Back to Journals » Cancer Management and Research » Volume 12
Study on Targeting Relationship Between miR-320b and FGD5-AS1 and Its Effect on Biological Function of Osteosarcoma Cells
Authors Song QH, Guo MJ, Zheng JS, Zheng XH, Ye ZH, Wei P
Received 28 May 2020
Accepted for publication 29 July 2020
Published 31 December 2020 Volume 2020:12 Pages 13589—13598
DOI https://doi.org/10.2147/CMAR.S264682
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Qing-Hua Song,1,* Ming-Jun Guo,1,* Jun-Shui Zheng,2 Xue-Hong Zheng,1 Zhao-Hui Ye,1 Peng Wei1
1Department of Repair and Reconstruction Surgery, The First Hospital of Ningbo, Ningbo, Zhejiang Province 315000, People’s Republic of China; 2Medical College, Ningbo University, Ningbo, Zhejiang Province 315000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Peng Wei
Department of Repair and Reconstruction Surgery, The First Hospital of Ningbo, No. 59 Liuting Street, Haishu District, Ningbo, Zhejiang Province 315000, People’s Republic of China
Tel +86-13265981587
Email [email protected]
Objective: To probe into the expression of FGD5-AS1 in osteosarcoma and its relationship with miR-320b.
Methods: The tissue and serum samples of 97 patients with osteosarcoma were collected, and the serum samples of 100 healthy subjects who concurrently underwent physical examination were selected as the control. FGD5-AS1 expression in tissues and serum was detected, and osteosarcoma cells were transfected to measure cell behaviors such as proliferation, invasion and apoptosis.
Results: FGD5-AS1 was highly expressed in osteosarcoma, and its elevated expression indicated poor survival of patients. Serum FGD5-AS1 was related to tumor size and clinical stage and could be used for the diagnosis of osteosarcoma. The study of osteosarcoma cell lines U2OS and SaOS-2 showed that after inhibiting FGD5-AS1, the viability and invasion capacity of osteosarcoma cells decreased statistically compared with the control group (CG), while the apoptosis ability could be improved by further regulating apoptotic proteins (P< 0.05). Detection of EMT-related proteins identified that E-cadherin increased while N-cadherin decreased significantly after FGD5-AS1 inhibition (P< 0.05). Correlation analysis revealed a negative correlation between miR-320b and FGD5-AS1 (r = − 0.410, P< 0.001). Overexpression of miR-320b significantly inhibited cell viability, invasion and EMT ability, and increased the apoptosis rate, while inhibiting miR-320b expression produced the opposite results. The targeting relationship between miR-320b and FGD5-AS1 was confirmed through the biological prediction website, luciferase assay and RNA binding protein immunoprecipitation (RIP) assay. Inhibition of miR-320b could reverse the regulatory effect of FGD5-AS1 knockdown on osteosarcoma cells.
Conclusion: FGD5-AS1 is highly expressed in osteosarcoma and is involved in the biological procession of osteosarcoma by targeting miR-320b.
Keywords: miR-320b, FGD5-AS1, osteosarcoma, biological behavior
Introduction
Osteosarcoma occurs predominantly in adolescents and is a primary bone malignancy.1 Metastasis, especially in the lungs and bones, is present in approximately 15% to 20% of the patients when diagnosed, and develops rapidly.2 Pain and swelling of the affected bones are the main clinical manifestations of the disease. Thanks to the continuous development of social medical technology and the implementation of comprehensive treatment methods including chemotherapy and limb salvage surgery, the survival rate of osteosarcoma patients has been increased in the past few decades.3 It is reported that the 5-year survival rate of patients with osteosarcoma is about 70%.4 Although our understanding of osteosarcoma has been improved with the deepening of research, there are still many patients with chemotherapy resistance and lung metastasis with a 5-year survival rate of less than 20% and an unfavorable prognosis.5,6 And the molecular mechanisms underlying the development of carcinogenesis and metastasis are still unknown. Numerous studies have shown that microRNAs (miRNA) are involved in the progression of osteosarcoma,7 and their abnormal expression profiles are related to the prognosis of patients.8 It is also widely acknowledged that miRNAs are indispensable in regulating the self-renewal ability, invasion and migration of osteosarcoma cells.9 Among them, miR-320 has been pointed out by a number of studies as a potential therapeutic target for cancer, and a prognostic indicator for multiple tumors.10,11 Moreover, in osteosarcoma, miR-320 is reduced in cell lines, and inhibits the heterologous expression of cell proliferation and induces cell cycle arrest.11 The study of Lv et al12 suggested that high levels of XIST were negatively correlated with miR-320b, and could promote osteosarcoma progression by regulating miR-320b. All the preceding evidence suggests that miR-320b participates in the biological process of osteosarcoma.
The critical role of long non-coding RNAs (lncRNAs) in osteosarcoma has been already well established, but the role of the key lncRNAs in the pathogenesis of osteosarcoma has not been fully clarified. As a new type of lncRNA, FGD5-AS1 has been shown to be effective in promoting non-small cell lung cancer13 and colorectal cancer.14 Based on previous studies, we hypothesized that FGD5-AS1 was vital in tumor progression. In this study, we detected FGD5-AS1 and explored its effects on osteosarcoma cells by evaluating abnormal biological behaviors such as cell viability, apoptosis, and migration. In addition, as numerous studies indicated that lncRNA played a competitive role in endogenous RNA (ceRNA) and regulation of miRNA in various diseases,15,16 the regulatory relationship between FGD5-AS1 and miR-320b was detected in this study, and later through luciferase reporter, we confirmed that FGD5-AS1 targeted the regulation of miR-320b. Overall, the purpose of our study is to demonstrate the role of FGD5-AS1 in osteosarcoma and its regulatory mechanism with miR-320b.
Materials and Methods
Sample Collection
A total of 97 patients with osteosarcoma treated in The First Hospital of Ningbo were collected. The ratio of male to female was 57:40, the age ranged from 12 to 28 years old, and the average age was 18.87±5.18 years old. Concurrently, 100 healthy subjects, aged 12–26 years, with an average age of 18.57±4.98 years old and the male-to-female ratio of 55:45, who underwent health examinations were selected as the control group (CG). There was no significant difference in general clinical data between the two groups (P>0.05). The protocol of this study was approved by the Clinical Research Ethics Committee of The First Hospital of Ningbo, conducted in accordance with the Declaration of Helsinki, and all the participants had signed the informed consent. In this study, patients were followed up in the form of telephone interview, outpatient reexamination and medical record inquiry, and the overall survival (OS) was from the end of the first treatment to death or the end of follow-up.
Cell Culture
Osteosarcoma cell lines U2OS, HOS, SaOS-2, SW1353 and human normal osteoblast cell line hFOB1.19 were all purchased from Aolu Biotechnology Co., Ltd., Shanghai, China. The cell lines were incubated in Dulbecco’s Modified Eagle’s Medium (DMEM: Chundu Biotechnology Co., Ltd., Wuhan, China) supplemented with 10% heat-inactivated fetal bovine serum in a 5% CO2 constant temperature incubator at 37°C.
Cell Transfection
Using Lipofectamine (Invitrogen, United States, Lipofectamine® 2000 Transfection Reagent), FGD5-AS1 inhibitor (si-FGD5-AS1), negative control (si-NC), miR-320b overexpression/inhibitor (miR-320b mimics, miR-320b inhibitor) and negative control (miR-NC) were transfected into U2OS and SaOS-2 cell lines, respectively, for observation and research.
Quantitative PCR Detection
Total RNA was extracted from the collected cell, tissue and serum samples using TRIZOL reagent (Invitrogen, CA, United States). High capacity cDNA reverse transcription kit (Applied Biosystems, CA, United States) was used for reverse transcription of RNA samples, and the specific operation was carried out according to the instructions. Quantitative RT-PCR was performed by Fast Start Universal SYBR Green Master (Roche, CA, United States). Primers are shown in Table 1. The relative fold change of candidate genes was analyzed using the 2-ΔΔCt method (Table 1).
 |
Table 1 Primer Sequences |
Western Blot Detection
The collected cells were lysed using RIPA buffer (Sigma-Aldrich, MO, United States). Protein concentration was determined by BCA (Beyotime Biotechnology, Shanghai, China), and the protein samples were diluted with buffer solution. The same amount of extracts were separated by SDS-PAGE and then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Shanghai, China) after ionization. Then, the membrane was blocked with 5% skimmed milk for 2h, immersed in primary antibodies Bax, Caspase3, Bcl-2, E-cadherin, N-cadherin, and β-Actin primary antibody (Invitrogen, CA, United States), and then blocked at 4 °C overnight. After that, the primary antibody was removed by washing the membrane, and the horseradish peroxidase-labeled goat anti-rabbit secondary antibody (Invitrogen, CA, United States) was added for 1h of incubation at 37 °C. After rinsing the membrane three times with TBST, the band was analyzed using ECL Luminescence Reagent (Thermo Fisher, MA, United States).
CCK-8 Determination for Cell Viability
Osteosarcoma cells were inoculated into 96-well plates (2×103) with complete medium, and then transfected with different cell lines. After transfection for 24h, 48h, 72h, and 96h, 10 μL CCK-8 solution (Biosharp, Shanghai, China) was added to each well for 2h of culture at 37 °C. The optical density (wavelength 450nm) of each well was measured by enzyme-linked immunosorbent assay (ELISA; Multiskan ™ FC microplate reader, Thermo Fisher Scientific, MA, USA), and the cell growth curve was plotted. The experiment was repeated in triplicate.
Apoptosis Assay
AnnexinV-FITC apoptosis kit (Keygen, Nanjing, China) was used to detect the apoptosis. After transfection, the cells were digested with trypsin, washed with PBS, and then re-suspended in 500 μL binding buffer. AnnexinV-FITC and PI were then added to incubate in the dark for 15–20 minutes, and FC500MCL flow cytometry system was utilized for detection. The experiment was repeated 3 times.
Invasion Experiment
The invasive ability of the cells was evaluated by transwell assay. The cells were inoculated into the upper chamber coated with matrix glue (BD Biosciences, CA, United States), and the medium with and without 10%FBS was added to the lower and upper wells, respectively, and cultured at 37 °C for 24 h. After wiping the stroma and cells of the upper chamber that did not pass through the membrane surface, the cells were immobilized and stained with crystal violet. The invasion of cells was observed under a microscope.
Luciferase Reporter Gene Detection
Osteosarcoma cell line was seeded in 24-well plates. After incubation, the pmirGLO reporter vectors with wild-type (FGD5-AS1-WT) and mutant (FGD5-AS1-MUT) were co-transfected into cells with miR-320b-mimics and miR-NC. The luciferase intensity was detected using a dual-luciferase reporter gene system (Solarbio, CA, China), and the steps were carried out in strict accordance with the instructions.
RNA Binding Protein Immunoprecipitation (RIP Experiment)
The RIP kit (Millipore, MA, United States) was used for detection, and the experimental procedures were carried out in strict accordance with the kit instructions. The whole cell protein extracts were incubated with RIP wash buffer containing magnetic beads bound to human anti-AgO2 antibody (Millipore, MA, USA) or mouse immunoglobulin G (IgG) controls. Protein in the sample was digested with protease K, and immunoprecipitation RNA was extracted. The purified RNA was subjected to qRT-PCR analysis to prove the presence of binding targets.
Statistical Analysis
All the experiments were carried out three times independently. GraphPad 7 was adopted to analyze and visualize the data. The counting data were expressed as percentage (%) and verified by chi-square test and denoted as X2. Student’s t-test was utilized for the analysis of the two groups, one-way analysis of variance (ANOVA) was used to confirm the differences among multiple groups, and LSD-t-test was adopted for pairwise post hoc comparisons. The diagnostic value was analyzed by receiver operator characteristic (ROC) curve. The overall survival of patients was plotted by K-M survival curve and analyzed by Log-rank test. Correlation analysis of gene expression was performed using Pearson correlation coefficients. When P<0.05, there was a statistical difference.
Results
FGD5-AS1 Expression in Osteosarcoma
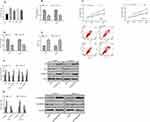
Through the detection of patient samples, it was found that the serum FGD5-AS1 of osteosarcoma patients was higher than that of normal controls (P<0.05), and FGD5-AS1 was higher in osteosarcoma tissues than in adjacent tissues (Figure 1). Correlation coefficient analysis showed that FGD5-AS1 in osteosarcoma tissue and serum was positively correlated, and serum FGD5-AS1 could be used in the diagnosis of osteosarcoma, with the AUC of 0.893. From Table 2, we can see that serum FGD5-AS1 was correlated with tumor size and clinical stage. Then, with the median value of FGD5-AS1 as the boundary, we classified patients into high and low expression groups, with 49 cases in each group. By analyzing the 5-year survival rate, it was found that the survival rate of patients with high FGD5-AS1 expression was significantly lower than that of patients with low expression (P<0.05). (Figure 1 and Table 2)
 |
Table 2 Relationship Between FGD5-AS1 and Pathological Characteristics of Patients with Osteosarcoma n [%] |
Effects of Inhibiting FGD5-AS1 on Tumor Cells
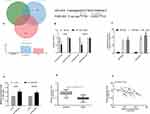
In this section, we conducted cytological experiments to further explore the role of FGD5-AS1 in osteosarcoma cells (Figure 2). It was found that FGD5-AS1 was higher in osteosarcoma cell lines U2OS, HOS, SaOS-2 and SW1353 than in human normal osteoblast cell line hFOB1.19 (P<0.05), so we selected U2OS and SaOS-2 cell lines for follow-up study to construct anti-FGD5-AS1 cell lines. The results showed that compared with the CG, FGD5-AS1 inhibition remarkably decreased the viability and invasion of osteosarcoma cells (P<0.05), and enhanced the apoptosis ability of osteosarcoma cells by further regulating the apoptotic proteins (P<0.05). Detection of EMT-related proteins revealed an increased E-cadherin and a decreased N-cadherin after FGD5-AS1 inhibition (P<0.05). (Figure 2)
Target Relationship Verification
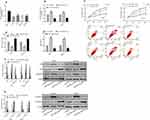
We searched for the target of FGD5-AS1 through starBase, lncRNASNP2, and Diana prediction websites, and found four common miRNA targets, among which, miR-320b had a strong binding site with FGD5-AS1. Through the starBase website, we found the binding site of miR-320b and FGD5-AS1 (Figure 4). Luciferase assay identified that the fluorescence activity of FGD5-AS1-WT was inhibited after miR-320b overexpression. Through RIP assay, it was observed that the miR-320b and FGD5-AS1 expression bound to Ago2 was higher than that bound to immunoglobulin (Ig) G. After transfection of osteosarcoma cells, it was noticed that miR-320b increased after inhibiting FGD5-AS1 (P<0.05). miR-320b presented low expression in osteosarcoma tissues, and correlation analysis revealed showed that miR-320b was negatively associated with FGD5-AS1 (r = −0.410, P<0.001). (Figure 3)
Effects of miR-320b Inhibition or Overexpression on Osteosarcoma Cells
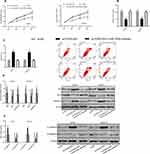
The miR-320b level of osteosarcoma cell lines U2OS, HOS, SaOS-2 and SW1353 was found to be lower than that of human normal osteoblast cell line hFOB1.19 (P<0.05). After constructing miR-320b inhibition/overexpression cell lines, it was found that compared with the CG, miR-320b overexpression significantly reduced the viability and invasion capacity of osteosarcoma cells (P<0.05), while increased the apoptosis ability by regulating the apoptotic protein (P<0.05). Detection of EMT-related proteins revealed that E-cadherin increased while N-cadherin decreased after miR-320b overexpression (P<0.05). However, the inhibition of miR-320b expression showed the opposite effect as that of overexpression (Figure 4).
Co-Transfection Experiment
To further explore the regulatory relationship between miR-320b and FGD5-AS1, we co-transfected si-FGD5-AS1 and miR-320b-inhibitor into cells. The results showed that there were no significant differences between si-FGD5-AS1 + miR-320b-inhibitor and si-NC in cell viability, invasion, apoptosis and EMT regulation (P>0.05). It can also be seen that si-FGD5-AS1 + miR-320b-inhibitor could reverse the effect of si-FGD5-AS1 on osteosarcoma cells (Figure 5).
Discussion
LncRNA is a class of non-protein transcript that can regulate various physiological and pathological functions.17 With the property of regulating signal transduction and protein transformation between protein complex tissues and cells, they are often used as feasible biomarkers for tumors.18 In osteosarcoma, lncRNAs participate in the occurrence of disease through epigenetic inheritance, transcription and post-transcription mechanisms.19 For example, Lu et al20 proposed that CASC2 could be used as a potential tumor suppressor lncRNA for osteosarcoma and participate in the regulation of malignant behavior of tumor cells. Upon review, it was found that the relationship between lncRNA and osteosarcoma proliferation, prognosis, chemotherapy resistance was introduced in detail, and it was pointed out that the role and regulatory mechanism of lncRNA in human osteosarcoma should be further studied.21
FGD5-AS1 is highly expressed in cancers and can function as a therapeutic target in oral squamous cell carcinoma.22 Since the current report on FGD5-AS1 in osteosarcoma has not been elaborated, we quantitatively detected by PCR to find that FGD5-AS1 was upregulated in osteosarcoma, which was consistent with the preceding research results, indicating its role as an oncogene. Combined with related literature,23,24 it is not difficult to find that serum biomarkers are widely used in the diagnosis of various diseases due to their easy access. For example, serum lncRNA XIST and HIF1A-AS1 enjoy favorable diagnostic and predictive value in non-small cell lung cancer.23 In this study, we found a correlation between the expression of serum and tissue through correlation coefficient analysis, and serum FGD5-AS1 can be used for the diagnosis of patients with osteosarcoma. In addition, the survival rate of osteosarcoma patients with high FGD5-AS1 expression was lower. These results suggest that FGD5-AS1 is a prognostic indicator for osteosarcoma, but the role of FGD5-AS1 in osteosarcoma remains unknown.
The occurrence and development of tumors is a process of gradual proliferation abnormality, while the spread mechanism of cancer cells includes cell invasion, migration and EMT.25,26 Therefore, we further conducted cytology experiments. First, the selected osteosarcoma cells were transfected. The results showed that the inhibition of FGD5-AS1 statistically reduced the viability and invasion of osteosarcoma cells, and increased the apoptosis ability by regulating apoptotic proteins. Changes in the biological traits of EMT will result in cells acquiring the ability to migrate and invade, while maintaining stem cell characteristics, tumor progression, and metastasis are all related to EMT.27,28 By detecting EMT-related proteins, we observed that FGD5-AS1 inhibition led to decreased EMT behavior, increased E-cadherin, and decreased N-cadherin (P<0.05), suggesting that FGD5-AS1 could be used as a therapeutic target for osteosarcoma to regulate the malignant behavior of tumor cells.
Increasing evidence has demonstrated that lncRNAs, highly expressed in the cytoplasm, can act as molecular sponges, competitively bind or adsorb with other regulatory proteins and miRNAs, thus participating in the biological processes of tumors.29,30 In this study, we predicted the lncRNA-miRNA binding sites through three biological target prediction sites: starBase, lncRNASNP2, and Diana. A total of 4 shared miRNAs were screened out, of which miR-320b belonged to the miR-320 family, which showed a correlation with osteosarcoma in previous reports.12 Further, the specific binding between FGD5-AS1 and miR-320b was confirmed by double luciferase report and RIP experiment, and there was a correlation between them. Recently, research has pointed out that miR-320b is involved in the regulation of the progression of a variety of cancers,31 and the inhibition of miR-320b can promote malignant behaviors such as cell proliferation in nasopharyngeal carcinoma32 and colorectal cancer.33 Therefore, we applied cytology to verify the effect of miR-320b on the biological behavior of osteosarcoma. The results showed that after miR-320b overexpression, the viability, invasion and EMT ability of osteosarcoma cells were decreased, and the apoptosis was increased, while the inhibition of miR-320b produced the opposite effects, further confirming the anti-tumor effect of miR-320b in osteosarcoma. Finally, through co-transfection experiments, we found that miR-320b inhibition could reverse the changes in cell behavior caused by silencing FGD5-AS1, suggesting that FGD5-AS1 could participate in the biological process of osteosarcoma by targeting miR-320b.
This study demonstrated the expression of FGD5-AS1 in osteosarcoma and the targeting relationship between miR-320b. However, as the experimental study of nude mice was absent in this study, the effect of FGD5-AS1 on the growth of solid tumors is not yet clear, which is one of the limitations. In addition, there are many network pathways involved in the regulation of osteosarcoma, and more experiments are needed to explore the involvement of FGD5-AS1 in mediating the specific downstream regulatory genes of miR-320b. What is more, the mechanism of drug resistance can also be investigated in future studies to better solve the limitations of clinical treatment. All in all, based on the comprehensive study, we conclude that FGD5-AS1 is involved in the biological process of osteosarcoma by targeting miR-320b.
Conclusion
This study demonstrated the expression of FGD5-AS1 in osteosarcoma and the targeting relationship between miR-320b. However, as the experimental study of nude mice was absent in this study, the effect of FGD5-AS1 on the growth of solid tumors is not yet clear, which is one of the limitations. In addition, there are many network pathways involved in the regulation of osteosarcoma, while miRNA is completed by regulating the downstream protein pathway, so the role of lncRNA/miRNA in osteosarcoma related pathways remains to be further investigated. What is more, the mechanism of drug resistance can also be investigated in future studies to better solve the limitations of clinical treatment. All in all, based on the comprehensive study, we conclude that FGD5-AS1 is involved in the biological process of osteosarcoma by targeting miR-320b.
Funding
This study was financially supported by The mechanism research of HIF-1 α regulating CXCR4 in lung metastasis of osteosarcoma Ningbo Natural Science Foundation, No.2019A610250.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lokau J, Schoeder V, Garbers C. [The role of interleukin-11 in osteosarcoma]. Pathologe. 2020;41:163–167. doi:10.1007/s00292-020-00756-1
2. Adamopoulos C, Gargalionis AN, Basdra EK, Papavassiliou AG. Deciphering signaling networks in osteosarcoma pathobiology. Exp Biol Med. 2016;241:1296–1305.
3. Birru B, Durthi CP, Kacham S, et al. Stem cells in tumour microenvironment aids for prolonged survival rate of cancer cells and developed drug resistance: major challenge in osteosarcoma treatment. Curr Drug Metab. 2020;21:44–52. doi:10.2174/1389200221666200214120226
4. Georges S, Calleja LR, Jacques C, et al. Loss of miR-198 and −206 during primary tumor progression enables metastatic dissemination in human osteosarcoma. Oncotarget. 2018;9:35726–35741. doi:10.18632/oncotarget.26284
5. Simpson S, Dunning MD, de Brot S, Grau-Roma L, Mongan NP, Rutland CS. Comparative review of human and canine osteosarcoma: morphology, epidemiology, prognosis, treatment and genetics. Acta Vet Scand. 2017;59:71. doi:10.1186/s13028-017-0341-9
6. Liu Y, Zhu ST, Wang X, et al. MiR-200c regulates tumor growth and chemosensitivity to cisplatin in osteosarcoma by targeting AKT2. Sci Rep. 2017;7:13598.
7. Fujii R, Osaka E, Sato K, Tokuhashi Y. MiR-1 suppresses proliferation of osteosarcoma cells by up-regulating p21 via PAX3. Cancer Genomics Proteomics. 2019;16:71–79. doi:10.21873/cgp.20113
8. Fujiwara T, Uotani K, Yoshida A, et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget. 2017;8:33375–33392. doi:10.18632/oncotarget.16498
9. Berlanga P, Munoz L, Piqueras M, et al. miR-200c and phospho-AKT as prognostic factors and mediators of osteosarcoma progression and lung metastasis. Mol Oncol. 2016;10:1043–1053. doi:10.1016/j.molonc.2016.04.004
10. Wang Y, Yu X, Wang L, Ma W, Sun Q. miR-320b is down-regulated in psoriasis and modulates keratinocyte proliferation by targeting AKT3. Inflammation. 2018;41:2160–2170. doi:10.1007/s10753-018-0859-7
11. Wu H, Li W, Zhang M, Zhu S, Zhang D, Wang X. Inhibitory roles of miR-320 in osteosarcoma via regulating E2F1. J Cancer Res Ther. 2016;12:68–71. doi:10.4103/0973-1482.191635
12. Lv GY, Miao J, Zhang XL. Long noncoding RNA XIST promotes osteosarcoma progression by targeting Ras-related protein RAP2B via miR-320b. Oncol Res. 2018;26:837–846. doi:10.3727/096504017X14920318811721
13. Fan Y, Li H, Yu Z, et al. Long non-coding RNA FGD5-AS1 promotes non-small cell lung cancer cell proliferation through sponging hsa-miR-107 to up-regulate FGFRL1. Biosci Rep. 2020;40. doi:10.1042/BSR20193309
14. Li D, Jiang X, Zhang X, Cao G, Wang D, Chen Z. Long noncoding RNA FGD5-AS1 promotes colorectal cancer cell proliferation, migration, and invasion through upregulating CDCA7 via sponging miR-302e. In Vitro Cell Dev Biol Anim. 2019;55:577–585. doi:10.1007/s11626-019-00376-x
15. Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol. 2019;13:46–60. doi:10.1002/1878-0261.12404
16. Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–1151. doi:10.1016/j.eururo.2013.12.003
17. Guo W, Jiang H, Li H, et al. LncRNA-SRA1 suppresses osteosarcoma cell proliferation while promoting cell apoptosis. Technol Cancer Res Treat. 2019;18:1533033819841438. doi:10.1177/1533033819841438
18. Sayad A, Hajifathali A, Hamidieh AA, Esfandi F, Taheri M. Fas-antisense long noncoding RNA and acute myeloid leukemia: is there any relation? Asian Pac J Cancer Prev. 2018;19:45–48. doi:10.22034/APJCP.2018.19.1.45
19. Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi:10.1186/s12943-018-0837-6
20. Lu L, Dai Z, Luo Q, Lv G. The long noncoding RNA cancer susceptibility candidate 2 inhibits tumor progression in osteosarcoma. Mol Med Rep. 2018;17:1947–1953. doi:10.3892/mmr.2017.8080
21. Yang Z, Li X, Yang Y, He Z, Qu X, Zhang Y. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis. 2016;7:e2389. doi:10.1038/cddis.2016.272
22. Liu L, Zhan Y, Huang Y, Huang L. LncRNA FGD5-AS1 can be predicted as therapeutic target in oral cancer. J Oral Pathol Med. 2020;49:243–252. doi:10.1111/jop.12989
23. Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887–7895.
24. Jin C, Shi W, Wang F, et al. Long non-coding RNA HULC as a novel serum biomarker for diagnosis and prognosis prediction of gastric cancer. Oncotarget. 2016;7:51763–51772. doi:10.18632/oncotarget.10107
25. Gholami MD, Falak R, Heidari S, et al. A truncated Snail1 transcription factor alters the expression of essential EMT markers and suppresses tumor cell migration in a human lung cancer cell line. Recent Pat Anticancer Drug Discov. 2019;14:158–169. doi:10.2174/1574892814666190527111429
26. Aiello NM, Maddipati R, Norgard RJ, et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev Cell. 2018;45:681–695 e684. doi:10.1016/j.devcel.2018.05.027
27. Natividad RJ, Lalli ML, Muthuswamy SK, Asthagiri AR. Golgi stabilization, not its front-rear bias, is associated with EMT-enhanced fibrillar migration. Biophys J. 2018;115:2067–2077. doi:10.1016/j.bpj.2018.10.006
28. Romeo E, Caserta CA, Rumio C, Marcucci F. The vicious cross-talk between tumor cells with an EMT phenotype and cells of the immune system. Cells. 2019;8:460. doi:10.3390/cells8050460
29. Laneve P, Po A, Favia A, et al. The long noncoding RNA linc-NeD125 controls the expression of medulloblastoma driver genes by microRNA sponge activity. Oncotarget. 2017;8:31003–31015. doi:10.18632/oncotarget.16049
30. Han Y, Wu N, Xia F, Liu S, Jia D. Long noncoding RNA GAS5 regulates myocardial ischemia-reperfusion injury through the PI3K/AKT apoptosis pathway by sponging miR5325p. Int J Mol Med. 2020;45:858–872. doi:10.3892/ijmm.2020.4471
31. Zhang S, Zhang X, Sun Q, et al. LncRNA NR2F2-AS1 promotes tumourigenesis through modulating BMI1 expression by targeting miR-320b in non-small cell lung cancer. J Cell Mol Med. 2019;23:2001–2011. doi:10.1111/jcmm.14102
32. Li Y, Tang X, He Q, et al. Overexpression of mitochondria mediator gene TRIAP1 by miR-320b loss is associated with progression in nasopharyngeal carcinoma. PLoS Genet. 2016;12:e1006183. doi:10.1371/journal.pgen.1006183
33. Wang H, Cao F, Li X, Miao H, Xing J, Fu CG. miR-320b suppresses cell proliferation by targeting c-Myc in human colorectal cancer cells. BMC Cancer. 2015;15:748. doi:10.1186/s12885-015-1728-5
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.