Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 15
Susceptibility of Genetic Variations in Methylation Pathway to Gastric Cancer
Authors Xiong M, Pan B, Wang X, Nie J, Pan Y, Sun H, Xu T, Cho WCS , Wang S, He B
Received 19 November 2021
Accepted for publication 17 February 2022
Published 4 May 2022 Volume 2022:15 Pages 441—448
DOI https://doi.org/10.2147/PGPM.S340941
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Martin H Bluth
Mengqiu Xiong,1,* Bei Pan,2,* Xuhong Wang,2 Junjie Nie,1 Yuqin Pan,1 Huiling Sun,1 Tao Xu,1 William CS Cho,3 Shukui Wang,1,4,5 Bangshun He1,4,5
1Clinical Laboratory, Nanjing First Hospital, Nanjing Medical University, Nanjing, 210006, People’s Republic of China; 2Medical College, Southeast University, Nanjing, 210006, People’s Republic of China; 3Department of Clinical Oncology, Queen Elizabeth Hospital, Hongkong SAR, People’s Republic of China; 4Collaborative Innovation Center for Cancer Personalized Medicine, Nanjing Medical University, Nanjing, 211166, People’s Republic of China; 5Helicobacter pylori Research Key Laboratory, Nanjing Medical University, Nanjing, 211166, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Bangshun He, Email [email protected]
Background: DNA methylation in the CpG island is associated with gastric cancer, genetic variations residue in genes involved in methylation pathway could contribute to the occurrence of gastric cancer. Here, we investigated the association between DNMTs (DNMT1/DNMT3A/DNMT3B), MTHFR genetic variations and gastric cancer risk and patients’ survival.
Patients and Methods: We recruited 490 gastric cancer patients and 488 age- and sex-matched healthy controls. The genotypes of the genetic variations were detected by a Mass-array platform. A commercial Helicobacter pylori (H. pylori) immunogold testing kit was used to determine the H. pylori infection.
Results: We found that carriers of DNMT1 rs2228612C allele was associated with decreased gastric cancer risk (CT vs. TT: adjusted OR = 0.70, 95% CI = 0.53– 0.94, P = 0.02; CT/CC vs.TT: adjusted OR = 0.73, 95% CI = 0.56– 0.96, P = 0.02). Further stratified analysis showed that DNMT1 rs2228612 CT/CC were associated with a decreased gastric cancer risk in the subgroups of age ≤ 64 years old (adjusted OR = 0.61, 95% CI = 0.41– 0.90, P = 0.01), male (adjusted OR = 0.72, 95% CI = 0.53– 0.98, P = 0.03), negative H. pylori infection (adjusted OR = 0.67, 95% CI = 0.45– 0.98, P = 0.04), tumor stage T3-T4 (adjusted OR = 0.69, 95% CI = 0.51– 0.92, P = 0.01), and non-gastric cardiac adenocarcinoma (NGCA) (adjusted OR = 0.72, 95% CI = 0.54– 0.97, P = 0.03). However, none of the genetic variations of this study was associated with overall survival.
Conclusion: We concluded that the DNMT1 rs2228612C genotype is a protective factor for gastric cancer in Han Chinese population.
Keywords: DNMTs, MTHFR, genetic variation, gastric cancer
Introduction
Gastric cancer is one of the most prevalent cancers in the world, ranking fifth among the most common cancers and third among cancer-related deaths, Helicobacter pylori (H. pylori) infection, age, living habits and diets (such as high salt intake, low fruit and vegetables), are proved as risk factors for gastric cancer.1 Specifically, H. pylori colonization in the stomach could result in chronic gastritis and may result in gastric cancer eventually. Therefore, clearance of H. pylori could reduce the risk of gastric cancer.2 In recent years, despite the decrease in global gastric cancer incidence, the incidence in East Asia is still high, especially in China.3 Therefore, to ascertain the risk of gastric cancer is of great significance.
Dysregulated gene expression in cancer caused by DNA methylation has been reported widely. Three main types of DNA methyltransferase (DNMTs: DNMT1, DNMT2 and DNMT3) are related with genomic methylation. For example, by activating the NF-κB pathway and regulating DNMT3b, H. pylori silenced NDRG2 (N-myc downstream-regulated gene 2) then promoting gastric cancer progression.4 Similarly, NDRG1 was down-regulated in gastric cancer by promoter DNA methylation.5 Methylation at CpG islands is a critical mechanism of gene silencing in gastric cancer.6 Besides, DNMT1 was reported to maintain these methylation patterns in the period of DNA replication. Act as de novo methyltransferases, DNMT3A and DNMT3B were reported to establish methylation patterns during embryogenesis.7 Several studies also indicated that upregulation of DNMTs can promote tumor progression, invasion and metastasis through down regulation of genes that play a role in proliferation inhibition and apoptosis-related pathway.8 Additionally, genetic variations in DNA methyltransferases, DNMT1, DNMT3A, DNMT3B were suggested to be associated with oral squamous cell carcinomas.9 In DNA methylation pathway, folate metabolism involves in DNA methylation, repair and synthesis, and methylenetetrahydrofolate reductase (MTHFR) is a key enzyme involved in the folate pathway.10 Increasing studies discovered that genetic variations in MTHFR may affect the enzymatic activity of the encoding protein and contribute to the development of cancers. MTHFR C677T (rs1801133) variations was also investigated for gastric cancer risk, but the results were inconsistent.11 Interestingly, studies have shown that MTHFR C677T polymorphism is associated with increased risk of gastric cancer and decreased risk of cardia gastric cancer in Chinese population.12
Based on the genetic variations in DNA methylation pathway involved in occurence of gastric cancer, here we conducted a case–control study on genotyped SNPs in a Chinese population to assess the association between variants in 4 genes (DNMT1, DNMT3A, DNMT3B and MTHFR) and susceptibility to gastric cancer. Nine genetic variations (DNMT1: rs16999593, rs10420321, rs2228612, rs7560488; DNMT3A: rs13420827, rs1550117; DNMT3B: rs1569686), in DNMTs and MTHFR (rs1476413, rs1801131) were selected to evaluate their susceptibility to the risk of gastric cancer, as well as their survival predictor role in gastric cancer patients.
Materials and Methods
Study Subjects
A total of 490 gastric cancer patients and 488 age- and sex-matched healthy controls were recruited in this study.13 All patients were histologically diagnosed with gastric cancer, the controls were individuals who came to the hospital for routine physical examination. The patients and healthy controls information were collected from the hospital records and questionnaire respectively. The clinical stages of gastric cancer were classified according to the 6th edition of the American Joint Commission for Cancer Staging Manual. The survival status of gastric cancer patients were obtained by on-site interviews, direct calling, or reviews of medical charts. The Institutional Review Board of the Nanjing First Hospital approved the study protocol, and written informed consent was obtained from all of the participants.
DNA Extraction and Genotyping
According to the manufacture’s protocol, the patient’s blood samples were collected to extract DNA by using GoldMag-Mini Whole Blood Genomic DNA Purification Kit (GoldMag Co. Ltd. Xi’an, China). The purity of the collected DNA was determined by spectroscopy (DU530UV/VIS spectrophotometer, Beckman Instruments, Fullerton, CA, USA). The Sequenom Mass-array platform was used to genotype all samples. SequenomTyper 4.0 Software was used for the data analysis.
We selected the DNMT1/DNMT3A/DNMT3B and MTHFR genetic variations to evaluate their associations with gastric cancer. For selecting the genetic variations, we retrieved the information from the National Center for Biotechnology Information dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP). Then, the following criteria were built for selecting the genetic variations: (1) the minor allele frequency (MAF) was ≥ 5% in the Han Chinese population; (2) the variation was located in an exon, promoter region (less than 2 kb apart from the transcription start), 5ʹuntranslated region (UTR), or 3ʹUTR; (3) the genetic variation has been reported that correlated with cancer risk. Finally, nine DNMT1/DNMT3A/DNMT3B genetic variations and two MTHFR genetic variations were selected to study further (Table S1).
H. pylori Assay on Serum
Commercial H. pylori immuno-gold testing kit (Kangmei Tianhong Biotech Co., Ltd, Beijing, China) was used to detect H. pylori antibodies in the sera of the participants. The sensitivity and specificity of the kit were 98.3% and 98.5% respectively.
Statistical Analysis
The difference of population characteristics between the case and control group was calculated by Chi square test (χ2) or t test, and the Hardy-Weinberg equilibrium (HWE) balance of the control group was calculated by Chi-square test of goodness of fit. In order to test the relationship between genetic variations and cancer, logistic regression of SAS software (Version 9.1; SAS Institute, Cary, NC, USA) was used to calculate odds ratios (ORs) and 95% confidence interval (CIs). Clinical and pathological characteristics subgroup analysis of cancer were used to prove whether genetic variations were still associated with cancer risk in subgroup. For patients with wild-type gene compared with other genes, the survival of cancer patients were used to calculate the hazard ratios (HR) and 95% confidence interval. The calculation method was the Cox regression model of SPSS (SPSS, Chicago, IL, USA) by using the log-rank test. A two-sided P value < 0.05 was considered statistically significant.
Results
Characteristics of the Study Population
The results of HWE analysis showed that the genotype results of nine genetic variations conformed to follow HWE (P > 0.05) (Table S1). The demographic and exposure data of all the participants are summarized in Table S2. There were no differences between the two groups for age (P = 0.748), gender (P = 0.916) and H. pylori infection (P = 0.055). The frequencies of cigarette smoking and alcohol consumption in the patients were higher than those in the controls. The distributions of the genetic variations in patients and the controls were showed in Table 1.
 |
Table 1 Associations Between DNMT1/DNMT3A/DNMT3B and MTHFR genetic variations and Gastric Cancer Risk |
Associations Between Genetic Variations and Gastric Cancer Risk
There was a significant difference in the distribution of the DNMT1 rs2228612 genotype between the case group and the control group. The result showed that the DNMT1 rs2228612CT (CT vs.TT: adjusted OR = 0.70, 95% CI = 0.53–0.94, P = 0.02) and CT/CC genotypes (CT/CC vs. TT: adjusted OR = 0.73, 95% CI = 0.56–0.96, P = 0.02) were associated with decreased gastric cancer risk, respectively. No significant association was observed between the other genetic variations and gastric cancer risk (Table 1).
To further assess the association between DNMT1 rs2228612 and the risk of gastric cancer, we performed a stratified-analysis by age, gender, H. pylori infection status, tumor stage, and tumor site using a co-dominant model (CT/CC vs. TT). The decreased risk of DNMT1 rs2228612C allele carriers (CT/CC) for gastric cancer remained significant in the following subgroups: age≤64 years old (adjusted OR = 0.61, 95% CI = 0.41–0.90, P = 0.01), male (adjusted OR = 0.72, 95% CI = 0.53–0.98, P = 0.03), negative for H. pylori infection (adjusted OR = 0.67, 95% CI = 0.45–0.98, P = 0.04), tumor stage T3-T4 (adjusted OR = 0.69, 95% CI = 0.51–0.92, P = 0.01), non-gastric cardiac adenocarcinoma (NGCA; adjusted OR = 0.72, 95% CI = 0.54–0.97, P = 0.03, Table 2).
 |
Table 2 Subgroup Analysis of rs2228612 to Gastric Cancer Risk |
Association Between Genetic Variations and Clinical Outcomes
In order to assess the relationship between patient survival and genetic variations, a total of 477 patients were followed up to five years for the overall survival (OS), and a Cox regression analysis is used to calculate HRs for patients to evaluate the predictive value of the genetic variations to patients’ survival. The comparison of wild type with those who with any mutantant allele revealed that no association between the genetic variations and OS (Table 3), indicating that these genetic variations have no predictive value for gastric cancer patients' survival.
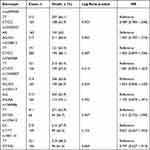 |
Table 3 Analysis of Associations Between Genetic Variations and Clinical Outcomes |
Discussion
In this population-based study, 490 gastric cancer patients and 488 age- and gender-matched healthy controls in a Chinese population were recruited. The result showed that the DNMT1 rs2228612C allele was related with decreased risk of gastric cancer and that such an association was maintained in the subgroups of age≤64 years old, male, negative for H. pylori infection, tumor stage T3-T4, non-gastric cardiac adenocarcinoma (NGCA). Whereas, all the enrolled nine genetic variations were not associated with gastric cancer patients' survival.
Previously, DNMT1 genetic variations has been reported to be associated with various diseases, such as autosomal dominant cerebellar ataxia-deafness and narcolepsy, hereditary sensory neuropathy with dementia and hearing loss.14 In addition, many studies have shown that DNMT1 is involved in the occurrence and development of tumors. Mechanically, knockdown of DNMT1 dysregulates tumor-suppressor P21 and the apoptosis inducer BIK (Bcl-2 interacting killer),15 and inhibits crosstalk of DNMT1 and oestrogen receptor-related receptor alpha (ERRα), resulting in breast cancer progression by regulating the expression of IRF4 (Interferon Regulatory Factor-4).16 Moreover, as a mediator, DNMT1 could promote carcinogenesis and progression of gastric cancer via various regulate networks.17–21 Meanwhile, the expression of DNMT1 could served as a survival biomarker for gastric cancer patients for that down regulation of DNMT1 could increase cisplatin sensitivity and high expression of DNMT1 predicted poor gastric cancer patients’ survival.22 Here, we observed that a genetic variation in DNMT1 (rs2228612) was susceptible to risk of gastric cancer. Actually, in recent years, several studies have shown that DNMT1 rs2228612G/A genotype was associated with decreased risk of breast cancer,23 which was consistent with our result. Whereas, an increased risk of DNMT1 rs2228612 GG genotype for breast cancer risk was also reported in a Chinese Guangdong population. For gastric cancer, three studies invested the DNMT1 rs2228612 in a Chinese population that, one reported no significant association, but they reported DNMT1 rs2228612 GG genotype acted as a protective factor for esophageal cancer, which was consistent with our results.24 Unfortunately, one study omitted the data due to fail to follow HWE,25 and another study reported it was not associated with gastric cancer patients’ survival,26 yet which was consistent with our result. Moreover, in this study, they reported the MAF (C/G allele) in DNMT1 rs2228612 was 0.431 in controls, which was consistent with the result (MAF=0.388) in Asian population of dbSNP database, and the study reported in a China population (MAF=0.450).27 The genotyping of this study was based on the Mass-array platform, which was reliable for genetic variation detection. Actually, due to limited published data regarding DNMT1 rs2228612 and gastric cancer, our result should be confirmed by further study with larger sample size .
DNMT1 rs2228611 is a synonymous genetic variation locates in exon 17, while DNMT1 rs2228612 locates in exon 12, whereas, these two genetic variations were not in linkage disequilibrium each other, according to previous report in a Chinese population.28 Studies have shown that substitution of phenylalanine by isoleucine at 327 amino acid in DNMT1 caused by DNMT1 rs2228612 (A/G) may affect the function of DNMT1 and involve in the carcinogenesis by regulating gene expression through effecting the CpG island hypermethylation statue. Additionally, we also found the significant association between DNMT1 rs2228612 and decreased risk of gastric cancer was maintained in the subgroup of those who with age ≤ 64 years old, male, tumor stage T3-T4, non-gastric cardiac adenocarcinoma or negative for H. pylori infection, which may be attribute to the fact that younger patients are less likely to be exposed to risk factors, that male are more likely to smoke and drink than female, and that the incidence of gastric cancer with negative H. pylori infection is lower, respectively.
Although DNMT1 rs2228612 was reported as an independent predictor of poor OS in melanoma patients;29 however, we found none of the genetic variations are associated with the prognosis of gastric cancer, which was partly consistent to the previous study reported.26 Admittedly, there is limitation in this study. The included subjects are from a single-centre, which may affect the study representation.
Conclusion
We concluded that DNMT1 rs2228612C allele may play a protective role in gastric cancer in Han Chinese population. On the other hand, nine genetic variations (DNMT1: rs16999593, rs10420321, rs2228612, rs7560488; DNMT3A: rs13420827, rs1550117; DNMT3B: rs1569686) in DNMTs and MTHFR (rs1476413, rs1801131) was not found to associate with the survival of gastric cancer patients.
Acknowledgments
This work was supported by the Jiangsu Provincial Key Research and Development Plan (BE2019614), Innovation Team of Jiangsu Provincial Health-Strengthening Engineering by Science and Education (CXTDB2017008); Jiangsu Youth Medical Talents Training Project to BH (QNRC2016066) and YP (QNRC2016074).
Author Contributions
The work presented here was carried out in collaboration among all authors. All authors made a signifcant contribution to the study, including the conception, study design, execution, acquisition of data, analysis and interpretation. Also, all authors took part in drafting, revising and critically reviewing the manuscript. All authors gave final approval of the version to be published, agreed on the journal to which the article has been submitted, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396(10251):635–648. doi:10.1016/S0140-6736(20)31288-5
2. den Hoed CM, Kuipers EJ. Gastric cancer: how can we reduce the incidence of this disease? Curr Gastroenterol Rep. 2016;18(7):34. doi:10.1007/s11894-016-0506-0
3. Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Przegladgastroenterologiczny. 2019;14(1):26–38. doi:10.5114/pg.2018.80001
4. Ling ZQ, Ge MH, Lu XX, et al. Ndrg2 promoter hypermethylation triggered by helicobacter pylori infection correlates with poor patients survival in human gastric carcinoma. Oncotarget. 2015;6(10):8210–8225. doi:10.18632/oncotarget.3601
5. Chang X, Ma J, Xue X, et al. DNMT family induces down-regulation of NDRG1 via DNA methylation and clinicopathological significance in gastric cancer. PeerJ. 2021;9:e12146. doi:10.7717/peerj.12146
6. Takano H, Shibata T, Nakamura M, et al. Effect of DNMT3A polymorphisms on CpG island hypermethylation in gastric mucosa. BMC Med Genet. 2020;21(1):205. doi:10.1186/s12881-020-01142-7
7. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi:10.1016/s0092-8674(00)81656-6
8. Toyota M, Yamamoto E. DNA methylation changes in cancer. Prog Mol Biol Transl Sci. 2011;101:447–457. doi:10.1016/B978-0-12-387685-0.00014-7
9. Supic G, Kozomara R, Zeljic K, Jovic N, Magic Z. Prognostic value of the DNMTs mRNA expression and genetic polymorphisms on the clinical outcome in oral cancer patients. Clin Oral Investig. 2017;21(1):173–182. doi:10.1007/s00784-016-1772-9
10. Levine AJ, Lee W, Figueiredo JC, et al. Variation in folate pathway genes and distal colorectal adenoma risk: a sigmoidoscopy-based case-control study. Cancer Causes Control. 2011;22(4):541–552. doi:10.1007/s10552-011-9726-7
11. Yan S, Xu D, Wang P, et al. MTHFR C677T polymorphism contributes to the risk for gastric cancer. Tumourbiology. 2014;35(3):2123–2132. doi:10.1007/s13277-013-1282-1
12. Si PR, Fang DC, Zhang H, Yang LQ, Luo YH, Liao HY. [The relationship between methylenetetrahydrofolate reductase gene polymorphism and microsatellite instability in gastric cancer]. Zhonghua liu xingbingxue za zhi. 2005;26(10):794–799. Chinese.
13. He B, Pan B, Pan Y, et al. IL-4/IL-4R and IL-6/IL-6R genetic variations and gastric cancer risk in the Chinese population. Am J Transl Res. 2019;11(6):3698–3706.
14. Winkelmann J, Lin L, Schormair B, et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;21(10):2205–2210. doi:10.1093/hmg/dds035
15. Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem. 2004;279(27):27915–27927. doi:10.1074/jbc.M312823200
16. Vernier M, McGuirk S, Dufour CR, et al. Inhibition of DNMT1 and ERRalpha crosstalk suppresses breast cancer via derepression of IRF4. Oncogene. 2020;39(41):6406–6420. doi:10.1038/s41388-020-01438-1
17. Tang H, Deng M, Tang Y, et al. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. 2013;19(20):5602–5612. doi:10.1158/1078-0432.CCR-13-1326
18. Yoon JH, Choi YJ, Choi WS, et al. GKN1-miR-185-DNMT1 axis suppresses gastric carcinogenesis through regulation of epigenetic alteration and cell cycle. Clin Cancer Res. 2013;19(17):4599–4610. doi:10.1158/1078-0432.CCR-12-3675
19. Ning X, Shi Z, Liu X, et al. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 2015;359(2):198–205. doi:10.1016/j.canlet.2015.01.005
20. Sun M, Nie F, Wang Y, et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76(21):6299–6310. doi:10.1158/0008-5472.CAN-16-0356
21. Wang HC, Chen CW, Yang CL, et al. Tumor-associated macrophages promote epigenetic silencing of gelsolin through DNA methyltransferase 1 in gastric cancer cells. Cancer Immunol Res. 2017;5(10):885–897. doi:10.1158/2326-6066.CIR-16-0295
22. Mutze K, Langer R, Schumacher F, et al. DNA methyltransferase 1 as a predictive biomarker and potential therapeutic target for chemotherapy in gastric cancer. Eur J Cancer. 2011;47(12):1817–1825. doi:10.1016/j.ejca.2011.02.024
23. Kullmann K, Deryal M, Ong MF, Schmidt W, Mahlknecht U. DNMT1 genetic polymorphisms affect breast cancer risk in the central European Caucasian population. Clin Epigenetics. 2013;5(1):7. doi:10.1186/1868-7083-5-7
24. Chang SC, Chang PY, Butler B, et al. Single nucleotide polymorphisms of one-carbon metabolism and cancers of the esophagus, stomach, and liver in a Chinese population. PLoS One. 2014;9(10):e109235. doi:10.1371/journal.pone.0109235
25. Yang XX, He XQ, Li FX, Wu YS, Gao Y, Li M. Risk-association of DNA methyltransferases polymorphisms with gastric cancer in the Southern Chinese population. Int J Mol Sci. 2012;13(7):8364–8378. doi:10.3390/ijms13078364
26. Jia Z, Wu X, Cao D, et al. Polymorphisms of the DNA Methyltransferase 1 gene predict survival of gastric cancer patients receiving tumorectomy. Dis Markers. 2016;2016:8578064. doi:10.1155/2016/8578064
27. Hu F, Li X, Li X, et al. Lack of association between DNMT1 gene polymorphisms and noise-induced hearing loss in a Chinese population. Noise Health. 2013;15(65):231–236. doi:10.4103/1463-1741.113517
28. Sun MY, Yang XX, Xu WW, Yao GY, Pan HZ, Li M. Association of DNMT1 and DNMT3B polymorphisms with breast cancer risk in Han Chinese women from South China. Gene Mol Res. 2012;11(4):4330–4341. doi:10.4238/2012.September.26.1
29. Maric H, Supic G, Kandolf-Sekulovic L, et al. DNMT1 and DNMT3B genetic polymorphisms affect the clinical course and outcome of melanoma patients. Melanoma Res. 2019;29(6):596–602. doi:10.1097/CMR.0000000000000612
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
