Back to Journals » Journal of Experimental Pharmacology » Volume 15
Synergistic Effects of Azithromycin and STING Agonist Promote IFN-I Production by Enhancing the Activation of STING-TBK1 Signaling
Authors Petcharat K, Munkong N, Thongboontho R, Chartarrayawadee W , Thim-Uam A
Received 25 August 2023
Accepted for publication 28 October 2023
Published 1 November 2023 Volume 2023:15 Pages 407—421
DOI https://doi.org/10.2147/JEP.S433181
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Roger Pinder
Kanoktip Petcharat,1 Narongsuk Munkong,2 Rungthip Thongboontho,1 Widsanusan Chartarrayawadee,3 Arthid Thim-Uam1
1Division of Biochemistry, School of Medical Sciences, University of Phayao, Phayao, 56000, Thailand; 2Department of Pathology, School of Medicine, University of Phayao, Phayao, 56000, Thailand; 3Division of Chemistry, School of Science, University of Phayao, Phayao, 56000, Thailand
Correspondence: Arthid Thim-Uam, Division of Biochemistry, School of Medical Sciences, University of Phayao, Phayao, 56000, Thailand, Tel +668 6 216 26 33, Email [email protected]; [email protected]
Background: Azithromycin (AZM) is a macrolide antibiotic that exhibits anti-inflammatory and anti-viral infection properties by enhancing type-I interferon (IFN-I) responses. The stimulator of interferon genes (STING) can directly induce IFN-I production. However, elevated IFN-I induces auto-immune phenotypes such as systemic lupus erythematosus (SLE). The effects of AZM and STING on the production of IFN-I are unclear.
Objective: Therefore, this study aims to evaluate the role of AZM and STING on IFN-I responses in macrophages.
Methods: RAW 264.7 macrophages were treated with AZM with and without a STING-agonist (DMXAA), and the maturation of macrophages was determined using flow cytometry. Gene expression and pro-inflammatory cytokines were analyzed using qPCR and ELISA, respectively. Moreover, protein expression was investigated using Western blot assays and immunofluorescence.
Results: Our results show that AZM significantly induced M1 phenotypes, promoting surface molecule expansion of CD80 and MHC-II and production of IL-6 and TNF-α cytokines on DMXAA-stimulated macrophages. Furthermore, we found that AZM-increased mRNA levels of interferon-stimulated genes (ISGs) could be due to the high expression of STNG-TBK1 signaling in the presence of DMXAA.
Conclusion: Our data suggest that AZM enhancement of IFN-I responses was STING dependent in DMXAA-stimulated macrophages. These data underline a novel approach to AZM action-mediated STING-TBK1 signaling for regulating IFN-I responses and may further augment the scientific basis and potential use of AZM in clinical applications.
Keywords: azithromycin, stimulator of interferon genes, TANK binding kinase 1, type-I interferon
Introduction
Type I interferon (IFN-I) is a type of inflammatory cytokine predominantly produced by innate immune cells, such as plasmacytoid dendritic cells (pDC) and macrophages,1,2 to combat viral infection.3 Furthermore, high circulation of IFN-I levels is associated with pathogenesis of SLE.4,5 The molecular mechanism of IFN-I induction is stimulated through several intracellular DNA, which is recognized by cytosolic DNA binding proteins such as cyclic GMP–AMP synthase (cGAS), DNA-dependent activator of IFN-regulatory factors (DAI), Interferon gamma inducible protein 16 (IFI16), Melanoma differentiation-associated gene 5 (MDA5), Retinoic acid-inducible gene I (RIG-I), and Toll-like receptor 9 (TLR9).6–8 Upon activation, DNA sensors bind to their ligand leading to IFN-I expression. The secreted IFN-I signals through an IFN-I receptor (IFNAR) to stimulate the phosphorylation of JAK-STAT signaling and regulate IFN-γ induction and expression of many interferon-stimulated genes (ISGs),9,10 such as MX Dynamin Like GTPase 1 (Mx1), C-X-C Motif Chemokine Ligand 10 (CXCL10) and Interferon-stimulated gene 15 (ISG15), which can fight against infection.11 Moreover, IFN-I plays a vital role in innate antiviral responses against early COVID-19 infection.12,13
Azithromycin (AZM) belongs to a group of macrolide antibiotics most commonly used for bacterial infection by targeting the 50S ribosomal RNA subunit, which can inhibit bacterial protein synthesis.14,15 It exhibits well-documented anti-inflammatory activity, which can reduce the activation of NF-kB and AP-1 activity in several immune cells, alleviating cytokine-mediated inflammation.16,17 Additionally, azithromycin may facilitate the M2 phenotype of macrophages under lupus stimulation by attenuating the PI3K/Akt signaling pathway in vitro.18,19 AZM also enhances the expression of type I and II interferons and ISGs, which can mitigate rhinoviral20 and Zika virus infection21 by upregulating expression of RIG-I and MDA5 and boosting the activation of downstream TBK1 and IRF3 proteins. Currently, several studies have shown the immunomodulatory effect of AZM on antiviral effects in patients with COVID-1922–24 by up-regulation of IFN-I production through IRF3 signaling.22 Likewise, AZM was demonstrated to suppress the binding between angiotensin-converting enzyme 2 (ACE2) and spike protein of SARS-CoV-2, which can reduce viral infection.25 Moreover, AZM can reduce the SARS-CoV-2 viral replication26 lysosomal pH change, leading to uncoating envelopes of viruses.24
The Stimulator of Interferon Genes (STING) is a cytosolic DNA sensor that is recognized by intracellular DNA-mediated IFN-I responses and inflammatory cytokines,27 and which signals through the phosphorylation of TANK-binding kinase 1 (TBK1) and interferon regulatory factor 3 (IRF3) to fight against microbial infection.28–30 STING activation-mediated IFN-I and ISG expression can also respond to DNA and RNA viral infection.31,32 It has been shown that STING is a major driver of IFN-I and ISG production, which can cause SLE through activation of dendritic cells.33 Moreover, activation of STING can induce the maturation of macrophages and produce cytokines, which can stimulate T cell activation.34 Several studies revealed that STING agonists, such as 5.6-dimethylxanthenone-4-acetic acid (DMXAA) and derivatives, interacted directly with STING and enhanced the IFN-I production to anti-cancer properties35,36 and against viral infection.37 Furthermore, activation of STING by DMXAA can promote severe SLE,33 Rheumatoid arthritis38,39, and Sjögren’s syndrome40 in vivo study by increasing IFN-I production. Therefore, the induction of IFN-I with the combination of AZM and STING agonists-DMXAA could be elucidated to understand the mechanism in autoimmune diseases. Interestingly, current the literature has demonstrated the relevance of the STING signaling pathway associated with type I IFN responses in COVID-19.41 Moreover, the STING agonist has shown potent inhibition of SARS-CoV-2 infection.42
The association of AZM with the STING-TBK1 signaling pathway regarding the regulation of IFN-I and ISGs responses is still lacking data. Therefore, we have focused on the role of AZM action via the STING signaling pathway by DMXAA activation to increase the scientific basis of information regarding AZM regulation of IFN-I exhibition. These data may be a profitable restoration target for STING-mediated IFN-I production, which could be used in clinical applications.
Materials and Methods
Chemicals
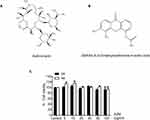
Azithromycin was purchased from Sigma (Sigma-Aldrich, Darmstadt, Germany). STING agonist (DMXAA), namely 5.6-Dimethylxanthenone-4-acetic acid, was purchased from Invitrogen (InvivoGen, San Diego, CA, USA). The chemical structure of these two reagents is presented in Figures 1A and B.
Cell Culture
RAW 264.7 macrophages were cultured in DMEM high glucose medium supplemented with 10% fetal bovine serum (HyCclone, Logan, UT, USA), and a final concentration of 1X anti-antibiotics (Gibco–Thermo Fisher Scientific, MA, USA). Cells were cultured in an incubator at 37°C with 5% CO2. For stimulation, 80 ug/mL of azithromycin and 10 ug/mL of DMXAA were added to the cells for 24 hours. DMSO-treated cells (Sigma-Aldrich, Darmstadt, Germany) were used as the control in the following experiments.
Determination of Cell Viability by the MTS Assay
200 µL (2x104 cells) of RAW 264.7 macrophages were cultured in 96 well-culture plates and maintained in an incubator at 37°C and 5% CO2 for 24 hr. The drug cytotoxicity assessment of AZM was performed using the MTS assay (Promega, cat. G3580, Madison, USA). In brief, RAW 264.7 macrophages were added at various doses of AZM between 5–100 ug/mL for 24 and 48 hours. Then, cells were washed with a complete medium. After washing, each sample was added with 100 µL of complete medium. Next, 20 µL of MTS reagents were added and incubated for 3 hours in an incubator. Then, the reaction signals were observed using a microplate reader at 490 nm (Thermo Fisher Scientific, MA USA). The percentage of cell viability was evaluated using DMSO (vehicle)-treated cells as the control.
Determination of Luciferase Activity
STING+/+ and STING−/− RAW-Lucia ISG cells (InvivoGen, San Diego, CA, USA) were cultured in a DMEM high glucose solution containing 10% fetal bovine serum, 2 Mm L-glutamine (HyCclone, Logan, UT, USA), 100 µg/mL Zeocin and 50 µg/mL Normocin (InvivoGen, San Diego, CA, USA) in an incubator, as described above. Next, STING wild-type and knockout cells were stimulated with 40 µg/mL and 80 µg/mL of AZM with and without 10 µg/mL of DMXAA for 3, 24, and 48 h. The luciferase activity in the culture medium was observed with the QUANTI-Luc luciferase detection kit according to the manufacturer’s protocol (InvivoGen, San Diego, CA, USA). In brief, 20 µL of culture medium from all experiments was collected in the black plate. Then, 50 µL of luciferase detector was added to each sample. Finally, luminescence activity was immediately measured by microplate reader (Thermo Fisher Scientific, MA USA).
Determination of Mature Macrophages
To determine mature macrophages, 1×106 cells of macrophages were cultured in a 6-well plate and stimulated with 80 µg/mL of AZM with and without 10 µg/mL of DMXAA for 24 h. Moreover, to confirm the effect of DMXAA on mature phenotypes of macrophages via STING signaling, 10 µg/mL of DMXAA and 1 µg/mL of LPS (Sigma-Aldrich, Darmstadt, Germany) were treated in STING-/- RAW-Lucia ISG cells for 24 h. Next, activated cells were collected and stained with anti-mouse F4/80 (clone: BM8), anti-mouse I-Ab (clone: AF6-120.1; cat. 116406), and anti-mouse CD80 (clone: 16–10A1; cat. 104733) (Bio Legend, San Diego, CA, USA). Fixable Viability Dye eFluor 780 (Thermo Fisher Scientific, MA USA) was used to stain excluded dead cells. Then, the maturation of stained macrophages was investigated using a BD LSR-II flow cytometer (BD Biosciences, North Brunswick, NJ, USA). The percentage of activated macrophages was evaluated with FlowJo software (version 10, USA).
Determination of Cytokines Production
To determine cytokine production, 1×106 STING+/+ and STING−/− RAW-Lucia ISG cells were cultured in the 6-well plate described above. Cells were treated with 80 µg/mL of AZM with or without 10 µg/mL of DMXAA in an incubator at 37°C at 5% CO2 for 24 h. IL-6 and TNF-α were then investigated in the supernatant using ELISA MAX Deluxe Set Mouse IL-6 (cat. 431304) and ELISA MAX Deluxe Set Mouse TNF-α (cat. 430904) (Bio Legend, San Diego, CA, USA) following the manufacturer’s protocol. Finally, the signals were measured using a microplate reader at 450 nm (Thermo Fisher Scientific, MA USA). The concentration of cytokines was calculated and presented as pg/mL.
Determination of Genes Expression
mRNA expression of interferon-inducible genes was investigated in 80 ug/mL of AZM-treated cells with and without 10 ug/mL of DMXAA. In brief, total RNA from all experiments was extracted using Trizol reagent (InvivoGen, San Diego, CA, USA) followed by RNA purification using the RNeasy mini kit (QIAGEN, Germantown, MD, USA) according to the manufacturer’s instructions. Next, cDNA synthesis from 1 µg of total RNA was performed using iScript RT Supermix (Bio-Rad, Hercules, CA, USA). The mRNA expression of genes of interest was conducted with quantitative PCR using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) and Applied Biosystems 7500 (Applied Biosystems). β-actin was used to normalize relative mRNA expression. The fold induction was calculated using the 2(−ddCt) formula. All primers are shown in Table 1.
 |
Table 1 The Primer Sequences for Quantitative Real Time-PCR |
Determination of Protein Expression
1 × 106 cells of RAW 264.7 macrophages were cultured in a 6-well plate, and then 80 ug/mL AZM was added with and without DMXAA for 180 min. Activated cells were lysed for 30 min on ice with a lysis buffer (10 mM Tris-HCL, 1% Triton-X100, 150 mM NaCl, 5 mM, and EDTA pH 8). Then, protein from the cell lysates was collected by centrifuging at 13,000 × g for 15 min at 4°C. The protein was collected from the supernatant. The amount of protein was measured using BCA assay (Thermo Fisher Scientific, Waltham, MA, USA). Next, total protein (25 µg) was separated on a 10% polyacrylamide gel. Then, proteins from the gel were transferred into nitrocellulose membranes and the antibodies’ unspecific binding was blocked with a blocking buffer for 1 h and then probed with Total TBK1 antibody (clone: D1B4 cat: 3504S, 1:1000) and Phospho-TBK1/NAK (Ser172) (clone: D52C2 cat: 5483S, 1:1000), Total STAT1 antibody (clone: 42H3 cat: 9175S, 1:1000), Phospho-Stat1 (Tyr701) (clone: D4A7 cat: 7649S, 1:1000) and β-ACTIN (clone: 13E5 cat: 4970S, 1:2500) (Cell Signaling, Danvers, MA, USA) overnight at 4°C. After washing with 1X TBS-T, membranes fluorescent secondary antibody IRDye 680RD donkey anti-rabbit IgG (1:10000) (LI-COR, Lincoln, NE, USA) was added for 1 h in the dark at room temperature. Finally, the targeted protein from the membrane was detected with ODYSSEY CLx (LI-COR, Lincoln, NE, USA). The quantitative intensity of the proteins was calculated with the ODYSSEY CLx program.
Immunofluorescence Staining
1 × 105 cells of macrophages were seeded in a cell culture slide. Cells were treated with AZM with and without DMXAA for 3 h. Then, 200 µL of 10% formalin (Sigma-Aldrich, Darmstadt, Germany) was added for 10 min at room temperature. Next, 200 µL of 0.2% Triton X-100 in PBS was added to the fixed cells for 10 min, followed by addition of 5% BSA in PBS for 1 h to block unspecific binding. Fixed macrophages were probed with STING antibodies (clone: D2P2F cat: 13647, 1:200) and Phospho-TBK1/NAK (Ser172)(clone: D52C2 cat: 5483S, 1:200) (Cell Signaling, Danvers, MA, USA) at 4°C for overnight. Then, secondary antibodies of Alexa Fluor 488 rabbit IgG (Thermo Fisher Scientific, Waltham, MA, USA) were added to stained cells in the dark for 1 h. Finally, DNA was stained with 1 µM DAPI in the dark (Thermo Fisher Scientific, Waltham, MA, USA) for 5 min, and fluorescence signals were observed by confocal microscope (Carl Zeiss LMS800, Oberkochen, Germany).
Statistical Analysis
All data are presented as mean ± standard error (SEM). Statistical analyses between multiple groups were performed with the one-way ANOVA analysis, and the comparison between groups was performed with a two-tailed Mann–Whitney using GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA). The statistically significant data with P < 0.05 were considered.
Results
Cell Cytotoxicity of Azithromycin
We first examined drug cytotoxicity effects in macrophages for 24 and 48 hr using MTS assay. The results indicate that cell viabilities were not significantly affected by azithromycin at different concentration between 5–100 µg/mL relative to the untreated cells, as shown in Figure 1C.
Azithromycin Increases Interferon Secretion in DMXAA-Induced Macrophages
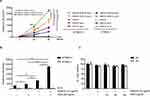
Next, we determined the effect of azithromycin on IFN-I production in DMXAA-stimulated macrophages, which was produced through activation of the STING/TBK1/IRF3 pathway. Our data indicate that AZM significantly increased the levels of Lucia luciferase in DMXAA-stimulated RAW-Lucia ISG cells relative to the DMXAA-treated and untreated cells (Figure 2A). However, AZM did not induce luciferase secretion in the culture medium of STING-/- RAW-Lucia ISG cells after 3 and 24 hr of stimulation. Interestingly, AZM alone was able to induce IFN-I production after 48 hr of activation (Figure 2B). Moreover, AZM-treated cells with DMXAA stimulation did not affect cell viability (Figure 2C). These results suggest that AZM-mediated IFN-I production by synergistic STING/TBK1/IRF3 signaling.
Azithromycin Enhances the Maturation of DMXAA-Stimulated Macrophages
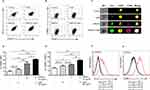
In order to determine whether AZM affects the maturation of macrophages after co-treatment with and without DMXAA, the surface molecules of macrophages were stained with CD80 and an I-Ab, indicating the mature macrophages. The results revealed that DMXAA-stimulated cells exhibited a significant expansion of I-Ab and CD80 relative to untreated cells. Interestingly, co-treatment with DMXAA and AZM significantly increased these activated markers compared to treatment with DMXAA alone, as shown in Figure 3A–E. Moreover, we confirmed the AZM-mediated maturation of macrophages using flow cytometry in STING-/- RAW-Lucia ISG cells. The results showed that AZM did not affect the activation of DMXAA-induced STING-/- cells, while LPS was able to induce this activation in STING-/- cells (Figure 3F and G). These data suggest that the synergy of AZM and DMXAA induced the maturation of macrophages.
AZM Increases Cytokines Production in DMXAA-Stimulated Macrophages
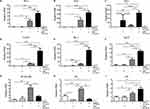
Levels of IL-6 and TNF-α were significantly higher in DMXAA-stimulated cells after 24 hours than in AZM- and untreated cells. Furthermore, co-treatment of AZM and DMXAA showed a significantly increase in these proinflammatory cytokines to DMXAA alone. However, inflammatory cytokine levels of STING-/- cells did not differ significantly from those of AZM with or without DMXAA- and untreated controls, as presented in Figures 4A and B.
Effects of AZM on Interferon Signature Genes (ISG) Expression
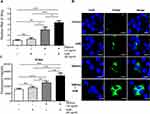
Our data show that co-treatment with AZM and DMXAA led to significant induction of ISG mRNA levels after 24 hours of activation in macrophages relative to DMXAA-treated and untreated controls, including Ifn-α, Ifn-β, Cxcl10, Mx-1, and Isg15. Nevertheless, AZM-treated cells did not show significant induction of ISG expression compared to untreated cells (Figure 5A–E). Interestingly, DMXAA-stimulated cells exhibited a significantly higher mRNA expression of NF-kβ p65 and Irf7 than DMXAA-treated cells with AZM (Figure 5G and H). In contrast, we found a significant increase in Irf3 expression in these groups, as presented in Figure 5I.
Effect of AZM on STING Activation
It is well established that STING is essential to initiation of IFN-I production. Here, an increase in IFN-I as a result of synergistic AZM and STING-TBK1 was observed. The data show that the mRNA level of Sting after co-treatment with AZM and DMXAA was significantly higher than in DMXAA-treated and untreated cells (Figure 6A). In addition, the activation of STING was confirmed by immunofluorescence staining using confocal microscopy. The results show a significantly higher expression of STING following co-treatment with AZM and DMXAA relative to DMXAA-treated and untreated cells, as shown in Figure 6B and C.
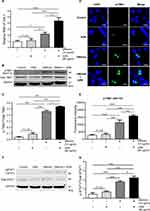
Effect of AZM on TBK-1 Activation
Next, the activation of TBK-1, which is downstream of STING signaling, was investigated. The data show a significantly higher transcription of mRNA levels of Tbk1 in co-treatment of AZM and DMXAA compared with DMXAA-treated and untreated cells (Figure 7A). Moreover, the phosphorylation of TBK1 (Ser172) after 3 hr of activation by AZM with or without DMXAA was observed. Cells treated with a combination of AZM and DMXAA showed a significantly higher expression of pTBK-1 relative to cells treated with DMXAA alone (Figure 7B and C). Additionally, we confirmed this activation by immunofluorescence using confocal microscopy. The data demonstrated a significantly higher expression of pTBK-1 in the co-treatment of AZM and DMXAA than in DMXAA-treated cells with Western blot assay (Figures 7D and E). These data suggested that AZM induced IFN-I production by activating the STING/TBK1 pathway.
Effect of AZM on STAT1 Signaling
To examine the mechanism of ISG expression, mainly caused by the activation of IFN- I. The phosphorylation of STAT1 (Tyr701) after 3 hr of activation by AZM with or without DMXAA was investigated. Macrophages treated with a combination of AZM and DMXAA exhibited a significantly higher expression of pSTAT-1 than in cells treated with DMXAA alone (Figure 7F and G). These results demonstrated that a combination of AZM and DMXAA induced IFN-I production through STING/TBK1/ signaling, leading to the expression of ISGs via the activation of STAT1 signaling.
Discussion
Azithromycin is an antibiotic agent14 exhibiting anti-inflammatory properties in macrophages.19,43 Moreover, AZM can induce IFN-I secretion against viral infection.21,44 It is well documented that the activation of cGAS-STING by intracellular DNA regulates IFN-I production.27 Our previous data have shown that STING may increase levels of IFN-I, leading to autoimmune phenotypes, such as SLE33 and rheumatoid arthritis.39 Thus, we further investigated whether STING, which is mainly expressed in macrophages, can be affected by treatment with AZM.
Our results showed that co-treatment with AZM and DMXAA increased IFN-I release in the culture medium. Moreover, we detected the expansion of CD80 and I-Ab (MHC-II) molecules, suggesting the maturation of macrophages as the antigen-presenting cells (APCs) in AZM with DMXAA-treated cells. These data suggest that AZM and DMXAA-treated cell-induced IFN-I responses and secrets into the culture medium, which binds to IFN-I receptors (IFNAR) and subsequently increases the expansion of co-stimulatory molecules CD8045 via regulation of interferon regulatory factor-1 (IRF1).46 Moreover, increasing IFN-I in a culture environment can induce MHC II expression10,33,47 by inducing the phosphorylation of STAT1, which is generally stimulated IFN-γ production,10,48 leading to enhancement of surface molecules of MHC-II on macrophages.49 Additionally, our results show that AZM enhanced mRNA levels of IFN-γ in DMXAA-treated cells. This may be due to IFN-γ activation via receptors (IFNGR) being dependent on IFNAR signaling.9,50 In contrast, AZM can reduce the expression of co-stimulatory molecules CD80 and MHC class II molecules in LPS-stimulated DCs by inhibiting the translocation of NF-κB.51 These data suggest that azithromycin alternatively promotes the maturation of macrophage phenotypes via STING-dependent IFN-I signaling.
The previous data have shown that AZM is an anti-inflammatory drug and attenuates pro-inflammatory cytokine production by reducing NF-κB and AP-1 activation in LPS-induced cells.16,17 Consistency with previous results indicated that AZM can reduce the expression of NF-κB17 and Irf752 in DMXAA-activated cells, which stimulates the induction of pro-inflammatory cytokines. However, we found that the pro-inflammatory cytokines IL-6 and TNF-α were significantly enhanced in the AZM and DMXAA-cotreated cells. These data might be due to elevated IFN-I and IFN-γ, which stimulate the polarization of M1 macrophages and enhance cytokine production through mediated IFN-γ /IRF1/IRF5 signaling.53–55 Additionally, the releasing of IFN-γ by AZM with DMXAA can activate the STAT1 signaling through the IFN-γ receptor (IFNGR), leading to the phosphorylation of STAT1 at Tyr701, resulting in translocation into the nucleus and initiating the induction of TNF-α and IL-6 production.56 Thus, our data suggest combining DMXAA and AZM enhances cytokine production through STING-dependent IFN- γ/STAT1 signaling. Moreover, AZM administration on P. aeruginosa-infected mice increased levels of IFN-γ, resulting in decreased bacterial infection.57 However, we also observed that AZM alone did not promote the levels of IL-6 and TNF-α in the culture medium. These results imply that a combination of AZM and DMXAA induces IFN-I and IFN-II responses that lead to establishment M1 phenotypes of macrophages that are STING-dependent.
Furthermore, we found high expression of Ifn-α, Ifn-β, Ifn- γ and interferon-inducible genes (ISGs), including Cxcl10, Mx1, and Isg15 in the AZM and DMXAA-cotreated cells, while cells treated with AZM alone did not affect the mRNA levels of these genes. Similarly, AZM can enhance IFN-I and ISGs gene expression in rhinovirus (RV) 1B-infected cells.20,58,59 These results might be due to the STING-dependent IFN-I and IFN-γ production, which induce ISGs expression through STAT1 signaling.10,60 AZM has also been shown to increase the expression of pathogen recognition receptors (PRRs), including RIG-I and MDA5, which induce ISGs expression in HT-29 cells after ZIKV infection.21 These results indicate that the effect of AZM-associated IFN-I and ISGs signaling could temporarily augment inflammatory response in the presence of DMXAA.
Nevertheless, the effects of AZM on STING signaling-mediated IFN-I responses in macrophages has not yet been clarified. Thus, we further investigated these effects. Our results showed that AZM could upregulate mRNA levels of Sting in DMXAA-stimulated cells consistently with high expression of STING by immunofluorescence. AZM has previously been reported to increase the levels of phosphorylated TBK1 and IRF3, downstream of RIG-I and MDA5 after stimulation by viral infection.20,21,26 Here, we found that AZM increased the phosphorylation of TBK1, which was activated by STING,29,61 leading to induction of IFN-I responses in the presence of DMXAA-stimulated macrophages. It is well-recognized that AZM is a weakly basic drug that accumulates in the lysosome, leading to lysosome function change, which can induce apoptosis62,63 and mitochondrial DNA leakage.64 These effects of AZM might stimulate the cGAS-STING pathway65 and increase the activation by combination with STING agonist (DMXAA). Our results showed that AZM-treated cell alone at 24 hr did not affect this mechanism. However, AZM-treated cells at 48 hr can enhance IFN-I luciferase activity via STING signaling compared with untreated cells (Figure 2B).
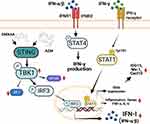
These results suggest that the combination of AZM and DMXAA may alternatively affect activation of IFN-I via STING/TBK1/IRF3 signaling, resulting in enhance the activation of interferon-stimulated genes and pro-inflammatory cytokines through STING-dependent IFN-γ/STAT1 pathway, as presented in Figure 8. In this study, we provide evidence for a probable new mechanism of azithromycin. Our study also showed that the efficacy of AZM on IFN-I response and ISG signaling on the regulation of macrophages was STING dependent.
Conclusion
In summary, Azithromycin is an antibiotic drug that exerts anti-inflammatory activity. We hypothesize that AZM could inhibit IFN-I via STING/TBK1/IRF3 signaling, which might be a targeted treatment for lupus. In contrast, the combination of AZM and DMXAA reveals the synergistic effects on the IFN-I responses by enhancing the activation of STING expression. This activation increases the phosphorylation of downstream TBK-1 and might stimulate IRF3 to translocate to the nucleus. Therefore, these effects increased interferon-stimulated gene expression, including Cxcl10, Isg15, and Mx1, through IFN-γ/STAT1 signaling. These data emphasize a novel approach to AZM use and may further augment the clinical application of AZM in terms of activating via the cGAS-STING pathway.
Ethical Approval
In this study, no ethics approval was required. RAW-Lucia ISG cells mouse macrophages (STING+/+; cat. Rawl-isg) and RAW-Lucia ISG-KO-STING cells mouse macrophages (STING-/-; cat. Rawl-kostg) were purchased from InvivoGen (InvivoGen, San Diego, CA, USA).
Acknowledgments
This research was funded by the Thailand Science Research and Innovation Fund and the University of Phayao, grant number FF65-RIM104.
Disclosure
The authors declare no conflict of interest.
References
1. Ali S, Mann-Nüttel R, Schulze A, Richter L, Alferink J, Scheu S. Sources of type I interferons in infectious immunity: plasmacytoid dendritic cells not always in the driver’s seat. Front Immunol. 2019;10:778. doi:10.3389/fimmu.2019.00778
2. Murira A, Lamarre A. Type-I interferon responses: from friend to foe in the battle against chronic viral infection. Front Immunol. 2016;7:609. doi:10.3389/fimmu.2016.00609
3. Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25(3):373–381. doi:10.1016/j.immuni.2006.08.007
4. Melissa N, Sarah J, Rachel K, et al. Type 1 interferon status in systemic lupus erythematosus: a longitudinal analysis. Lupus Sci Med. 2022;9(1):e000625. doi:10.1136/lupus-2021-000625
5. Psarras A, Emery P, Vital EM. Type I interferon–mediated autoimmune diseases: pathogenesis, diagnosis and targeted therapy. Rheumatology. 2017;56(10):1662–1675. doi:10.1093/rheumatology/kew431
6. Keating SE, Baran M, Bowie AG. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32(12):574–581. doi:10.1016/j.it.2011.08.004
7. Lukhele S, Boukhaled GM, Brooks DG. Type I interferon signaling, regulation and gene stimulation in chronic virus infection. Semin Immunol. 2019;43:101277. doi:10.1016/j.smim.2019.05.001
8. Zahid A, Ismail H, Li B, Jin T. Molecular and structural basis of DNA sensors in antiviral innate immunity. Front Immunol. 2020;11:613039. doi:10.3389/fimmu.2020.613039
9. Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. doi:10.1038/nri1604
10. Lee AJ, Ashkar AA. The dual nature of type I and type II interferons. Front Immunol. 2018;9:2061. doi:10.3389/fimmu.2018.02061
11. Ashley CL, Abendroth A, McSharry BP, Slobedman B. Interferon-independent upregulation of interferon-stimulated genes during human cytomegalovirus infection is dependent on IRF3 expression. Viruses. 2019;11(3):246. doi:10.3390/v11030246
12. Park A, Iwasaki A. Type I and type III interferons – induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27(6):870–878. doi:10.1016/j.chom.2020.05.008
13. Lucas C, Wong P, Klein J, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi:10.1038/s41586-020-2588-y
14. Heidary M, Ebrahimi Samangani A, Kargari A, et al. Mechanism of action, resistance, synergism, and clinical implications of azithromycin. J Clin Lab Anal. 2022;36(6):e24427. doi:10.1002/jcla.24427
15. Jednačak T, Mikulandra I, Novak P. Advanced methods for studying structure and interactions of macrolide antibiotics. Int J Mol Sci. 2020;21(20):7799. doi:10.3390/ijms21207799
16. Cigana C, Nicolis E, Pasetto M, Assael BM, Melotti P. Anti-inflammatory effects of azithromycin in cystic fibrosis airway epithelial cells. Biochem Biophys Res Commun. 2006;350(4):977–982. doi:10.1016/j.bbrc.2006.09.132
17. Aghai ZH, Kode A, Saslow JG, et al. Azithromycin suppresses activation of nuclear factor-kappa b and synthesis of pro-inflammatory cytokines in tracheal aspirate cells from premature infants. Pediatr Res. 2007;62(4):483–488. doi:10.1203/PDR.0b013e318142582d
18. Wang J, Chen Q, Zhang Z, et al. Azithromycin alleviates systemic lupus erythematosus via the promotion of M2 polarisation in lupus mice. Cell Death Discov. 2021;7(1):82. doi:10.1038/s41420-021-00466-4
19. Wang J, Xie L, Wang S, Lin J, Liang J, Xu J. Azithromycin promotes alternatively activated macrophage phenotype in systematic lupus erythematosus via PI3K/Akt signaling pathway. Cell Death Dis. 2018;9(11):1080. doi:10.1038/s41419-018-1097-5
20. Gielen V, Johnston SL, Edwards MR. Azithromycin induces anti-viral responses in bronchial epithelial cells. Eur Respir J. 2010;36(3):646–654. doi:10.1183/09031936.00095809
21. Li C, Zu S, Deng YQ, et al. Azithromycin protects against zika virus infection by upregulating virus-induced type I and III interferon responses. Antimicrob Agents Chemother. 2019;63(12):e00394–19. doi:10.1128/AAC.00394-19
22. Venditto VJ, Haydar D, Abdel-Latif A, et al. Immunomodulatory effects of azithromycin revisited: potential applications to COVID-19. Front Immunol. 2021;12:574425. doi:10.3389/fimmu.2021.574425
23. Sultana J, Cutroneo PM, Crisafulli S, Puglisi G, Caramori G, Trifirò G. Azithromycin in COVID-19 patients: pharmacological mechanism, clinical evidence and prescribing guidelines. Drug Saf. 2020;43(8):691–698. doi:10.1007/s40264-020-00976-7
24. Kournoutou GG, Dinos G. Azithromycin through the lens of the COVID-19 treatment. Antibiotics. 2022;11(8):1063. doi:10.3390/antibiotics11081063
25. Khezri MR, Zolbanin NM, Ghasemnejad-berenji M, Jafari R. Azithromycin: immunomodulatory and antiviral properties for SARS-CoV-2 infection. Eur J Pharmacol. 2021;905:174191. doi:10.1016/j.ejphar.2021.174191
26. Oliver ME, Hinks TSC. Azithromycin in viral infections. Rev Med Virol. 2021;31(2):e2163. doi:10.1002/rmv.2163
27. Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi:10.1038/nature08476
28. Zhang C, Shang G, Gui X, Zhang X, X-c B, Chen ZJ. Structural basis of STING binding with and phosphorylation by TBK1. Nature. 2019;567(7748):394–398. doi:10.1038/s41586-019-1000-2
29. Yum S, Li M, Fang Y, Chen ZJ. TBK1 recruitment to STING activates both IRF3 and NF-κB that mediate immune defense against tumors and viral infections. Proc Natl Acad Sci U S A. 2021;118(14):e2100225118. doi:10.1073/pnas.2100225118
30. Ahn J, Barber GN. STING signaling and host defense against microbial infection. Exp Mol Med. 2019;51(12):1–10. doi:10.1038/s12276-019-0333-0
31. Singh RS, Vidhyasagar V, Yang S, et al. DDX41 is required for cGAS-STING activation against DNA virus infection. Cell Rep. 2022;39(8):110856. doi:10.1016/j.celrep.2022.110856
32. Franz KM, Neidermyer WJ, Tan Y-J, Whelan SPJ, Kagan JC. STING-dependent translation inhibition restricts RNA virus replication. Proc Natl Acad Sci U S A. 2018;115(9):E2058–E2067. doi:10.1073/pnas.1716937115
33. Thim-uam A, Prabakaran T, Tansakul M, et al. STING mediates lupus via the activation of conventional dendritic cell maturation and plasmacytoid dendritic cell differentiation. iScience. 2020;23(9):101530. doi:10.1016/j.isci.2020.101530
34. Vieira R, Nascimento MS, Noronha IH, et al. STING signaling drives production of innate cytokines, generation of CD8+ T cells and enhanced protection against Trypanosoma cruzi infection. Front Immunol. 2022;12:775346. doi:10.3389/fimmu.2021.775346
35. Luo K, Li N, Ye W, Gao H, Luo X, Cheng B. Activation of Stimulation of Interferon Genes (STING) signal and cancer immunotherapy. Molecules. 2022;27(14):4638. doi:10.3390/molecules27144638
36. Corrales L, Glickman Laura H, McWhirter Sarah M, et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11(7):1018–1030. doi:10.1016/j.celrep.2015.04.031
37. Cerón S, North BJ, Taylor SA, Leib DA. The STING agonist 5,6-dimethylxanthenone-4-acetic acid (DMXAA) stimulates an antiviral state and protects mice against herpes simplex virus-induced neurological disease. Virology. 2019;529:23–28. doi:10.1016/j.virol.2019.01.006
38. Shen H, Jin L, Zheng Q, et al. Synergistically targeting synovium STING pathway for rheumatoid arthritis treatment. Bioact Mater. 2023;24:37–53. doi:10.1016/j.bioactmat.2022.12.001
39. Tansakul M, Thim-uam A, Saethang T, et al. Deficiency of STING promotes collagen-specific antibody production and B cell survival in collagen-induced arthritis. Front Immunol. 2020;11:1101. doi:10.3389/fimmu.2020.01101
40. Papinska J, Bagavant H, Gmyrek GB, et al. Activation of Stimulator of Interferon Genes (STING) and sjögren syndrome. J Dent Res. 2018;97(8):893–900. doi:10.1177/0022034518760855
41. Domizio JD, Gulen MF, Saidoune F, et al. The cGAS–STING pathway drives type I IFN immunopathology in COVID-19. Nature. 2022;603(7899):145–151. doi:10.1038/s41586-022-04421-w
42. Li M, Ferretti M, Ying B, et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci Immunol. 2021;6(59):eabi9007. doi:10.1126/sciimmunol.abi9007
43. Banjanac M, Munić Kos V, Nujić K, et al. Anti-inflammatory mechanism of action of azithromycin in LPS-stimulated J774A.1 cells. Pharmacol Res. 2012;66(4):357–362. doi:10.1016/j.phrs.2012.06.011
44. Menzel M, Akbarshahi H, Uller L. Azithromycin exhibits interferon-inducing properties in an experimental mouse model of asthma exacerbation. Eur Respir J. 2015;46(suppl 59):PA5095.
45. Marckmann S, Wiesemann E, Hilse R, Trebst C, Stangel M, Windhagen A. Interferon-β up-regulates the expression of co-stimulatory molecules CD80, CD86 and CD40 on monocytes: significance for treatment of multiple sclerosis. Clin Exp Immunol. 2004;138(3):499–506. doi:10.1111/j.1365-2249.2004.02624.x
46. Bauvois B, Nguyen J, Tang R, Billard C, Kolb JP. Types I and II interferons upregulate the costimulatory CD80 molecule in monocytes via interferon regulatory factor-1. Biochem Pharmacol. 2009;78(5):514–522. doi:10.1016/j.bcp.2009.05.005
47. Karimi Y, Giles EC, Vahedi F, et al. IFN-β signalling regulates RAW 264.7 macrophage activation, cytokine production, and killing activity. Innate Immun. 2020;26(3):172–182. doi:10.1177/1753425919878839
48. McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15(2):87–103. doi:10.1038/nri3787
49. Fulton SA, Reba SM, Pai RK, et al. Inhibition of major histocompatibility complex II expression and antigen processing in murine alveolar macrophages by Mycobacterium bovis BCG and the 19-kilodalton mycobacterial lipoprotein. Infect Immun. 2004;72(4):2101–2110. doi:10.1128/IAI.72.4.2101-2110.2004
50. Takaoka A, Mitani Y, Suemori H, et al. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science. 2000;288(5475):2357–2360. doi:10.1126/science.288.5475.2357
51. Iwamoto S, Kumamoto T, Azuma E, et al. The effect of azithromycin on the maturation and function of murine bone marrow-derived dendritic cells. Clin Exp Immunol. 2011;166(3):385–392. doi:10.1111/j.1365-2249.2011.04480.x
52. Huang S-W, Chen Y-J, Wang S-T, et al. Azithromycin impairs TLR7 signaling in dendritic cells and improves the severity of imiquimod-induced psoriasis-like skin inflammation in mice. J Dermatol Sci. 2016;84(1):59–70. doi:10.1016/j.jdermsci.2016.07.007
53. Wang F, Zhang S, Jeon R, et al. Interferon gamma induces reversible metabolic reprogramming of M1 macrophages to sustain cell viability and pro-inflammatory activity. EBioMedicine. 2018;30:303–316. doi:10.1016/j.ebiom.2018.02.009
54. Xie C, Liu C, Wu B, et al. Effects of IRF1 and IFN-β interaction on the M1 polarization of macrophages and its antitumor function. Int J Mol Med. 2016;38(1):148–160. doi:10.3892/ijmm.2016.2583
55. Zhu X, Guo Q, Zou J, et al. MiR-19a-3p suppresses M1 macrophage polarization by inhibiting STAT1/IRF1 pathway. Front Pharmacol. 2021;12:614044. doi:10.3389/fphar.2021.614044
56. Cohen Katsenelson K, Stender JD, Kawashima AT, et al. PHLPP1 counter-regulates STAT1-mediated inflammatory signaling. eLife. 2019;8:e48609. doi:10.7554/eLife.48609
57. Tsai WC, Hershenson MB, Zhou Y, Sajjan U. Azithromycin increases survival and reduces lung inflammation in cystic fibrosis mice. Inflamm Res. 2009;58(8):491–501. doi:10.1007/s00011-009-0015-9
58. Menzel M, Akbarshahi H, Tufvesson E, Persson C, Bjermer L, Uller L. Azithromycin augments rhinovirus-induced IFNβ via cytosolic MDA5 in experimental models of asthma exacerbation. Oncotarget. 2017;8(19):31601–31611. doi:10.18632/oncotarget.16364
59. Schögler A, Kopf BS, Edwards MR, et al. Novel antiviral properties of azithromycin in cystic fibrosis airway epithelial cells. Eur Respir J. 2015;45(2):428–439. doi:10.1183/09031936.00102014
60. Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32(1):513–545. doi:10.1146/annurev-immunol-032713-120231
61. Mosallanejad K, Kagan JC. Control of innate immunity by the cGAS-STING pathway. Immunol Cell Biol. 2022;100(6):409–423. doi:10.1111/imcb.12555
62. Nujić K, Banjanac M, Munić V, Polančec D, Eraković Haber V. Impairment of lysosomal functions by azithromycin and chloroquine contributes to anti-inflammatory phenotype. Cell Immunol. 2012;279(1):78–86. doi:10.1016/j.cellimm.2012.09.007
63. Toriyama K, Takano N, Kokuba H, et al. Azithromycin enhances the cytotoxicity of DNA-damaging drugs via lysosomal membrane permeabilization in lung cancer cells. Cancer Sci. 2021;112(8):3324–3337. doi:10.1111/cas.14992
64. Tian A-L, Wu Q, Liu P, et al. Lysosomotropic agents including azithromycin, chloroquine and hydroxychloroquine activate the integrated stress response. Cell Death Dis. 2021;12(1):6. doi:10.1038/s41419-020-03324-w
65. Kim J, Kim H-S, Chung JH. Molecular mechanisms of mitochondrial DNA release and activation of the cGAS-STING pathway. Exp Mol Med. 2023;55(3):510–519. doi:10.1038/s12276-023-00965-7
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.