Back to Journals » Infection and Drug Resistance » Volume 13
Species Diversity, Molecular Characterization, and Antimicrobial Susceptibility of Opportunistic Actinomycetes Isolated from Immunocompromised and Healthy Patients of Markazi Province of Iran
Authors Azadi D , Motallebirad T , Ghaffari K, Shokri D, Rezaei F
Received 11 October 2019
Accepted for publication 13 December 2019
Published 7 January 2020 Volume 2020:13 Pages 1—10
DOI https://doi.org/10.2147/IDR.S234292
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Davood Azadi,1,2 Tahereh Motallebirad,1 Kazem Ghaffari,1 Dariuosh Shokri,3 Fatemeh Rezaei1
1Department of Laboratory Sciences, Khomein University of Medical Sciences, Khomein, Iran; 2Molecular Medicine Research Center, Faculty of Medicine, Arak University of Medical Sciences, Arak, Iran; 3Department of Biology and Microbiology, Shahid Ashrafi Isfahani University, Isfahan, Iran
Correspondence: Davood Azadi
Khomein University of Medical Sciences, Gods Street, Khomein, Iran
Tel +98-8646224143
Email [email protected]
Background: Actinomycetes widely exist in nature and these species cause infections in immunocompromised and healthy patients, although they are frequently found as members of the normal microbiota of humans and animals. These subsequent infections are often misdiagnosed as malignancy and tuberculosis. Due to this issue, the present study aimed to determine the presence and diversity of actinomycetes species causing infections in Iranian patients.
Materials and Methods: A total of 79 clinical samples collected from five hospitals in Markazi province were analyzed for the existence of actinomycetes using standard protocols for isolation and characterization of the isolates. The conventional tests were used for preliminary identification, the PCR amplification of hsp65 gene, the specific region of the 16S rRNA, and sequence analyses of 16S rRNA were applied for the genus and species identification. MICs of the antimicrobial agent were determined by the broth microdilution method and interpreted according to the NCCLS guidelines.
Results: A total of 17 (21.51%) actinomycetes isolates were recovered from clinical samples. In other analyzed samples, eight (10.12%) gram-positive, 12 (15.18) gram-negative bacteria, and six (7.6) fungi isolates were recovered. The most prevalent actinomycetes species were M. fortuitum (17.64%), N. Mexicana and S. heliomycini (11.76% each), and 10 species, ie, N. farcinica, M. lehmannii, M. flavescens, Arthrobacter crystalopoetis, N. neocaledoniensis, M. phocaicum, M. abscessus, M. arupense, M. setense, and N. cyriacigeorgica made up the single isolates. Results of DST illustrated that all of the isolates were susceptible to Amikacin, Levofloxacin, Ofloxacin, and Ciprofloxacin, whereas all of them were resistant to Rifampicin and Doxycycline.
Conclusion: In conclusion, increasing isolation of actinomycetes found in various clinical cases merits special attention by health authorities in developing countries. In health centers, action should be taken to increase awareness of appropriate diagnostic criteria and management guidelines for actinomycetes diseases. Furthermore, an increase in the number as well as the quality of national and regional reference laboratories may facilitate more accurate diagnosis of actinomycetes diseases.
Keywords: actinomycetes, 16SrRNA, drug susceptibility pattern, immunocompromised
Introduction
Actinomycetes are the general term for a heterogeneous group of gram-positive bacteria that grow as anaerobic facultative or aerobic rods with various degrees of branching.1 Actinomycetes species are widely distributed in nature and are found in soil, water, and decaying vegetation,2 and also frequently found as members of the normal microbiota in open cavities, especially the oropharynx, upper respiratory tract, gastrointestinal tract, and female genital tract.1
To date, the group actinomycetes comprise over eight genera and 500 species. Although M. tuberculosis is one of the most common species, the major genera of the actinomycetes group are the mycobacterium, corynebactenium, nocardioform actinomycetes, actinomyces, and streptomyces. Among the different species of actinomycetes identified, only a few lead to diseases such as mycobacterium, n ocardia, and actinomyces, moreover nowadays several other species are being increasingly recognized as significant human pathogens.3,4 These species have been cited as responsible pathogens for opportunistic infections in immunocompromised patients such as those with neoplastic diseases, diabetic patients, patients on immunosuppressive therapy, and those with autoimmune disorders.5,7 However, several recent reports indicated that nocardia and actinomyces can produce infections in patients with no preexisting illness, trauma, or immunosuppressive therapy.8,9 The major pathologic appearance of the disease by these bacteria is a granulomatous inflammatory reaction, which may progress to abscess formation.8,9
The conventional diagnosis of actinomycetes infections has been done by a combination of clinical and microbiological methods, however they are often non-specific and require invasive diagnostic biopsy procedures, though isolation and accurate characterization of isolates are difficult.10 Recent advances in microbiological and molecular methods in disease diagnosis and identification of bacterial isolates improve the outcomes of infected patients, helping clinicians and clinical microbiology personnel with the earliest possible diagnosis; and the evaluation of newer effective drug therapies for these patients.10,11 Due to this issue nowadays for isolation and identification of actinomycetes species used the various molecular method such as sequence analysis of housekeeping gene included 16SrRNA and rpoB gene etc, DNA–DNA hybridization and direct amplification of specific gene such as hsp65 and ITS gene.12,14
Despite the abundance of reports in the scientific literature regarding the isolation of actinomycetes from both clinical samples and non-hospitalized patients around the world,15,17 few studies have investigated the isolation and identification of actinomycetes species in clinical samples from Iran.18,19
Drug susceptibility testing results among the actinomycetes species is very different. For example, some mycobacterium, nocardia, and other actinomycetes species are sensitive to macrolides, aminoglycosides, and tigecycline, while some of them are resistant to this antibiotic.20,22 Due to this issue, antibiotic susceptibility testing should be performed In order to determine the susceptibility of the infectious strains for proper treatment of patients.
The aim of the current study was to assess the frequency, diversity, and drug susceptibility testing (DST) of actinomycetes species causing infection in hospitalized and non-hospitalized patients, by applying molecular and conventional microbiologic approaches in order to provide a deeper insight into their role in pathogenesis, nosocomial infections, and achieve a better diagnosis and effective drug therapies for infected patients.
Materials and Methods
Sampling and Isolation
From April 2018 to September 2019, there were a total of 79 clinical samples, including 10 sputum samples, 10 pleural fluid/pus samples, eight dental abscess samples, eight bronchial wash samples, 22 wound samples, nine blood samples, and 12 abscesses were collected from immunocompromised (patients with underlying disease such as malignancy, diabetes mellitus, HIV, organ transplantation, inherent immune deficiency, etc.) and immunocompetent patients (patients with no underlying disease) from five hospitals in Markazi province of Iran. The sources and clinical details of the isolates were summarized (Table 1). The sampling and pretreatment were carried out based on standard methods for isolation and identification of actinomycetes.19,23 In summary, each sample was transported at 4°C to the laboratory and processed within a maximum period of 24 hours. Initially non-sterile samples such as wound, pus, and sputum processed by NaOH 4% and HCL 1% and other sterile samples include blood, tracheal fluid, and pleural fluid condensed with centrifugation in 3,000 ×g, then inoculated in Lowenstein Jensen (LJ) (Merk, Germany), carbon free broth, blood agar (Merk, Germany), and Sauton’s media supplemented with antifungal and antibacterial antibiotics [kanamycin, nalidixic, and nystatin acid (each at 50 μg/mL)] and incubated for 8 weeks at 25, 32, and 37°C.24
 |
Table 1 Isolation Source, Underlying Disease of Patients, and Phenotypic and Biochemical Features of Actinomycetes Isolated from Clinical Samples |
This study was approved by the ethics committee of Arak University of Medical Sciences and Social Welfare Organization under which the private and public daycare nurseries or kindergartens are organized and operate (Grant No. 3005). Any patient’s participant provided an informed consent on their behalf.
Conventional Bacterial Identification
The clinical isolates were identified primarily as actinomycetes such as mycobacterium, nocardia, and streptomyces by conventional phenotypic and biochemical tests including acid-fast staining, semi-quantitative, and heat-stable (68°C) catalase production, tween opacity, nitrate reduction, urease activity, tellurite reduction, niacin accumulation, growth at 25, 32, 37, and 42°C, pigment production, resistance to lysozyme, hydrolysis of tyrosine, xanthine, and hypoxanthine tests.2,23 The recognition was further pursued by molecular tests as follows.
Molecular Identification
The method of Pitcher et al25 was used for DNA extraction with some modifications. In brief, for the lysis of actinomycetes cells, a pretreatment with lipase (2 mg/mL) and sonication was carried out followed by cell wall disruption with a high concentrations of lysozyme (200 mg/mL final concentration) and proteinase K (300 mg/mL final concentration) in the presence of sodium dodecyl sulfate (SDS) and additional treatment with guanidium isothiocyanate solution. The extracted DNA was purified with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) and chloroform-isoamyl alcohol (24:1, vol/vol) and precipitated with sodium acetate and ethanol at 20°C. Precipitated DNA was then washed with 70% ethanol and resuspended in 200 mL of Milli-Q water.
The isolates identified phenotypically as actinomycetes were further analyzed in terms of genus and species levels using a panel of molecular tests that included a genus-specific PCR based on a 596 bp region of the 16S rRNA for Nocardia and Streptomyces, as recommended by Laurent et al,26 and a genus-specific PCR based on a 618-bp fragment of the 65-kDa heat shock protein for mycobacterium, as recommended by Khan and Yadav,27 followed by the amplification and direct analysis of almost complete 16SrRNA sequencing for species identification, as described by Shojaei et al.28 Sequencing was performed by the Bioneer Company (South Korea), and the sequence data received were aligned manually with existing sequences of actinomycetes retrieved from the GenBank database and analyzed using the Blast program in GenBank and the jPhydit program.29
Drug Susceptibility Testing (DST)
To perform DST for clinical actinomycetes isolates, we selected amikacin, ciprofloxacin, cefoxitin, rifampicin, doxycycline, Imipenem, linezolid, and streptomycin based on Clinical & Laboratory Standards Institute (CLSI) 2017.30 The stock solutions of each agent were prepared by dissolving pure powder of individual drugs in sterile distilled water or another suitable solvent. All isolates were subcultured in Sauton’s media with 0.5% Tween 80 and subsequently incubated at 37°C for 7 days. For each isolate, a suspension in broth was prepared from isolates grown in agar medium and the inoculum was standardized for susceptibility testing to a turbidity equivalent to a 0.5 McFarland standard. Suspensions were then diluted and inoculated into 96-well microtiter plates.
Minimum inhibitory concentrations (MICs) of the antimicrobial agents were determined by the broth microdilution method and interpreted based on the guidelines established by the NCCLS.31 Microdilution plates were prepared manually for each antibiotic. Serial double dilutions of antimicrobial agents were prepared ranging from 0.06–512 mg/L, added to Mueller–Hinton broth plus 10% tween 80, and then 0.1 mL was dispensed into the wells of microdilution plates. Following inoculation, all cultures were incubated at 37°C. Growth was examined up to 7 days. The MIC was defined as the lowest concentration of the antibiotic that inhibited bacterial growth. Susceptible and resistant breakpoints were defined according to the NCCLS recommendations.31 Quality control of MICs was performed by testing NCCLS recommended reference strains, including Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212, and Mycobacterium peregrinum ATCC 700686.
Nucleotide Sequence Accession Numbers
The GenBank accession numbers for the 16S rRNA sequencing of clinical isolates of Iranian actinomycetes determined in this study are; N. farcinica MK478806, M. lehmannii MK478802, M. flavescens MK478804, M. fortuitum MK478942, N. mexicana MK478947, S. heliomycini MK478949, A. crystalopoetis MK483330, N. neocaledoniensis MK483335, M. phocaicum MK483337, M. arupense MK493306, M. setense KF019693, and N. cyriacigeorgica MF437315.
Result
In this study of the 79 patients referred to our laboratory, 32 (40.5%) were female and 47 (59.5%) were male, 18 (22.78%) were between the ages of 30 and 40 years, 16 (20.25%) were between the ages of 50 and 60 years, 14 (17.72%) were between the ages of 40 and 50 years, 12 (15.2%) were over the age of 70 years, 10 (12.65%) were between the ages of 20 and 30 years, five (6.32%) were between the ages of 60 and 70 years, and four (5%) were between the ages of 10 and 20 years. Forty-five patients were found to have various immunosuppressive syndromes, including malignancy (14 cases), diabetes mellitus (10 cases), HIV infection (5 cases), chronic obstructive pulmonary diseases (6 cases), hepatitis (6 cases), and autoimmune diseases (4 case), while the remainder of the patients had no apparent history of immune disorder.
In the present study, a total of 17 (21.51%) actinomycetes isolates were recovered from 79 clinical samples collected from immunocompromised and immunocompetent patients from Markazi province hospitals. Among all actinomycetes isolates, a total of 14 isolates were recovered from immunocompromised patients and three isolates were recovered from immunocompetent patients. In other analyzed samples, eight (10.12%) gram-positive, 12 (15.18%) gram-negative bacteria, and six (7.6%) fungi isolates were recovered. Of the total isolates, five isolates were from an abscess, four from a wound, three from a bronchial wash, two from a dental abscess and blood of each, and one isolate was from sputum.
On the basis of morphological, culture, and biochemical properties, and the genus specific marker, ie, presence of a 596 bp fragment of the 16S rRNA and a 618-bp fragment of the hsp65, all 17 isolates were identified as actinomycetes (nocardia, Streptomyces, and mycobacterium), of which nine isolates were identified as mycobacterium, five isolates were identified as Nocardia, two isolates were identified as Streptomyces, and one isolate was identified as Arthrobacter.
16S rRNA gene sequences’ analysis of the isolates revealed that all isolates had nucleotide signatures of actinomycetes, at positions 70–98 (U-A), 139–224 (G-C), 843 (C), 1008–1021 (C-G), 1189 (C), 1244–129 (C-G), and 1308–1329 (C-G) for mycobacterium and at positions 70–98 (A-T), 293–304 (G-T), 307 (C), 328 (T), 614–626 (A-T), 631(G), 661–744 (G-C), 824e876 (T-A), 825–875 (A-T), 843 (C), and 1122–1151 (A-T) for Nocardia and Streptomyces.32
The most prevalent actinomycetes species isolated from Iranian clinical samples were M. fortuitum (3 isolates, 17.64%), N. Mexicana and Streptomyces heliomycini (2 isolates, 11.76%) each and 10 species, ie, N. farcinica, M. lehmannii, M. flavescens, Arthrobacter crystalopoetis, N. neocaledoniensis, M. phocaicum, M. abscessus, M. arupense, M. setense, and N. cyriacigeorgica, made up the single isolates. The almost complete 16S rRNA gene sequences obtained for the species’ accurate identification of clinical isolates showed that all isolates had 100% similarity with the nearest standard type strain of actinomycetes.
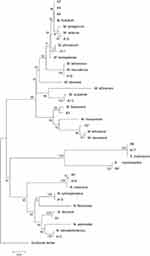
The phylogenic relationship between our isolates and the validated actinomycetes species was supported by a phylogenetic tree of 16S rRNA and by the high bootstrap value obtained using the neighbor-joining method with arithmetic mean using the matrix of pairwise differences (Figure 1).
Drug Susceptibility Pattern
Table 2 shows that the MIC of the tested isolates and the interpretation of activity of 10 antimicrobial agents against the 17 actinomycetes isolates. MIC lowest rates of antimicrobial agents against tested isolates belonged to Levofloxacin, Ofloxacin, and Ciprofloxacin, respectively, with a MIC rate of 0.25–2 µg/mL and the highest rates of MIC of antimicrobial agents belonged to Doxycycline and Isoniazid with MIC rate 4 (Table 2).
 |
Table 2 MIC Range and Susceptibility Pattern of Iranian Actinomycetes Using the Broth Microdilution Method |
Actinomycetes isolates recovered from immunocompromised patients had a higher MIC rate to Rifampicin, Doxycycline, Streptomycin, and Imipenem; however they had a low MIC rate to Ciprofloxacin, Ofloxacin, and Cefoxitin. Actinomycetes isolates recovered from immunocompetent patients had a higher MIC rate to Amikacin, Doxycycline, and Cefoxitin, but also had a low MIC rate to Streptomycin, Levofloxacin, and Kanamycin.
Discussion
Many species of actinomycetes such as nocardia, mycobacterium, and ie, are increasingly being recognized as important human pathogens in both immunocompetent and immunocompromised patients.17,33,34 The clinical presentation and severity of disease and the prognosis in an infected patient are extremely variable and may be determined by several factors such as the route of infection and the functioning level of the immune system.5,35 Likewise, accurate identification and DST of actinomycetes clinical isolates is a significant decision support tool which may help clinicians in order to select more appropriate therapy and, thus, improve the management and outcomes of some important actinomycetes diseases.
In low-income countries like Iran, where the microbiological diagnosis of infection agents relies mostly on microscopy, and also where access to culture and drug susceptibility testing is virtually non-existent, a clinician may neglect to consider the possibility that infections found in patients.28,36 Isolation and identification of actinomycetes species from clinical samples can often require specific preparatory procedures, while extensive microbial identification of this group of bacteria comprises a minimum of 10 phenotypic tests used to identify the entire spectrum of actinomycetes. These tests are difficult to interpret and generally lead to inconclusive results.36 Consequently, attempts to isolate and identify actinomycetes species by conventional phenotypic tests alone may result in erroneous or incomplete identification if a sufficient number of discriminative tests are not available.5 However, in response to the need for a more rapid and accurate identification of clinically relevant actinomycetes, appropriate use of molecular tests, in conjunction with the key phenotypic tests, has shown significant promise.28,36 Increasing recovery of actinomycetes species from environmental and clinical sources in different parts of the world17,37,39 prompted us to design the present study to determine the extent of the actinomycetes diversity in clinical samples of Markazi province of Iran. We envisaged that optimization of methods in isolation and accurate characterization of actinomycetes species from clinical samples by application of conventional microbiologic and molecular approaches is a key step towards the description of the emerging infections that they cause but are often ignored or missed.
In the current study, we isolated and characterized 17 (21.51%) actinomycetes species from 79 clinical samples collected from five educational hospitals. The actinomycetes species recovered from clinical samples in our studies included nine mycobacterial species, including M. fortuitum, M. phocaicum, M. lehmannii, M. abscessus, M. arupense, and M. setense, four nocardia species, N. Mexicana, N. farcinica, N. neocaledoniensis, and N. cyriacigeorgica, two Streptomyces heliomycini, and one Arthrobacter crystalopoetis. However, there are no comprehensive reports about isolation of actinomycetes from clinical samples. The results of this study compared with other reports showed that the M. fortuitum, N. farcinica, and N. cyriacigeorgica are the most common actinomycetes species isolated from the clinical samples in Iran and various parts of the world.28,40,41 In the current study we reported the first isolation of M. lehmannii, N. neocaledoniensis, S. heliomycini, and A. crystalopoetis from clinical samples.
In our study M. fortuitum was the most encountered actinomycete, including three isolates (17.64%). This organism is a rapidly growing mycobacteria initially isolated and characterized in 1938 from a stomach lavage specimen.42 These findings are in accordance with another report from various parts of the world, which showed the M. fortuitum is one of the most common actinomycetes species isolated from clinical samples.35 In the current study, M. fortuitum was isolated from lung secretion, abscess, and wound of cancerous patients.
N. Mexicana and S. heliomycini ranked second, which included 11.76% of the isolates. Based on literature, N. mexicana is a human opportunistic pathogen isolated from various clinical samples including bronchial secretions, mycetoma, and cutaneous botryomycosis, pulmonary, and cerebral abscess in humans and tenosynovitis and arthritis in animals.43,45 In the present study N. mexicana was isolated from a breast wound in immunocompetent patients. S. heliomycini is an antibiotics producer of bacteria that was first isolated and characterized in 1958 from soil.46 Based on the literature, there are no reports about isolation of S. heliomycini from clinical samples, nevertheless in the present study we reported the first isolation and characterization of S. heliomycini from a breast abscess of cancerous patient and a foot wound of diabetic patients.
In this study, ten species, ie, N. cyriacigeorgica, N. neocaledoniensis, N. farcinica, M. lehmannii, M. flavescens, A. crystalopoetis, M. phocaicum, M. abscessus, M. arupense, and M. setense consisted of the single isolates recovered from immunocompromised patients.
N. cyriacigeorgica is a human opportunistic pathogen that was first isolated from the bronchial secretions of a patient with chronic bronchitis in 2001.47 In our study, this organism was isolated from a brain abscess of a malignancy patient. N. farcinica is one of the first historically identified Nocardia species and has been the most commonly reported isolate from clinical and environmental samples worldwide.48,49 In the current study, N. farcinica was isolated from the breast abscess of a cancerous patient.
In this study M. setense, M. abscessus, M. phocaicum, M. flavescens, and M. arupense were recovered from foot wounds of immunocompetent patients, skin abscesses of organ recipient patients, foot wounds of diabetic patients, wounds of malignancy patients, and dental abscesses of malignancy patients, respectively. These mycobacteria are rare emerging organisms originally isolated from clinical specimens.50 Our study reports the first isolation of this organism from clinical samples in Iran.
In the present study N. neocaledoniensis, M. lehmannii, and A. crystalopoetis were recovered from surgery wounds of immunocompetent patients, abscesses of AIDS patients, and foot wounds of diabetic patients, respectively. These organisms were originally isolated from environmental resources of actinomycetes and there were no reports on the isolation of these bacteria from clinical samples.51,53 Here, we reported the first isolation and characterization of N. neocaledoniensis, M. lehmannii, and A. crystalopoetis from clinical specimens.
DST of actinomycetes clinical isolates may be even more critical in our region due to use of a traditional method for identification, leading to misdiagnosis with tuberculosis, mycetoma, mycosis, etc.28,54 In developing countries, for example, all smear positive patients are automatically given first line anti-TB agents, and the majority of NTM diseases in Iran are resistant to this treatment regimen.19,55 Likewise, DST of actinomycetes clinical isolates is an important decision tool which may help clinicians in order to select a suitable and efficient therapy and, thus, improve the management and outcomes of some actinomycetes diseases. Due to this issue in the present study DST for all actinomycetes isolates were done by applying the broth microdilution methods.
According to an antimicrobial susceptibility test of actinomycetes, clinical isolates N. neocaledoniensis and N. mexicana had low resistance, whiles S. heliomycini and N. cyriacigeorgica had a high resistance profile to our selected antibiotics. All of the isolates were susceptible to Amikacin, Levofloxacin, Ofloxacin, and Ciprofloxacin, whereas all of them were resistant to Rifampicin and Doxycycline.
Conclusion
In conclusion, increasing isolation of actinomycetes found in various clinical cases of healthy and immunocompromised patients and the high diversity of species of isolated actinomycetes merit special attention by health authorities, microbiologists, and clinicians in developing countries. In health centers, action should be taken to increase awareness of appropriate diagnostic criteria and management guidelines for actinomycetes diseases. Furthermore, an increase in the number as well as the quality of national and regional reference laboratories may cause more accurate diagnosis of actinomycetes diseases. Although the role of in vitro DST in the treatment of the majority of actinomycetes species has not been evaluated, it may play an essential role in the management of patients suffering from actinomycetes-related disease.
Acknowledgment
The authors are grateful to the Office of Vice-chancellor for Research of Khomein University of Medical Sciences for financial support of the current study (Grant No. 3005).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pierre I, Zarrouk V, Noussair L, Molina J-M, Fantin B. Invasive actinomycosis: surrogate marker of a poor prognosis in immunocompromised patients. Int J Infect Dis. 2014;29:74–79. doi:10.1016/j.ijid.2014.06.012
2. Saubolle MA. Aerobic actinomycetes. In: Diagnostic Microbiology of the Immunocompromised Host. American Society of Microbiology; ASM Press, Washington DC, 2009:269–281.
3. Dilip CV, Mulaje S, Mohalkar R. A review on actinomycetes and their biotechnological application. Int J Pharm Sci Res. 2013;4(5):1730.
4. from GM-BNU-t-D–r. 2019. Available from: https://www.dsmz.de/bacterial-diversity/prokaryotic-nomenclature-up-to-date/prokariotic-nomenclature-up-to-date.html.
5. Conville PS, Witebsky FG. Nocardia, Rhodococcus, Gordonia, Actinomadura, Streptomyces, and Other Aerobic Actinomycetes. Manual of Clinical Microbiology, 10th Edition. American Society of Microbiology; 2011:443–471.
6. Carr JM, Hagan G, Guest P, Gompertz S. A “not so superficial” skin infection in a patient with diabetes. BMJ Case Rep. 2012;2012:. doi:10.1136/bcr-2012-007062
7. Bang D, Herlin T, Stegger M, et al. Mycobacterium arosiense sp. nov., a slowly growing, scotochromogenic species causing osteomyelitis in an immunocompromised child. Int J Syst Evol Microbiol. 2008;58(10):2398–2402. doi:10.1099/ijs.0.65503-0
8. Corti ME, Fioti MEV. Nocardiosis: a review. Int J Infect Dis. 2003;7(4):243–250. doi:10.1016/S1201-9712(03)90102-0
9. Wong V, Turmezei T, Weston V. Actinomycosis. BMJ. 2011;343:d6099. doi:10.1136/bmj.d6099
10. Ahmed AA, van de Sande W, Fahal AH. Mycetoma laboratory diagnosis. PLoS Negl Trop Dis. 2017;11(8):e0005638. doi:10.1371/journal.pntd.0005638
11. Mencarini J, Antonelli A, Scoccianti G, et al. Madura foot in Europe: diagnosis of an autochthonous case by molecular approach and review of the literature. New Microbiol. 2016;39(2):156–159.
12. Rajivgandhi G, Ramachandran G, Maruthupandy M, Vaseeharan B, Manoharan N. Molecular identification and structural characterization of marine endophytic actinomycetes Nocardiopsis sp. GRG 2 (KT 235641) and its antibacterial efficacy against isolated ESBL producing bacteria. Microb Pathog. 2019;126:138–148. doi:10.1016/j.micpath.2018.10.014
13. Betrán A, Villuendas M, Rezusta A, Pereira J, Revillo M, Rodríguez-Nava V. Clinical significance, antimicrobial susceptibility and molecular identification of Nocardia species isolated from children with cystic fibrosis. Braz J Microbiol. 2016;47(3):531–535. doi:10.1016/j.bjm.2016.01.029
14. Madhaiyan M, Poonguzhali S, Saravanan VS, et al. Streptomyces pini sp. nov., an actinomycete isolated from phylloplane of pine (Pinus sylvestris L.) needle-like leaves. Int J Syst Evol Microbiol. 2016;66(10):4204–4210. doi:10.1099/ijsem.0.001336
15. Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207.
16. Fong P, Francis MJ, Hamblin JF, Korman TM, Graham M. Identification and diversity of Actinomyces species in a clinical microbiology laboratory in the MALDI-TOF MS era. Anaerobe. 2018;54:151–158. doi:10.1016/j.anaerobe.2018.09.007
17. Boyanova L, Kolarov R, Mateva L, Markovska R, Mitov I. Actinomycosis: a frequently forgotten disease. Future Microbiol. 2015;10(4):613–628. doi:10.2217/fmb.14.130
18. Irandoost M, Ghanbari MZ, Sakhaee F, et al. High rates of Mycobacterium fortuitum isolation in respiratory samples from Iranian patients with suspected tuberculosis: is it clinically important? J Med Microbiol. 2018;67(9):1243–1248. doi:10.1099/jmm.0.000814
19. Hashemi-Shahraki A, Heidarieh P, Bostanabad SZ, et al. Genetic diversity and antimicrobial susceptibility of Nocardia species among patients with nocardiosis. Sci Rep. 2015;5:17862. doi:10.1038/srep17862
20. Cowman S, Burns K, Benson S, Wilson R, Loebinger M. The antimicrobial susceptibility of non-tuberculous mycobacteria. J Infect. 2016;72(3):324–331. doi:10.1016/j.jinf.2015.12.007
21. McTaggart LR, Doucet J, Witkowska M, Richardson SE. Antimicrobial susceptibility among clinical Nocardia species identified by multilocus sequence analysis. Antimicrob Agents Chemother. 2015;59(1):269–275. doi:10.1128/AAC.02770-14
22. Njeru FM, Ndungu P, Bii C. Characterization and antimicrobial susceptibility of actinomycetes from TB smear negative and retreatment patients in Nairobi, Kenya. J Biosci Med (Irvine). 2019;7(8):1–12. doi:10.4236/jbm.2019.78001
23. Saubolle MA, Sussland D. Nocardiosis review of clinical and laboratory experience. J Clin Microbiol. 2003;41(10):4497–4501. doi:10.1128/JCM.41.10.4497-4501.2003
24. Azadi D, Shojaei H, Mobasherizadeh S, Naser AD. Screening, isolation and molecular identification of biodegrading mycobacteria from Iranian ecosystems and analysis of their biodegradation activity. AMB Express. 2017;7(1):180. doi:10.1186/s13568-017-0472-4
25. Pitcher D, Saunders N, Owen R. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8(4):151–156. doi:10.1111/j.1472-765X.1989.tb00262.x
26. Laurent FJ, Provost F, Boiron P. Rapid identification of clinically relevant Nocardia species to genus level by 16S rRNA gene PCR. J Clin Microbiol. 2000;38(7):2805.
27. Khan IU, Yadav JS. Development of a single-tube, cell lysis-based, genus-specific PCR method for rapid identification of mycobacteria: optimization of cell lysis, PCR primers and conditions, and restriction pattern analysis. J Clin Microbiol. 2004;42(1):453–457. doi:10.1128/JCM.42.1.453-457.2004
28. Shojaei H, Heidarieh P, Hashemi A, Feizabadi M, Daei NA. Species identification of neglected nontuberculous mycobacteria in a developing country. Jpn J Infect Dis. 2011;64(4):265–271.
29. Jeon Y-S, Chung H, Park S, Hur I, Lee J-H, Chun J. jPHYDIT: a JAVA-based integrated environment for molecular phylogeny of ribosomal RNA sequences. Bioinformatics. 2005;21(14):3171–3173. doi:10.1093/bioinformatics/bti463
30. Wayne P. Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute (CLSI); 2017:20.
31. Woods GL, Brown-Elliott B, Desmond EP, et al. Susceptibility testing of mycobacteria, Nocardia, and other aerobic actinomycetes; approved standard. NCCLS Document M24-A. 2003;23:18.
32. Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Sys Evol Microbiol. 1997;47(2):479–491.
33. Cortez-Escalante JJ, Santos A, Garnica G, Sarmento AL, Castro C, Romero GAS. Mediastinitis and pericardial effusion in a patient with AIDS and disseminated Mycobacterium avium infection: a case report. Rev Soc Bras Med Trop. 2012;45(3):407–409. doi:10.1590/S0037-86822012000300027
34. Ambrosioni J, Lew D, Garbino J. Nocardiosis: updated clinical review and experience at a tertiary center. Infection. 2010;38(2):89–97. doi:10.1007/s15010-009-9193-9
35. Prevots DR, Marras TK. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin Chest Med. 2015;36(1):13–34. doi:10.1016/j.ccm.2014.10.002
36. Azadi D, Motallebirad T, Ghaffari K, Shojaei H. Mycobacteriosis and tuberculosis: laboratory diagnosis. Open Microbiol J. 2018;12:41. doi:10.2174/1874285801812010041
37. DeWitt JP, Stetson CL, Thomas KL, Carroll BJ. Extensive cutaneous botryomycosis with subsequent development of nocardia-positive wound cultures. J Cutan Med Surg. 2018;22(3):344–346. doi:10.1177/1203475418755762
38. Kim S, Lee KL, Lee DM, et al. Nocardia brain abscess in an immunocompetent patient. Infect Chemother. 2014;46(1):45–49. doi:10.3947/ic.2014.46.1.45
39. Raby E, Hiew V, Arthur I. A case of Nocardia mexicana cerebral abscess highlights deficiencies in susceptibility testing and the utility of direct molecular identification. Pathology. 2016;48(5):508–510. doi:10.1016/j.pathol.2016.03.016
40. Bhargava A, Kombade S, Dash D, Jain Y. Disseminated nocardiasis by Nocardia farcinica: review and first case report from Central India. Med J Armed Forces India. 2019;75(1):106–111. doi:10.1016/j.mjafi.2018.11.006
41. Kandi V. Human Nocardia infections: a review of pulmonary nocardiosis. Cureus. 2015;7:8. doi:10.7759/cureus.268
42. Okamori S, Asakura T, Nishimura T, et al. Natural history of Mycobacterium fortuitum pulmonary infection presenting with migratory infiltrates: a case report with microbiological analysis. BMC Infect Dis. 2018;18(1):1. doi:10.1186/s12879-017-2892-9
43. Rodriguez-Nava V, Couble A, Molinard C, Sandoval H, Boiron P, Laurent F. Nocardia mexicana sp. nov., a new pathogen isolated from human mycetomas. J Clin Microbiol. 2004;42(10):4530–4535.
44. Ikeda T, Nakanishi H, Morishita Y, Houdai K, Ito J, Gonoi T. First case report of pulmonary nocardiosis caused by Nocardia mexicana. JMM Case Rep. 2016;3(4):e005054.
45. Owen H, Buckle K, Olm J, et al. Isolation of Nocardia mexicana from focal proliferative tenosynovitis and arthritis in a steer. Aust Vet J. 2015;93(5):170–173. doi:10.1111/avj.v93.5
46. Avila-Flores R, Medellin RA. Ecological, taxonomic, and physiological correlates of cave use by mexican bats. J Mammal. 2004;85(4):675–687. (). doi:10.1644/BOS-127
47. Yassin A, Rainey F, Steiner U. Nocardia cyriacigeorgici sp. nov. Int J Syst Evol Microbiol. 2001;51(4):1419–1423. doi:10.1099/00207713-51-4-1419
48. Elsayed S, Kealey A, Coffin CS, Read R, Megran D, Zhang K. Nocardia cyriacigeorgica septicemia. J Clin Microbiol. 2006;44(1):280–282. doi:10.1128/JCM.44.1.280-282.2006
49. Kageyama A, Hoshino Y, Yazawa K, et al. Nocardia cyriacigeorgica is a significant pathogen responsible for nocardiosis in Japan and Thailand. Mycopathologia. 2005;160(1):15–19. doi:10.1007/s11046-005-3050-2
50. Nasiri MJ, Dabiri H, Darban-Sarokhalil D, Shahraki AH. Prevalence of non-tuberculosis mycobacterial infections among tuberculosis suspects in Iran: systematic review and meta-analysis. PLoS One. 2015;10(6):e0129073. doi:10.1371/journal.pone.0129073
51. Saintpierre-Bonaccio D, Maldonado LA, Amir H, Pineau R, Goodfellow M. Nocardia neocaledoniensis sp. nov., a novel actinomycete isolated from a new-caledonian brown hypermagnesian ultramafic soil. Int J Syst Evol Microbiol. 2004;54(Pt 2):599–603. doi:10.1099/ijs.0.02881-0
52. Nouioui I, Sangal V, Carro L, et al. Two novel species of rapidly growing mycobacteria: mycobacterium lehmannii sp. nov. and Mycobacterium neumannii sp. nov. Int J Syst Evol Microbiol. 2017;67:4948–4955. doi:10.1099/ijsem.0.002350
53. Busse HJ, Wieser M, Buczolits S. Arthrobacter. In: Bergey’s Manual of Systematics of Archaea and Bacteria. 2nd ed., vol. 1, Springer-Verlag, New York, NY, 2015:1–70.
54. Nai-Yang L, Qi Z. Canalicular inflammatory etiology and the common misdiagnosis study. Int Eye Sci. 2016;16(11):2154–2156.
55. Mirsaeidi SM, Tabarsi P, Khoshnood K, et al. Treatment of multiple drug-resistant tuberculosis (MDR-TB) in Iran. Int J Infect Dis. 2005;9(6):317–322. doi:10.1016/j.ijid.2004.09.012
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.