensitivity of cancer cells to several anti-cancer drugs. Mendoza et al reported that the IL-1β-induced upregulation of genes potentiates the acquisition of cisplatin resistance in breast cancer cells.56 PTK2B has been demonstrated to play a pivotal role in integrin signaling in resistance to metformin, cisplatin, and proteasome inhibitors in multiple cancers, including breast cancer, ovarian cancer, myeloma, and non-small-cell lung cancer.47,53,63,64 SMAD7 has also been found to be associated with chemoresistance to HDAC4/5 inhibitor, cisplatin, and sorafenib in leukemia, lung cancer, and hepatocellular carcinoma.65–67 In a previous study by Hideya et al, PPFIA4 inhibition sensitized SBC-5 cells to cisplatin under hypoxia.50 However, evidence is lacking for the involvement of the four key genes in cervical cancer chemoresistance, which might provide critical information on drug selection in the MNP delivery system in cervical cancer treatment.
To achieve personalized medicine, the systematic investigation of the interface between the MNP and biological environments of different cancer types is necessary owing to the highly heterogeneous tumor biology and microenvironments.68–70 It should be noted that key genes of MNP in cancer cells and their roles may vary among different cancer types. A full understanding of the genomic alterations upon MNP treatment in different tumor types might have implications for specific cancer treatments. Furthermore, it would be interesting to investigate the genomic alterations in non-cancer cells. Consequently, delivery drug selection decisions will be based on not only the tumor molecular profiles in response to drugs but also the delivery carriers.
Conclusion
The results demonstrated that treatment of bare MNPs in cervical cancer cells significantly reduced the mRNA expression levels of PTK2B, PPFIA4, SMAD7, and IL1B, which play crucial roles in cervical cancer development and progression. The significance of the four key genes was validated by performing multi-omics data analysis, clinical relevance estimation, and qRT-PCR. Our study suggested that interactions of the key genes with anti-cancer drugs must be considered in the rational design of an MNP drug delivery system. Thus, this study could pave a novel way toward a systems pharmacology approach, characterizing the most potential molecular reaction to MNP in cervical cancer, which may help in further research on drug selection in MNP delivery systems to exert synergetic or additive anti-cancer effects.
Abbreviations
AO, acridine orange; C6, coumarin 6; CTSB, cathepsin-B; DEG, differentially expressed gene; GSEA, Gene Set Enrichment Analysis; IL, interleukin; LTR, LysoTracker Red; MNP, magnetic iron oxide nanoparticle; MTT, 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide; PI, propidium iodide; PPFIA4, liprin-alpha-4; PPI, protein–protein interaction; PTK2B, protein tyrosine kinase 2 beta; qRT-PCR, quantitative reverse transcription PCR; SMAD7, mothers against decapentaplegic homolog 7; TCGA, The Cancer Genome Atlas; TIMER, Tumor Immune Estimation Resource.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81772794 (Sun), 81672948 (Su), and 81472419 (Sun)), Jilin Provincial Industrial Innovation Project (2018C052-7), Jilin Provincial Research Foundation for the Development of Science and Technology Projects (20191004004TC), and “the Fundamental Research Funds for the Central Universities, JLU”, Education Department of Jilin Province (JJKH20201014KJ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Dissanayake S, Denny WA, Gamage S, Sarojini V. Recent developments in anticancer drug delivery using cell penetrating and tumor targeting peptides. J Control Release. 2017;250:62–76. doi:10.1016/j.jconrel.2017.02.006
2. Hurria A, Togawa K, Mohile SG, et al. Predicting chemotherapy toxicity in older adults with cancer: a prospective multicenter study. J Clin Oncol. 2011;29(25):3457–3465. doi:10.1200/JCO.2011.34.7625
3. Jahangirian H, Kalantari K, Izadiyan Z, Rafiee-Moghaddam R, Shameli K, Webster TJ. A review of small molecules and drug delivery applications using gold and iron nanoparticles. Int J Nanomedicine. 2019;14:1633–1657. doi:10.2147/IJN.S184723
4. Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2017;17(1):20–37. doi:10.1038/nrc.2016.108
5. Peng XH, Qian X, Mao H, et al. Targeted magnetic iron oxide nanoparticles for tumor imaging and therapy. Int J Nanomedicine. 2008;3(3):311–321. doi:10.2147/ijn.s2824
6. Talluri S, Malla RR. Superparamagnetic Iron Oxide Nanoparticles (SPIONs) for diagnosis and treatment of breast, ovarian and cervical cancers. Curr Drug Metab. 2019;20:942–945. doi:10.2174/1389200220666191016124958
7. Wahajuddin AS. Superparamagnetic iron oxide nanoparticles: magnetic nanoplatforms as drug carriers. Int J Nanomedicine. 2012;7:3445–3471. doi:10.2147/IJN.S30320
8. Douziech-Eyrolles L, Marchais H, Herve K, et al. Nanovectors for anticancer agents based on superparamagnetic iron oxide nanoparticles. Int J Nanomedicine. 2007;2(4):541–550.
9. Zhou Z, Sun Y, Shen J, et al. Iron/iron oxide core/shell nanoparticles for magnetic targeting MRI and near-infrared photothermal therapy. Biomaterials. 2014;35(26):7470–7478. doi:10.1016/j.biomaterials.2014.04.063
10. Jeon M, Halbert MV, Stephen ZR, Zhang M. Iron oxide nanoparticles as T1 contrast agents for magnetic resonance imaging: fundamentals, challenges, applications, and prospectives. Adv Mater. 2021;33(23):1906539. doi:10.1002/adma.201906539
11. Lee C-W, Wu P-C, Hsu I-L, et al. New templated Ostwald ripening process of mesostructured FeOOH for third-harmonic generation bioimaging. Small. 2019;15(20):1805086. doi:10.1002/smll.201805086
12. Liao MY, Wu CH, Lai PS, et al. Surface state mediated NIR Two‐Photon fluorescence of iron oxides for nonlinear optical microscopy. Adv Funct Mater. 2013;23(16):2044–2051. doi:10.1002/adfm.201202676
13. Di Marco M, Sadun C, Port M, Guilbert I, Couvreur P, Dubernet C. Physicochemical characterization of ultrasmall superparamagnetic iron oxide particles (USPIO) for biomedical application as MRI contrast agents. Int J Nanomedicine. 2007;2(4):609–622.
14. Jain TK, Reddy MK, Morales MA, Leslie-Pelecky DL, Labhasetwar V. Biodistribution, clearance, and biocompatibility of iron oxide magnetic nanoparticles in rats. Mol Pharm. 2008;5(2):316–327. doi:10.1021/mp7001285
15. Ansari MO, Ahmad MF, Shadab GG, Siddique HR. Superparamagnetic iron oxide nanoparticles based cancer theranostics: a double edge sword to fight against cancer. J Drug Deliv Sci Technol. 2018;45:177–183. doi:10.1016/j.jddst.2018.03.017
16. Chang T-W, Ko H, Huang W-S, et al. Tannic acid-induced interfacial ligand-to-metal charge transfer and the phase transformation of Fe3O4 nanoparticles for the photothermal bacteria destruction. Chem Eng J. 2022;428:131237. doi:10.1016/j.cej.2021.131237
17. Pyataev NA, Petrov PS, Minaeva OV, et al. Amylase-sensitive polymeric nanoparticles based on dextran sulfate and doxorubicin with anticoagulant activity. Polymers. 2019;11(5):921. doi:10.3390/polym11050921
18. Ashrafizadeh M, Ahmadi Z, Kotla NG, et al. Nanoparticles targeting STATs in cancer therapy. Cells. 2019;8(10):1158. doi:10.3390/cells8101158
19. Sun C, Du K, Fang C, et al. PEG-mediated synthesis of highly dispersive multifunctional superparamagnetic nanoparticles: their physicochemical properties and function in vivo. ACS Nano. 2010;4(4):2402–2410. doi:10.1021/nn100190v
20. Kheirkhah P, Denyer S, Bhimani AD, et al. Magnetic drug targeting: a novel treatment for intramedullary spinal cord tumors. Sci Rep. 2018;8(1):11417. doi:10.1038/s41598-018-29736-5
21. Srisa-Nga K, Mankhetkorn S, Okonogi S, Khonkarn R. Delivery of superparamagnetic polymeric micelles loaded with quercetin to hepatocellular carcinoma cells. J Pharm Sci. 2019;108(2):996–1006. doi:10.1016/j.xphs.2018.08.008
22. Nagesh PKB, Johnson NR, Boya VKN, et al. PSMA targeted docetaxel-loaded superparamagnetic iron oxide nanoparticles for prostate cancer. Colloids Surf B Biointerfaces. 2016;144:8–20. doi:10.1016/j.colsurfb.2016.03.071
23. Alarifi S, Ali D, Alkahtani S, Alhader MS. Iron oxide nanoparticles induce oxidative stress, DNA damage, and caspase activation in the human breast cancer cell line. Biol Trace Elem Res. 2014;159(1–3):416–424. doi:10.1007/s12011-014-9972-0
24. Zanganeh S, Hutter G, Spitler R, et al. Iron oxide nanoparticles inhibit tumour growth by inducing pro-inflammatory macrophage polarization in tumour tissues. Nat Nanotechnol. 2016;11(11):986–994. doi:10.1038/nnano.2016.168
25. Casalini T, Limongelli V, Schmutz M, et al. Molecular modeling for nanomaterial-biology interactions: opportunities, challenges, and perspectives. Front Bioeng Biotechnol. 2019;7:268. doi:10.3389/fbioe.2019.00268
26. Wist AD, Berger SI, Iyengar R. Systems pharmacology and genome medicine: a future perspective. Genome Med. 2009;1(1):11. doi:10.1186/gm11
27. Frueh FW, Huang SM, Lesko LJ. Regulatory acceptance of toxicogenomics data. Environ Health Perspect. 2004;112(12):A663–664. doi:10.1289/ehp.112-1277121
28. Kann MG. Advances in translational bioinformatics: computational approaches for the hunting of disease genes. Brief Bioinform. 2010;11(1):96–110. doi:10.1093/bib/bbp048
29. Overby CL, Tarczy-Hornoch P. Personalized medicine: challenges and opportunities for translational bioinformatics. Per Med. 2013;10(5):453–462. doi:10.2217/pme.13.30
30. Tenenbaum JD. Translational bioinformatics: past, present, and future. Genomics Proteomics Bioinformatics. 2016;14(1):31–41. doi:10.1016/j.gpb.2016.01.003
31. Massart R. Preparation of aqueous magnetic liquids in alkaline and acidic media. IEEE Trans Magn. 2003;17(2):1247–1248. doi:10.1109/TMAG.1981.1061188
32. Sun X, Dong B, Xu H, et al. Amphiphilic silane modified multifunctional nanoparticles for magnetically targeted photodynamic therapy. ACS Appl Mater Interfaces. 2017;9(13):11451–11460. doi:10.1021/acsami.7b00647
33. Song X-B, Liu G, Liu F, et al. Autophagy blockade and lysosomal membrane permeabilization contribute to lead-induced nephrotoxicity in primary rat proximal tubular cells. Cell Death Dis. 2017;8(6):e2863–e2863. doi:10.1038/cddis.2017.262
34. Repnik U, Hafner Cesen M, Turk B. Strategies for assaying lysosomal membrane permeabilization. Cold Spring Harb Protoc. 2016;2016(6):
35. Shen Y, Gong S, Li J, et al. Co-loading antioxidant N-acetylcysteine attenuates cytotoxicity of iron oxide nanoparticles in hypoxia/reoxygenation cardiomyocytes. Int J Nanomedicine. 2019;14:6103. doi:10.2147/IJN.S209820
36. Wu Q, Jin R, Feng T, et al. Iron oxide nanoparticles and induced autophagy in human monocytes. Int J Nanomed. 2017;12:3993–4005. doi:10.2147/IJN.S135189
37. Assaraf YG, Brozovic A, Goncalves AC, et al. The multi-factorial nature of clinical multidrug resistance in cancer. Drug Resist Updates. 2019;46:100645. doi:10.1016/j.drup.2019.100645
38. Groth-Pedersen L, Jaattela M. Combating apoptosis and multidrug resistant cancers by targeting lysosomes. Cancer Lett. 2013;332(2):265–274. doi:10.1016/j.canlet.2010.05.021
39. Arami H, Khandhar A, Liggitt D, Krishnan KM. In vivo delivery, pharmacokinetics, biodistribution and toxicity of iron oxide nanoparticles. Chem Soc Rev. 2015;44(23):8576–8607. doi:10.1039/c5cs00541h
40. Frtús A, Smolková B, Uzhytchak M, et al. Analyzing the mechanisms of iron oxide nanoparticles interactions with cells: a road from failure to success in clinical applications. J Controlled Release. 2020;328:59–77. doi:10.1016/j.jconrel.2020.08.036
41. Malvindi MA, De Matteis V, Galeone A, et al. Toxicity assessment of silica coated iron oxide nanoparticles and biocompatibility improvement by surface engineering. PLoS One. 2014;9(1):e85835. doi:10.1371/journal.pone.0085835
42. Singh N, Jenkins GJS, Asadi R, Doak SH. Potential toxicity of superparamagnetic iron oxide nanoparticles (SPION). Nano Rev. 2010;1(1):5358. doi:10.3402/nano.v1i0.5358
43. Huang H, Zhou M, Ruan L, et al. AMPK mediates the neurotoxicity of iron oxide nanoparticles retained in mitochondria or lysosomes. Metallomics. 2019;11(7):1200–1206. doi:10.1039/c9mt00103d
44. Ren X, Chen Y, Peng H, et al. Blocking autophagic flux enhances iron oxide nanoparticle photothermal therapeutic efficiency in cancer treatment. ACS Appl Mater Interfaces. 2018;10(33):27701–27711. doi:10.1021/acsami.8b10167
45. Zhu L, Zhou Z, Mao H, Yang L. Magnetic nanoparticles for precision oncology: theranostic magnetic iron oxide nanoparticles for image-guided and targeted cancer therapy. Nanomedicine. 2017;12(1):73–87.
46. Khan JA, Mandal TK, Das TK, Singh Y, Pillai B, Maiti S. Magnetite (Fe3O4) nanocrystals affect the expression of genes involved in the TGF-beta signalling pathway. Mol Biosyst. 2011;7(5):1481–1486. doi:10.1039/c0mb00192a
47. Sarin N, Engel F, Rothweiler F, et al. Key players of cisplatin resistance: towards a systems pharmacology approach. Int J Mol Sci. 2018;19(3):767. doi:10.3390/ijms19030767
48. Mattauch S, Sachs M, Behrens J. Liprin-alpha4 is a new hypoxia-inducible target gene required for maintenance of cell-cell contacts. Exp Cell Res. 2010;316(17):2883–2892. doi:10.1016/j.yexcr.2010.06.022
49. Yamasaki A, Nakayama K, Imaizumi A, et al. Liprin-α4 as a possible new therapeutic target for pancreatic cancer. Anticancer Res. 2017;37(12):6649–6654. doi:10.21873/anticanres.12122
50. Onishi H, Yamasaki A, Nakamura K, et al. Liprin-α4 as a new therapeutic target for SCLC as an upstream mediator of HIF1α. Anticancer Res. 2019;39(3):1179–1184. doi:10.21873/anticanres.13227
51. Chen YF, Chiu WT, Chen YT, et al. Calcium store sensor stromal-interaction molecule 1-dependent signaling plays an important role in cervical cancer growth, migration, and angiogenesis. Proc Natl Acad Sci USA. 2011;108(37):15225–15230. doi:10.1073/pnas.1103315108
52. Naser R, Aldehaiman A, Diaz-Galicia E, Arold ST. Endogenous control mechanisms of FAK and PYK2 and their relevance to cancer development. Cancers. 2018;10(6):196. doi:10.3390/cancers10060196
53. Al-Juboori SI, Vadakekolathu J, Idri S, et al. PYK2 promotes HER2-positive breast cancer invasion. J Exp Clin Cancer Res. 2019;38(1):210. doi:10.1186/s13046-019-1221-0
54. Lim ST, Miller NL, Nam JO, Chen XL, Lim Y, Schlaepfer DD. Pyk2 inhibition of p53 as an adaptive and intrinsic mechanism facilitating cell proliferation and survival. J Biol Chem. 2010;285(3):1743–1753. doi:10.1074/jbc.M109.064212
55. Stanzione R, Picascia A, Chieffi P, et al. Variations of proline-rich kinase Pyk2 expression correlate with prostate cancer progression. Lab Invest. 2001;81(1):51–59. doi:10.1038/labinvest.3780211
56. Mendoza-Rodriguez MG, Ayala-Sumuano JT, Garcia-Morales L, Zamudio-Meza H, Perez-Yepez EA, Meza I. IL-1beta inflammatory cytokine-induced TP63 isoform NP63alpha signaling cascade contributes to cisplatin resistance in human breast cancer cells. Int J Mol Sci. 2019;20(2):270. doi:10.3390/ijms20020270
57. De Simone V, Bevivino G, Sedda S, et al. Smad7 knockdown activates protein kinase RNA-associated eIF2α pathway leading to colon cancer cell death. Cell Death Dis. 2017;8(3):e2681–e2681. doi:10.1038/cddis.2017.103
58. Kaczorowski M, Biecek P, Donizy P, Pieniazek M, Matkowski R, Halon A. SMAD7 is a novel independent predictor of survival in patients with cutaneous melanoma. Transl Res. 2019;204:72–81. doi:10.1016/j.trsl.2018.09.002
59. Tyagi N, Song YH, De R. Recent progress on biocompatible nanocarrier-based genistein delivery systems in cancer therapy. J Drug Target. 2019;27(4):394–407. doi:10.1080/1061186X.2018.1514040
60. Si HY, Li DP, Wang TM, et al. Improving the anti-tumor effect of genistein with a biocompatible superparamagnetic drug delivery system. J Nanosci Nanotechnol. 2010;10(4):2325–2331. doi:10.1166/jnn.2010.1913
61. Patel P, Meghani N, Kansara K, Kumar A. Nanotherapeutics for the treatment of cancer and arthritis. Curr Drug Metab. 2019;20(6):430–445. doi:10.2174/1389200220666181127102720
62. Pund S, Pawar S, Gangurde S, Divate D. Transcutaneous delivery of leflunomide nanoemulgel: mechanistic investigation into physicomechanical characteristics, in vitro anti-psoriatic and anti-melanoma activity. Int J Pharm. 2015;487(1):148–156. doi:10.1016/j.ijpharm.2015.04.015
63. Muz B, Buggio M, Azab F, et al. PYK2/FAK inhibitors reverse hypoxia-induced drug resistance in multiple myeloma. Haematologica. 2019;104(7):e310–e313. doi:10.3324/haematol.2018.194688
64. Pasquier J, Gosset M, Geyl C, et al. CCL2/CCL5 secreted by the stroma induce IL-6/PYK2 dependent chemoresistance in ovarian cancer. Mol Cancer. 2018;17(1):47. doi:10.1186/s12943-018-0787-z
65. Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58(2):629–641. doi:10.1002/hep.26369
66. Guo Y, Fang Q, Ma D, et al. Up-regulation of HO-1 promotes resistance of B-cell acute lymphocytic leukemia cells to HDAC4/5 inhibitor LMK-235 via the Smad7 pathway. Life Sci. 2018;207:386–394. doi:10.1016/j.lfs.2018.06.004
67. Jeon WK, Hong HY, Seo WC, et al. Smad7 sensitizes A549 lung cancer cells to cisplatin-induced apoptosis through heme oxygenase-1 inhibition. Biochem Biophys Res Commun. 2012;420(2):288–292. doi:10.1016/j.bbrc.2012.02.151
68. Yuan Y. Spatial heterogeneity in the tumor microenvironment. Cold Spring Harb Perspect Med. 2016;6(8):a026583. doi:10.1101/cshperspect.a026583
69. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–628. doi:10.1016/j.cell.2017.01.018
70. Swetha KL, Roy A. Tumor heterogeneity and nanoparticle-mediated tumor targeting: the importance of delivery system personalization. Drug Deliv Transl Res. 2018;8(5):1508–1526. doi:10.1007/s13346-018-0578-5
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.