Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 16
Study on the Key Genes and Molecular Mechanisms of IL-27 Promoting Keratinocytes Proliferation Based on Transcriptome Sequencing
Authors Wu Z, Yang Q, Xu K, Wu J, Yang B
Received 28 March 2023
Accepted for publication 31 May 2023
Published 7 June 2023 Volume 2023:16 Pages 1457—1472
DOI https://doi.org/10.2147/CCID.S414633
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Zijun Wu,1,2,* Qin Yang,3,* Kai Xu,2 Juanjuan Wu,2 Bin Yang1,2
1The First School of Clinical Medicine, Southern Medical University, Guangzhou, People’s Republic of China; 2Department of Burn and Plastic Surgery, General Hospital of Central Theater Command, Wuhan, People’s Republic of China; 3Department of Laboratory Medicine, General Hospital of Central Theater Command, Wuhan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Bin Yang, The First School of Clinical Medicine, Southern Medical University, Nos.1023-1063, Shatai South Road, Baiyun District, Guangzhou, Guangdong, 510515, People’s Republic of China, Tel +86-18062601971, Email [email protected]
Background: IL-27 involves psoriasis pathogenesis potentially by promoting excessive keratinocyte proliferation. However, the underlying mechanisms remain unclear. This study aims to explore the key genes and molecular mechanisms of IL-27-induced keratinocyte proliferation.
Methods: Primary keratinocytes and immortalized human keratinocyte HaCaT cells were treated with different concentrations of IL-27 for 24 h and 48 h respectively. CCK-8 assay was used to detect cell viability and Western blot was used to detect the expression of CyclinE and CyclinB1. Primary keratinocytes and HaCaT cells were treated with IL-27, and their differentially expressed (DE) genes were obtained by transcriptome sequencing. Then Kyoto Encyclopedia of Genes and Genomes enrichment analysis was performed to predict related pathways, and the long non-coding RNA-microRNA-messenger RNA network and protein-protein interaction network were constructed to screen key genes. Biochemical experiments were performed to assess the content of glucose (Glu), lactic acid (LA), and ATP. Flow cytometry and Mito-Tracker Green staining were used to detect mitochondrial membrane potential and the number of mitochondria respectively. Western blot was performed to assess the expression of glucose transporter 1 (GLUT1), hexokinase 2 (HK2), lactate dehydrogenase A (LDHA), phosphoglycerate kinase 1 (PGK1), phosphorylated dynamin-related protein 1 (p-DRP1) (s637) and mitofusin 2 (MFN2).
Results: IL-27 concentration-dependently increased keratinocyte viability and the expression of CyclinE and CyclinB1. Bioinformatics analysis showed that the enriched pathways of DE genes were closely associated with cellular metabolism. miR-7-5p, EGFR, PRKCB, PLCB1 and CALM3 were key genes. IL-27 increased the content of LA, mitochondrial membrane potential, and the expression of GLUT1, HK2, LDHA, PGK1, p-DRP1 (s637), and MFN2, accompanied by decreased contents of Glu and ATP (P< 0.001).
Conclusion: IL-27 potentially promotes keratinocyte proliferation by enhancing glycolysis, mitochondrial function, and mitochondrial fusion. The findings of this study may be conducive to revealing the role of IL-27 in the pathogenesis of psoriasis.
Keywords: psoriasis, RNA-seq, glycolysis, mitochondria
Introduction
Psoriasis is a chronic inflammatory skin disease with a long-term course, which affects the quality of life in about 3% of the global population. In recent years, its incidence rate has been on the rise.1–3 The pathogenic mechanism of psoriasis is sophisticated, associated with genetic, environmental, infectious, psychiatric, and endocrine factors, and remains uncertain.4 But it is commonly accepted that the crosstalk between keratinocytes and immune cells plays a significant role in the progression of psoriasis. Stimulated by the cytokines released by immune cells in the lesion, keratinocytes excessively proliferate and produce various proinflammatory mediators which further exaggerate inflammation, thus leading to the occurrence of psoriasis.5
Interleukin-27 (IL-27), a member of the IL-6 and IL-12 cytokines families, composed of Epstein–Barr virus-induced gene protein 3 and p28 subunits, is produced by activated macrophages and dendritic cells.6,7 It is reported that in psoriatic patients, compared with the healthy control, the level of serum IL-27 is significantly increasing (318±239 vs 452±254 pg/mL), which is positively correlated with the illness severity. Besides, the infiltration of IL-27-positive cells can be detected in the papillary dermis of the psoriatic lesions, but not in the lesions with atopic dermatitis and normal skin.8 What’ more, in the imiquimod-induced psoriasis-like mice model, the messenger RNA (mRNA) expression level of IL-27 is significantly increased in the lesions and IL-27 injection exacerbates skin inflammation.9 These suggest that IL-27 is closely associated with the pathogenesis of psoriasis. Furthermore, we previously found that IL-27 upregulates the expressions of Ki67, keratin (K) 5, and K14 (markers of basal keratinocyte, relevant to proliferation) in keratinocytes,10 indicating that IL-27 possibly involves in the psoriatic pathogenesis by promoting keratinocytes hyperproliferation. However, the underlying mechanism is incompletely understood.
In this study, we obtained the expression profiles of differentially expressed (DE) genes in IL-27-treated keratinocytes. Then we performed the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, constructed the long non-coding RNA (lncRNA)-microRNA (miRNA)-mRNA network and protein-protein interaction (PPI) network, and carried out in vitro experiments to investigate the key genes and molecular mechanisms in IL-27 promoting keratinocytes proliferation. This work can provide new insight into the role of IL-27 in keratinocyte proliferation in psoriasis.
Materials and Methods
Primary Keratinocytes Isolated
Human primary keratinocytes were isolated from eyebrow skin tissues acquired by cosmetic surgery at the Department of Burn and Plastic Surgery, General Hospital of Central Theater Command. Informed consents were received from the participants and the study was approved by the Ethics Committee of the Wuhan General Hospital of Guangzhou Command (Wuhan, China; No. [2018]002–1). The skin tissues of the eyebrow were excised under aseptic conditions, and incubated with 0.4% neutral protease (D195752, Aladdin, China) for 12 h at 4 °C. Epidermic was separated, digested with 0.25% trypsin at 37 °C for 5 min, filtered by the 200-mesh sieve and centrifuged (1500 rpm, 3 min). Then, the harvested keratinocytes were resuspended with EpiLife™ medium (MEPICF500, Thermo Fisher Scientific, USA) and inoculated at 1.25×104 cells /mL in the culture flasks.
Immunofluorescence
Primary cells were washed with Phosphate-Buffered Saline (P1010, Solarbio, China) and fixed with 4% paraformaldehyde for 30 min, followed by treatment with 0.5% Triton X-100 for 20 min at room temperature. Next, the cells were blocked in 5% BSA (A8010, Solarbio, China) for 1 h at 37 °C and incubated with primary antibodies against Keratin (1:200, ab8068, Abcam, UK), K19 (1:200, PAB 30070, Bioswamp, China), and K15 (1:200, ab52816, Abcam, UK) at 4 °C overnight. The next day, the cells were incubated with Alexa Fluor 594-conjugated Goat Anti-Rabbit antibody (1:200, SAB43732, Bioswamp, China) and Alexa Fluor 594-conjugated Goat Anti-Mouse IgG (H+L) (1:200, SAB45897, Bioswamp, China) for 1 h at 37 °C and the nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI, S2110, Solarbio, China). Immunofluorescence was observed under a fluorescence microscope (DMIL LED, Leica, Germany).
Cell Culture and Grouping
Primary cells and HaCaT cells (CL-0090, Procell Life Science&Technology Co.,Ltd., China) were cultured in DMEM medium (SH30022.01B, Hyclone, USA) containing 10% fetal bovine serum (10,270–106, Gibco, USA) in an incubator at 37 °C with 5% CO2. Cells at the logarithmic growth stage were taken for experiments. Cells were intervened with varying concentrations (0,10,20,50,100 ng/mL) of IL-27 (200–38, PEPROTECH, USA) for 24 h and 48 h. Cell viability was detected by CCK-8 assay and the protein levels of CyclinE and CyclinB1 were detected by Western blot. The subsequent experiments selected “50 ng/mL, 24 h” as the condition of IL-27 intervention. Both two types of cells were divided into control group (0 ng/mL IL-27) and IL-27 group (50 ng/mL IL-27), respectively.
Cell Counting Kit-8 (CCK-8) Assay
Cell viability was assessed by CCK-8 assay. With a density of 3×103 cells/well, cells were seeded in 96-well plates and treated with varying concentrations of IL-27 (0, 10, 20, 50, 100 ng/mL) for 24 h and 48 h. Then, 10 μL CCK-8 (CA1210, Solarbio, China) solution was added to each well and the cells were cultured at 37 °C for 4 h. Optical density was measured using a microplate reader (AMR-100, ALLSHENG, China) at 450 nm.
RNA Sequencing and Analysis
Total RNAs were extracted from HaCaT cells, and ribosomal RNAs were removed to construct strand-specific libraries. Illumina Novaseq™ 6000 sequencing was performed for library quality control, with 2×150 bp at both ends. Sequences were filtered out ineligible ones by Cutadapter and then were compared with Homo sapiens genomes. Log2|Fold Change|≥1 and q-value <0.05 were used as the screening criteria for DE lncRNAs and DE mRNAs. Log2|Fold Change|≥1 and p-value <0.05 were used as the screening criteria for DE miRNAs. Volcano plots about DE genes were visualized using the gglpot2 package in RStudio. KEGG enrichment analysis was performed to define the signaling pathway enriched in DE genes.
As for the construction of lncRNA-miRNA-mRNA network, the miRNAs binding with DE lncRNAs were predicted using the miRcode website (http://www.mircode.org/). Target Scan (v5.0), miRanda (v3.3a), and miRDB (http://mirdb.org/) were used to predict the target genes of miRNAs, with the threshold values of TargetScan_score ≥80, miranda_Energy < −20 and miRDB score ≥ 80. The intersections of the predicted target genes from these three databases were considered the final target genes of the DE miRNAs. Cytoscape (3.7.0) was used to establish the lncRNA-miRNA-mRNA network. PPI network about DE mRNAs in the lncRNA-miRNA-mRNA network was constructed using the STRING database. The PPI network was visualized excluding isolated proteins. The Degree values of genes in two networks were calculated using the cytoHubba plugin.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNAs were extracted from cells with TRIzol™ reagent (15,596,018, Thermo Fisher Scientific, USA) and reverse transcribed into cDNA with a reverse transcription kit (TAKARA Biotechnology Co., Ltd., China). The cDNA was subjected to PCR amplification with the SYBR Green PCR Kit (KAPA Biosystems, USA) using the following primer sequences: microRNA-7-5p (miR-7-5p): forward 5’-GGGGTGGAAGACTAGTGATTT-3’and reverse 5’-CTGGTGTCGTGGAGTCGG-3’, U6: forward 5’-CTCGCTTCGGCAGCACA-3’and reverse 5’-AACGCTTCACGAATTTGCGT-3’, lncRNA MIR31HG: forward 5’-CTAGCCTCCAGTTTTACCCT-5’and reverse 5’-TAACCATCAACGTCTTCTGTG-3’, lncRNA NEAT1: forward 5’-GCAGATCAGCATCCTTCG-5’and reverse 5’-ACAAGACACCTGTGACAAATG-3’, epidermal growth factor receptor (EGFR): forward 5’-GCTATGAGATGGAGGAAGACG-5’and reverse 5’-CACTGATGGAGGTGCAGTTTT-3’, GAPDH: forward 5’-CCACTCCTCCACCTTTG-3’and reverse 5’-CACCACCCTGTTGCTGT-3’. The analysis of data was conducted with the 2-ΔΔCt method.
Western Blot
Total protein was extracted from cells using RIPA lysis buffer (R0010, Solarbio, China) and quantified using a Bicinchoninic Acid Kit (PC0020, Solarbio, China). The extracted proteins were separated and transferred onto polyvinylidene fluoride membranes (IPVH00010, Millipore, USA). The membranes were blocked with 5% skim milk for 1 h and incubated overnight with primary antibodies against CyclinE (1:1000, ab33911, Abcam, UK), phosphorylated dynamin-related protein 1 (s637) (p-DRP1, 1:1000, ab193216, Abcam, UK), phosphorylated epidermal growth factor receptor (p-EGFR, 1:1000, ab40815, Abcam, UK); CyclinB1 (1:1000, PAB32796, Bioswamp, China), glucose transporter 1 (GLUT1, 1:1000, PAB34949, Bioswamp, China), hexokinase 2 (HK2, 1:1000, PAB30271, Bioswamp, China), lactate dehydrogenase A (LDHA, 1:1000, PAB30703, Bioswamp, China), phosphoglycerate kinase 1 (PGK1, 1:1000, PAB31853, Bioswamp, China), dynamin-related protein 1 (DRP1, 1:1000, PAB33409, Bioswamp, China), mitofusin 2 (MFN2, 1:1000, PAB37988, Bioswamp, China), calmodulin-3 (CALM3, 1:1000, PAB37540, Bioswamp, China), protein kinase C beta type (PRKCB, 1:1000, PAB31677, Bioswamp, China), epidermal growth factor receptor (EGFR, 1:1000, PAB30733, Bioswamp, China), and GAPDH (1:1000, PAB36269, Bioswamp, China). Then, the membranes were incubated with Goat Anti-Rabbit IgG (1:10,000, SAB43714, Bioswamp, China) secondary antibody for 1 h. GAPDH served as an endogenous control.
Biochemical Tests
The glucose (Glu), ATP, and lactic acid (LA) contents in the cells were determined according to the corresponding instruction of the biochemical kit: Glu biochemical kit (F006), ATP biochemical kit (A095-1-1), and LA biochemical kit (A019-2), all purchased from Nanjing Jiancheng Bioengineering Institute, China.
JC-1 Assay
Cells at a density of 1×106 were collected, resuspended, and added with 0.5 mL of JC-1 staining buffer (C2006, Beyotime, China), then incubated for 20 min in the incubator. By centrifuging at 4 °C (3 min, 400 g), the precipitate was collected and resuspended with JC-1 staining buffer. The Fluorescence was analyzed by NovoCyte software (ACEA Biosciences Inc., USA).
Mito-Tracker Green Staining
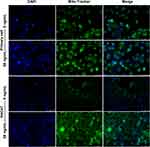
Mito-Tracker Green staining was applied to detect the number of mitochondria. Mito-Tracker Green working reagent (C1048, Beyotime, China), at a final concentration of 100 nM, was prewarmed for 10 min at 37 °C and added to the cells incubated for 25 min at 37 °C in darkness. Cells were incubated with DAPI at room temperature for 10 min in darkness. The fluorescence was observed by fluorescence microscope.
Statistical Analysis
SPSS 26.0 software was applied for statistical analysis and GraphPad Prism 8.3.0 software was used for data visualization. The measurement data were presented as mean ± standard deviation. T-test was used for comparison between the two groups. One-way ANOVA followed by Tukey’s post hoc test was used for comparison between multiple groups. P<0.05 was considered statistically significant.
Results
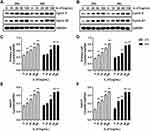
IL-27 Enhanced Viability and Protein Levels of CyclinE and CyclinB1 in Keratinocytes
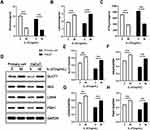
Primary keratinocytes were validated by immunofluorescence staining (Figure 1). Primary cells and HaCaT cells were treated with varying concentrations of recombinant human IL-27 (0, 10, 20, 50, and 100 ng/mL) for 24 h and 48 h, respectively. IL-27 concentration-dependently upregulated the viability (Figure 2A and B) and protein levels of CyclinE (Figure 3A–C and E) and CyclinB1 (Figure 3A, B, D and F) in keratinocytes. Compared with the 0ng/mL IL-27 group, cell viability and protein levels of CyclinE and CyclinB1 in the 50 ng/mL IL-27 group were significantly increased (P<0.001) and approached maximum. In addition, little differences were found in the cell viability and protein levels of CyclinE and CyclinB1 induced by IL-27 for 24 h or 48 h. Therefore, 50 ng/mL and 24 h were selected as the intervention conditions for IL-27 in subsequent experiments.
Differentially Expressed RNAs Analysis and KEGG Enrichment Analysis
In order to clarify the effect of IL-27 on keratinocytes transcriptome, HaCaT cells were divided into IL-27 group and control group. Total RNAs were extracted from the cells of two groups for transcriptome sequencing. We found that 146 lncRNAs (including 38 up-regulated and 108 down-regulated), 12 miRNAs (including 10 up-regulated and 2 down-regulated), and 1486 mRNAs (including 1097 up-regulated and 389 down-regulated) were statistically differentially expressed between the IL-27 group and control group. The DE RNAs were shown in the volcano plots (Figure 4).
To investigate the signaling pathways related to IL-27-treated keratinocytes, KEGG enrichment analysis was conducted for DE RNAs. KEGG analysis indicated that DE lncRNAs were mainly concentrated in “Central carbon metabolism in cancer” and “Choline metabolism in cancer” (Figure 5A). DE miRNAs were mainly enriched in “PI3K/Akt signaling pathway”, “Pathways in cancer”, “HIF-1 signaling pathway”, “ErbB signaling pathway”, “Ras signaling pathway”, “Calcium signaling pathway” and “Hippo signaling pathway” (Figure 5B). DE mRNAs were mainly enriched in “Glycolysis/gluconeogenesis”, “p53 signaling pathway”, “Pentose phosphate pathway”, “Glycerolipid metabolism”, “HIF-1 signaling pathway” and “PPAR signaling pathway” (Figure 5C). The vast majority of these signaling pathways above were closely associated with cellular metabolism, suggesting that IL-27 presumably induced keratinocytes proliferation by mediating metabolic pathways.
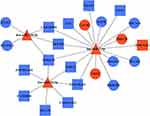
Construction of lncRNA-miRNA-mRNA Network and PPI Network
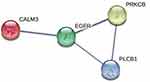
To further elucidate the association among DE RNAs, we established the lncRNA-miRNA-mRNA network and PPI network. The lncRNA-miRNA-mRNA network consists of 13 DE lncRNAs (including 1 up-regulated and 12 down-regulated), 3 DE miRNAs (including 3 up-regulated), and 10 DE mRNAs (including 3 up-regulated and 7 down-regulated) (Figure 6). According to the centrality analysis of network nodes, we found that miR-7-5p, with the maximal degree value of 17, maybe the key non-coding RNA (ncRNA) in the lncRNA-miRNA-mRNA network. The PPI network was composed of 4 interacting edges and 4 node proteins, including EGFR, PRKCB, phospholipase C Beta 1 (PLCB1), and CALM3 (Figure 7). These proteins were closely interacted, which might play a key role in the proliferation of keratinocytes due to IL-27 stimulation. In the PPI network, EGFR with the highest degree value of 6, was potentially the most important protein.
Verification of the Accuracy of Transcriptome Sequencing Using qRT-PCR and Western Blot
To ascertain the reliability of transcriptome sequencing, we selected miR-7-5p, lncRNA MIR31HG, lncRNA NEAT1, EGFR, PRKCB, and CALM3 from lncRNA-miRNA-mRNA network for qRT-PCR or Western Blot. The RNA expression profiles of the experimental assays were consistent with those of the transcriptome sequencing (Figure 8), which confirmed the authenticity of the transcriptome sequencing.
IL-27 Strengthened Glycolysis in Keratinocytes
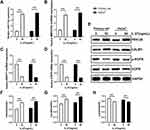
Studies have reported that metabolic activity is crucial to determine the state of cell proliferation.11 Among various metabolic pathways, glycolysis and mitochondrial metabolism are the main pathways supporting cell proliferation.12 Based on the results of the KEGG enrichment analysis, we assessed whether IL-27 affected the glycolytic pathway and thus stimulated the hyperproliferation of keratinocytes. Both primary cells and HaCaT cells were divided into control group and IL-27 group. Compared with the control group, the IL-27 group showed significantly lower Glu (Figure 9A) and ATP (Figure 9C) contents (P<0.001), and significantly higher LA (Figure 9B) content and glycolysis-related proteins expression, including GLUT1 (Figure 9D and E), HK2 (Figure 9D and F), LDHA (Figure 9D and G) and PGK1 (Figure 9D and H) (P<0.001). These results indicated that IL-27 enhanced glycolysis in keratinocytes to maintain cell proliferation.
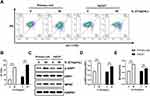
IL-27 Enhanced Mitochondrial Function and Mitochondrial Fusion in Keratinocytes
Mitochondrial function and dynamics play an important role in the regulation of cellular growth and proliferation.13,14 For this reason, we evaluated the effect of IL-27 on mitochondria in keratinocytes. Both primary cells and HaCaT cells were divided into control group and IL-27 group. Compared with the control group, IL-27 increased mitochondrial membrane potential (Figure 10A and B) (P<0.001) and mitochondrial number (Figure 11). Meanwhile, p-DRP1 (s637) (Figure 10C and D) and MFN2 (Figure 10C and E) protein levels were upregulated by IL-27 (P<0.001). These results indicated that IL-27 enhanced mitochondrial function and mitochondrial fusion in keratinocytes.
Discussion
IL-27 is closely associated with the pathogenesis of psoriasis. Based on our previous study,10 we speculated that IL-27 potentially participated in psoriasis pathogenesis by stimulating keratinocytes hyperproliferation. Whereas, the underlying mechanism remains unclear. We aimed to investigate the key genes and molecular mechanisms in IL-27 promoting keratinocyte proliferation.
In the present study, we first found that IL-27 concentration-dependently upregulated cell viability and expression of CyclinE and CyclinB1 to promote keratinocyte proliferation. Besides, we determined “50 ng/mL, 24 h” as the optimal condition for IL-27 intervening keratinocytes. Then, we applied transcriptome sequencing to obtain expression profiles of DE RNAs in keratinocytes treated with IL-27. We identified 146 DE lncRNAs, 12 DE miRNAs, and 1486 DE mRNAs.
KEGG analysis showed that “PI3K/Akt signaling pathway”, “ErbB signaling pathway”, “Hippo signaling pathway”, “Glycolysis/gluconeogenesis”, “p53 signaling pathway” and “HIF-1 signaling pathway”, were significantly enriched by DE RNAs. Glycolysis is one of the important pathways that foster cell proliferation. PI3K/Akt signaling pathway, p53 signaling pathway, ErbB signaling pathway, and Hippo signaling pathway, all of which can mediate glycolysis by directly or indirectly regulating the activities of glycolytic enzymes.15,16 The HIF-1 signaling pathway not only increases LA production by upregulating LDHA expression but also elevates the level of pyruvate dehydrogenase kinases, decreasing pyruvate entry into mitochondria, and thus enhancing glycolysis.16–18 The signaling pathways predicted by KEGG analysis may mediate IL-27-induced hyperproliferation in keratinocytes, but the precise regulatory mechanisms remain to be further investigated.
In order to establish DE RNAs interactions and discover key genes, we constructed the lncRNA-miRNA-mRNA networks and PPI networks. Based on the centrality analysis, miR-7-5p was identified as the key ncRNA in the lncRNA-miRNA-mRNA network. miR-7-5p is a tumor suppressor in a variety of cancers. In some tumors, it regulates cell growth and cycle progression mainly by mediating the EGFR signaling pathway.19 It has been reported that miR-7-5p negatively regulates the EGFR signaling pathway to mediate the proliferation of many cancers, such as prostate cancer, glioma, and glioblastoma.20–22 The constructed PPI network was composed of EGFR, PRKCB, PLCB1, and CALM3. These proteins are relevant to cellular proliferation, differentiation, and cycle regulation. PRKCB is one of the isoforms of protein kinase C involved in a variety of cellular functions and signals transduction pathways and can mediate Glu metabolism and mitochondrial pathways.23,24 PLCB1 is a member of the phospholipase family, which mediates cell proliferation through influences cyclin, cyclin-dependent kinase, and cell cycle progression.25,26 CALM3 is a multifunctional and calcium-binding protein associated with the repairment of the epidermal barrier in keratinocytes.27 According to the bioinformatic analysis, these three proteins are regulated by miR-7-5p and interact with EGFR. They possibly play a key role in IL-27-induced keratinocyte proliferation. Furthermore, EGFR was likely to be the most important gene of the PPI network. It is reported that EGFR plays a crucial role in controlling the processes of both proliferation and differentiation in the epidermis.28 Recent studies have revealed that EGFR regulates a complex signaling network within cells and its role in regulating cellular metabolism is emerging as one of its important functions.29 Among the various metabolic pathways, glycolysis and mitochondrial metabolism are the main pathways that support cell proliferation.12 Several studies have found that the EGFR signaling pathway regulates the cellular uptake of Glu and the activities of various transcription factors and enzymes, thereby mediating aerobic glycolysis.12,16 What’s more, it can also mediate mitochondria, including mitochondrial biogenesis, metabolic processes, and dynamics. However, it remains to be explored whether the EGFR signaling pathway regulates glycolysis and mitochondrial metabolism thus involved in IL-27 promoting keratinocyte proliferation. Finally, the authenticity of transcriptome sequencing was determined by qRT-PCR and Western blot.
It has been suggested that the “Warburg effect” exists in proliferating cells, a shift in energy metabolism from mitochondrial oxidative phosphorylation to aerobic glycolysis.30 The mitosis process requires double cellular components. Compared with mitochondrial oxidative phosphorylation, aerobic glycolysis produces more NADPH and synthetic substrates and rapidly generates more ATP to satisfy the needs of proliferating cells.31,32 GLUT1 is an important protein initiating cellular Glu uptake. It is reported that GLUT1 expression is upregulated in the psoriatic epidermis and correlated with the degree of acanthosis and the percentage of Ki-67 expression.33 Additionally, a recent study found that the metabolic transition of aerobic glycolysis participates in the proliferation of psoriasis keratinocytes.34 In line with this, we found that IL-27 decreased the contents of Glu and ATP, increasing LA content and the levels of glycolysis-related proteins including GLUT1, HK2, LDHA, and PGK1. These results suggested that IL-27 may maintain the proliferation of keratinocytes by promoting glycolysis.
Glycolysis and mitochondrial metabolism are two interactive and closely coupled processes. Cancer cells depend on enhanced glycolysis due to defective mitochondria, as Warburg initially proposed.35 However, though some cancer cells do have defects in mitochondrial metabolic enzymes, functional mitochondria exist in most proliferating cells (including cancer cells) as well.36 In the present study, we found that IL-27 elevated mitochondrial membrane potential and the number of mitochondria, increasing the levels of p-DRP1 (s637) and MFN2. Mitochondria are highly dynamic organelles, constantly undergoing fission and fusing states to adapt to cellular demands in different situations. These two processes are also known as mitochondrial dynamics. The former can be mediated by DRP1, while the latter can be mediated by MFN1 and MFN2. Mitochondrial fusion entails an intact mitochondrial membrane potential facilitating cell proliferation. In accordance with Yao et al36 they find enhanced glycolysis in proliferating fibroblasts accompanied by elevated levels of mitochondrial oxidative phosphorylation, which is mediated by mitochondrial fusion. Similarly, the growth rate of mouse embryonic fibroblast was significantly reduced after the knockdown of MFN1 and MFN2.37 Furthermore, Mitra38 finds that mitochondrial dynamics components modulate cell cycle regulators. The decrease of DRP1 sustains mitochondria in the fused state with the increased level of Cyclin E, promoting entry into the S phase, thus leading to excessive cell proliferation.39 These suggested that IL-27 enhanced mitochondrial function and fusion to promote keratinocyte proliferation.
There are several limitations to this study. Firstly, we have not explored the exact mechanisms by which the identified key genes, such as miR-7-5p, EGFR, PRKCB, PLCB1, CALM3, and other pathways predicted by KEGG analysis, such as p53 signaling pathway, HIF-1 signaling pathway, and Hippo signaling pathway, participate in IL-27-stimulated keratinocyte proliferation. Secondly, the specific mechanisms by which IL-27 promotes keratinocyte proliferation through glycolytic and mitochondrial pathways remain to be further investigated. Finally, the mechanism of action of IL-27 on keratinocyte proliferation in a psoriasis model has not been examined.
Conclusions
In summary, this study revealed the expression profiles and enrichment pathways of DE genes in IL-27-stimulated keratinocytes. We found that miR-7-5p, EGFR, PRKCB, PLCB1, and CALM3 were key genes. The signaling pathways of IL-27 promoting keratinocytes proliferation were closely associated with cellular metabolism. Furthermore, through performing in vitro experiments, we demonstrated that IL-27 potentially enhanced glycolysis, mitochondrial function, and mitochondrial fusion, thereby contributing to keratinocytes hyperproliferation. The findings of this study may be conducive to revealing the role of IL-27 in the pathogenesis of psoriasis. In the future, we will further focus on the effect of IL-27 on keratinocyte proliferation in psoriasis, so as to provide new insight into the pathogenesis of psoriasis.
Abbreviations
IL-27, interleukin-27; lncRNA, long non-coding RNA; miRNA, microRNA; mRNA, messenger RNA; ncRNA, non-coding RNA; miR-7-5p, microRNA-7-5p; EGFR, epidermal growth factor receptor; p-EGFR, phosphorylated epidermal growth factor receptor; PRKCB, protein kinase C beta type; PLCB1, phospholipase C Beta 1; CALM3, calmodulin 3; Glu, glucose; LA, lactic acid; GLUT1, glucose transporter 1; HK2, hexokinase 2; LDHA, lactate dehydrogenase A; PGK1, phosphoglycerate kinase 1; DRP1, dynamin-related protein 1; p-DRP1, phosphorylated dynamin-related protein 1; MFN2, mitofusin 2; DE, differentially expressed; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction; K, keratin; qRT-PCR, quantitative real-time polymerase chain reaction; DAPI, 4’,6-diamidino-2-phenylindole.
Ethics Approval and Consent to Participate
The studies involving human participants were reviewed and approved by the Declaration of Helsinki and the Ethics Committee of the Wuhan General Hospital of Guangzhou Command (Wuhan, China; No. [2018]002-1). Informed consents were received from the participants.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No.81872536).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Deng Y, Chang C, Lu Q. The inflammatory response in psoriasis: a comprehensive review. Clin Rev Allergy Immunol. 2016;50(3):377–389. doi:10.1007/s12016-016-8535-x
2. Martins AM, Ascenso A, Ribeiro HM, Marto J. The brain-skin connection and the pathogenesis of psoriasis: a review with a focus on the serotonergic system. Cells. 2020;9(4):796. doi:10.3390/cells9040796
3. Takeshita J, Wang S, Shin DB, et al. Comparative effectiveness of less commonly used systemic monotherapies and common combination therapies for moderate to severe psoriasis in the clinical setting. J Am Acad Dermatol. 2014;71(6):1167–1175. doi:10.1016/j.jaad.2014.08.003
4. Chandra A, Ray A, Senapati S, Chatterjee R. Genetic and epigenetic basis of psoriasis pathogenesis. Mol Immunol. 2015;64(2):313–323. doi:10.1016/j.molimm.2014.12.014
5. Gao J, Chen F, Fang H, Mi J, Qi Q, Yang M. Daphnetin inhibits proliferation and inflammatory response in human HaCaT keratinocytes and ameliorates imiquimod-induced psoriasis-like skin lesion in mice. Biol Res. 2020;53(1):48. doi:10.1186/s40659-020-00316-0
6. Pflanz S, Timans JC, Cheung J, et al. IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4+ T cells. Immunity. 2002;16(6):779–790. doi:10.1016/s1074-7613(02)00324-2
7. Beizavi Z, Zohouri M, Asadipour M, Ghaderi A. IL-27, a pleiotropic cytokine for fine-tuning the immune response in cancer. Int Rev Immunol. 2021;40(5):319–329. doi:10.1080/08830185.2020.1840565
8. Shibata S, Tada Y, Kanda N, et al. Possible roles of IL-27 in the pathogenesis of psoriasis. J Invest Dermatol. 2010;130(4):1034–1039. doi:10.1038/jid.2009.349
9. Shibata S, Tada Y, Asano Y, et al. IL-27 activates Th1-mediated responses in imiquimod-induced psoriasis-like skin lesions. J Invest Dermatol. 2013;133(2):479–488. doi:10.1038/jid.2012.313
10. Yang B, Suwanpradid J, Sanchez-Lagunes R, et al. IL-27 facilitates skin wound healing through induction of epidermal proliferation and host defense. J Invest Dermatol. 2017;137(5):1166–1175. doi:10.1016/j.jid.2017.01.010
11. Huang X, Chen J, Zeng W, Wu X, Chen M, Chen X. Membrane-enriched solute carrier family 2 member 1 (SLC2A1/GLUT1) in psoriatic keratinocytes confers sensitivity to 2-deoxy-D-glucose (2-DG) treatment. Exp Dermatol. 2019;28(2):198–201. doi:10.1111/exd.13850
12. Chandel NS. Metabolism of Proliferating Cells. Cold Spring Harb Perspect Biol. 2021;13(10):a040618. doi:10.1101/cshperspect.a040618
13. Xin Y, Zhao L, Peng R. HIF-1 signaling: an emerging mechanism for mitochondrial dynamics. J Physiol Biochem. 2023. doi:10.1007/s13105-023-00966-0
14. Kumar Sharma R, Chafik A, Bertolin G. Mitochondrial transport, partitioning, and quality control at the heart of cell proliferation and fate acquisition. Am J Physiol Cell Physiol. 2022;322(2):C311–c325. doi:10.1152/ajpcell.00256.2021
15. Zhang X, Zhao H, Li Y, et al. The role of YAP/TAZ activity in cancer metabolic reprogramming. Mol Cancer. 2018;17(1):134. doi:10.1186/s12943-018-0882-1
16. Icard P, Simula L, Fournel L, et al. The strategic roles of four enzymes in the interconnection between metabolism and oncogene activation in non-small cell lung cancer: therapeutic implications. Drug Resist Updat. 2022;63:100852. doi:10.1016/j.drup.2022.100852
17. Cong LN, Chen H, Li Y, et al. Physiological role of Akt in insulin-stimulated translocation of GLUT4 in transfected rat adipose cells. Mol Endocrinol. 1997;11(13):1881–1890. doi:10.1210/mend.11.13.0027
18. Nagao A, Kobayashi M, Koyasu S, Chow CCT, Harada H. HIF-1-dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. Int J Mol Sci. 2019;20(2):238. doi:10.3390/ijms20020238
19. Giles KM, Brown RA, Ganda C, et al. microRNA-7-5p inhibits melanoma cell proliferation and metastasis by suppressing RelA/NF-κB. Oncotarget. 2016;7(22):31663–31680. doi:10.18632/oncotarget.9421
20. Zhang X, Niu W, Mu M, Hu S, Niu C. Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J Exp Clin Cancer Res. 2020;39(1):196. doi:10.1186/s13046-020-01695-8
21. Ma X, Ren H, Zhang Y, Wang B, Ma H. LncRNA RHPN1-AS1 inhibition induces autophagy and apoptosis in prostate cancer cells via the miR-7-5p/EGFR/PI3K/AKT/mTOR signaling pathway. Environ Toxicol. 2022;37(12):3013–3027. doi:10.1002/tox.23656
22. Wang H, Feng J, Ao F, et al. Tumor-derived exosomal microRNA-7-5p enhanced by verbascoside inhibits biological behaviors of glioblastoma in vitro and in vivo. Molecul ther Oncol. 2021;20:569–582. doi:10.1016/j.omto.2020.12.006
23. Lee EE, Ma J, Sacharidou A, et al. A protein kinase C phosphorylation motif in GLUT1 affects glucose transport and is mutated in GLUT1 deficiency syndrome. Mol Cell. 2015;58(5):845–853. doi:10.1016/j.molcel.2015.04.015
24. Tsui C, Martinez-Martin N, Gaya M, et al. Protein Kinase C-β Dictates B cell fate by regulating mitochondrial remodeling, metabolic reprogramming, and heme biosynthesis. Immunity. 2018;48(6):1144–1159.e5. doi:10.1016/j.immuni.2018.04.031
25. Lee SJ, Leoni G, Neumann PA, Chun J, Nusrat A, Yun CC. Distinct phospholipase C-β isozymes mediate lysophosphatidic acid receptor 1 effects on intestinal epithelial homeostasis and wound closure. Mol Cell Biol. 2013;33(10):2016–2028. doi:10.1128/mcb.00038-13
26. Piazzi M, Blalock WL, Bavelloni A, et al. PI-PLCβ1b affects Akt activation, cyclin E expression, and caspase cleavage, promoting cell survival in pro-B-lymphoblastic cells exposed to oxidative stress. FASEB J. 2015;29(4):1383–1394. doi:10.1096/fj.14-259051
27. Liu S, Guo C, Wu D, Ren Y, Sun MZ, Xu P. Protein indicators for HaCaT cell damage induced by UVB irradiation. J Photochem Photobiol B. 2012;114:94–101. doi:10.1016/j.jphotobiol.2012.05.015
28. Fukuyama T, Nakamura Y, Kanemaru K, et al. Phospholipase Cγ1 is required for normal irritant contact dermatitis responses and sebaceous gland homeostasis. Exp Dermatol. 2019;28(9):1051–1057. doi:10.1111/exd.14009
29. Orofiamma LA, Vural D, Antonescu CN. Control of cell metabolism by the epidermal growth factor receptor. Biochim Biophys Acta Mol Cell Res. 2022;1869(12):119359. doi:10.1016/j.bbamcr.2022.119359
30. Wackerhage H, Vechetti IJ, Baumert P, et al. Does a hypertrophying muscle fibre reprogramme its metabolism similar to a cancer cell? Sports Med. 2022;52(11):2569–2578. doi:10.1007/s40279-022-01676-1
31. Izosimova AV, Shirmanova MV, Shcheslavskiy VI, et al. FLIM of NAD(P)H in lymphatic nodes resolves T-cell immune response to the tumor. Int J Mol Sci. 2022;23(24):15829. doi:10.3390/ijms232415829
32. Sousa MI, Rodrigues AS, Pereira S, Perestrelo T, Correia M, Ramalho-Santos J. Mitochondrial mechanisms of metabolic reprogramming in proliferating cells. Curr Med Chem. 2015;22(20):2493–2504. doi:10.2174/0929867322666150514095718
33. Abdou AG, Maraee AH, Eltahmoudy M, El-Aziz RA. Immunohistochemical expression of GLUT-1 and Ki-67 in chronic plaque psoriasis. Am J Dermatopathol. 2013;35(7):731–737. doi:10.1097/DAD.0b013e3182819da6
34. Liu YZ, Xu MY, Dai XY, et al. Pyruvate Kinase M2 mediates glycolysis contributes to psoriasis by promoting keratinocyte proliferation. Front Pharmacol. 2021;12:765790. doi:10.3389/fphar.2021.765790
35. Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. doi:10.1126/science.124.3215.269
36. Yao CH, Wang R, Wang Y, Kung CP, Weber JD, Patti GJ. Mitochondrial fusion supports increased oxidative phosphorylation during cell proliferation. eLife. 2019;8. doi:10.7554/eLife.41351
37. Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280(28):26185–26192. doi:10.1074/jbc.M503062200
38. Mitra K. Mitochondrial fission-fusion as an emerging key regulator of cell proliferation and differentiation. BioEssays. 2013;35(11):955–964. doi:10.1002/bies.201300011
39. Mitra K, Rikhy R, Lilly M, Lippincott-Schwartz J. DRP1-dependent mitochondrial fission initiates follicle cell differentiation during Drosophila oogenesis. J Cell Biol. 2012;197(4):487–497. doi:10.1083/jcb.201110058
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.