Back to Journals » OncoTargets and Therapy » Volume 13
SP1-Mediated Upregulation of lncRNA LINC01614 Functions a ceRNA for miR-383 to Facilitate Glioma Progression Through Regulation of ADAM12
Received 18 December 2019
Accepted for publication 26 March 2020
Published 18 May 2020 Volume 2020:13 Pages 4305—4318
DOI https://doi.org/10.2147/OTT.S242854
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Hao Wang,1 Jiang Wu,2 Wei Guo1
1Department of Neurosurgery, Shenzhen People’s Hospital, The Second Clinical Medical College of Jinan University, Shenzhen, Guangdong, People’s Republic of China; 2Department of Neurosurgery, Hebei General Hospital, Shijiazhuang, Hebei, People’s Republic of China
Correspondence: Wei Guo
Department of Neurosurgery, Shenzhen People’s Hospital, The Second Clinical Medical College of Jinan University, No. 1017, Dongmen North Road, Shenzhen, Guangdong, People’s Republic of China
Email [email protected]
Background: Long non-coding RNAs (lncRNAs) play an imperative role in tumorigenesis, but few lncRNAs have been functionally characterized in glioma. The aim of the present study was to identify the role of long non-coding RNA LINC01614 (LINC01614) in glioma development and explore the underlying mechanisms of LINC01614/miR-383/ADAM12 axis.
Patients and Methods: LncRNA expression in glioma specimens was measured by lncRNA microarray and qRT-PCR. The prognostic value of LINC01614 expression was statistically analyzed in 112 glioma patients. Loss-of-function experiments were conducted to investigate the biological functions of LINC01614 in vitro. Luciferase analyses, ChIP assays, and RNA pull-down were performed to determine the underlying LINC01614 mechanisms.
Results: We identified a novel glioma-related lncRNA LINC01614 by analyzing TCGA datasets. The distinct upregulation of LINC01614 was observed in both glioma specimens and cell lines using RT-PCR. We also observed that LINC01614 upregulation was induced by nuclear transcription factor SP1. Clinical assays revealed that high levels of LINC01614 were associated with KPS, WHO grade and shorter overall survival of glioma patients. Multivariate analysis further confirmed that LINC01614 was an independent prognostic marker for glioma patients. Besides, functional assays displayed that silence of LINC01614 knockdown distinctly inhibited cell growth, migration and invasion and promoted cell apoptosis in glioma cells. LINC01614 expression was enriched in the cytoplasm of glioma cells. Mechanistic investigation revealed that LINC01614 functioned as a competing endogenous RNA to upregulate a disintegrin and metalloproteinase 12 (ADAM12) by sponging miR-383.
Conclusion: Overall, these findings showed that SP1-induced upregulation of LINC01614 promoted glioma malignant progression via modulating the miR-383/ADAM12 axis, which may provide a promising therapy for glioma.
Keywords: lncRNA LINC01614, miR-383, ADAM12, prognosis, metastasis, glioma
Introduction
Gliomas are the most frequent type of primary brain cancer (45–55%) in adults and encompass a spectrum of neoplasms which vary in aggressiveness and differentiation.1 Based on the cellular behaviors and histopathological features of gliomas, they can be divided into four grades (I, II, III and IV).2,3 Especially, glioblastoma (III and IV in grade) are the aggressive malignant neoplasms.4 Despite surgical treatment combined with radiotherapy, chemotherapy, rising targeted therapy and immunotherapy, the overall survival for patients with glioblastoma remains unfavorable.5,6 The poor clinical outcome of glioblastoma patients is due to distant metastasis and the limited understanding involved in its molecular mechanisms.7,8 Thus, it is critical to develop efficient biomarkers and novel therapeutic reagents to prolong survivals among glioma patients.
Long non-coding RNAs (lncRNAs) refer to t RNA transcripts with more than 200 nucleotides with no protein-coding capacity and are poorly conserved.9 Previously, lncRNAs are considered “transcription noise,” meaning that they are useless in the biological progression.10 However, growing evidences highlight the fact that lncRNAs play an imperative role in the regulation of various genes through some complex mechanisms, such as epigenetic modification.11,12 Abnormal content or functions of lncRNAs have been demonstrated to be associated with tumorigenesis and metastasis, suggesting that lncRNAs may be novel biomarkers for tumor diagnosis and potential therapeutic targets.13,14 Although many functional lncRNAs have been identified using bioinformatics platform and various cellular functional experiments, a large number of lncRNAs related to glioma progression remain to be further studied.15,16 Thus, analyzing the clinical significance, biological function and molecular mechanisms of lncRNAs in glioma is of great importance.
LncRNA LINC01614 (LINC01614), located in 2q35, was firstly identified to be a tumor promoter in lung cancer by Liu et al.17 In their study, upregulation of LINC01614 was observed in both lung cancer tissues and cells. Functional assays revealed that knockdown of this lncRNA suppressed cell growth and accelerate apoptosis via modulating miRNA-217/FOXP1 axis. Then, LINC01614 was also reported to be overexpressed in breast cancer and was proved to have great prognostic values in patients with breast cancer.18 However, the expression and biological function of LINC01614 in glioma are unclear.
Patients and Methods
Patients and Specimens
Glioma tumor samples and matched normal specimens were collected from 112 patients who underwent surgical resection at Shenzhen People’s Hospital between June 2011 and May 2014. Tissue samples were obtained immediately after the surgical resection and stored in liquid nitrogen. None of 112 patients receive anti-cancer therapy prior to surgical resection. All clinical experiments were carried out in accordance with the Declaration of Helsinki and undertaken with the approval and written consent of all patients. The study was approved by the Ethics Committee of Shenzhen People’s Hospital.
Public Data Collection and Bioinformatics Analyses
Microarray data and their clinical data were downloaded from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov). “R” software was used for the assays of the collected data. An online tool GEPIA (Gene Expression Profiling Interactive Analysis) was used to analyze the possible influence of genes on the long-term survival of Glioma patients. LncBook algorithm (https://bigd.big.ac.cn/lncbook/index) was used to study the methylation levels of LINC01614 promoter region in glioma specimens. cBioPortal algorithm (https://www.cbioportal.org/) was also employed to analyze the genetic alternation of the potential genes in glioma specimens.
Cell Transfection
Glioma cells (LN18, U251, T98G, LN229 and A172) and NHAs cells (as control cells) were bought from QiBio Biotechnology company (Qingdao, Shandong, China). The cells were cultured in flasks containing RPMI-1640 media (with 10% FBS). All the cells were maintained in an incubator with 37°C and 5% CO2. Cell transfection was conducted by using Lipofectamine 2000 reagent kits (Dongyuan, Ningbo, Zhejiang, China). The siRNAs targeting SP1 (si-SP1), negative control siRNAs (si-NC), siRNAs targeting LINC01614 (si-LINC01614#1, si-LINC01614#2) were synthesized by Biotek Biological company (Xiamen, Fujian, China). miRNA mimics or inhibitors were obtained from Shanghai Shenggong Biological company (Songjiang, Shanghai, China). The overexpressing plasmids including SP1 (ov-SP1), LINC01614 (ov-LINC01614) and ADAM12 (ov-ADAM12) were constructed by MAS Biological company (Pudong, Shanghai, China).
Real-Time PCR
Total RNA extraction was performed using Trizol reagents (Qikun, Kunming, Yunnan, China). Reverse transcription was conducted using the cDNA synthesis kits (Takara, Dalian, Liaoning, China). The qPCR detection for SP1, LINC01614 and ADAM12 was conducted by using SYBR Green qPCR kits (MCE, Hangzhou, Zhejiang, China) according to the kits’ protocols. For miR-383 detection, total miRNAs were extracted using Qiagen miRNeasy mini kits (Kewei, Hefei, Anhui, China), and qPCR for miR-383 detection was carried out using miRcute miRNA qPCR (SYBR Green) kits (Tiangen, Xuhui, Shanghai, China) in accordance with the kits’ protocols. The relative levels of lncRNAs and miRNAs were calculated using the 2−ΔΔCt methods with the U6 or GAPDH as the endogenous reference. Table 1 shows the primers used in this study.
 |
Table 1 The Primers Used in This Study for RT-PCR |
Cell Viability Detection
Cellular viabilities were determined using Cell Counting Kit-8 (CCK-8) assays by applying CCK-8 kits (Hongjun, Nanjing, Jiangsu, China). Briefly speaking, GLIOMA cells after LINC01614 siRNAs transfection at a density of 2500 cells/well were grown in plates (ninety-six-well). At indicated time points (0, 1, 2, 3 and 4 days, respectively), 25 μL CCK8 reagents were placed into the plates (per well). The plates were then kept in an incubator (37°C with 5% CO2) for 2.5 h, followed by being examined using a microplate reader apparatus at an optical density (OD) 450 nm.
Colony Formation Assays
GBM cells after LINC01614 siRNAs transfection were seeded into six-well plates (800 cells/well). Colonies were allowed to form for about two weeks in RPMI-1640 media (with 10% FBS) in an incubator with 37°C and 5% CO2. Then, the colonies were washed and treated with methanol (100%) and crystal violet (0.2%) for a quarter. After washing with PBS twice, the colonies were photographed by a microscope.
EdU Assays
Cell-Light EdU assay kits (Ruibo, Guangzhou, Guangdong, China) were also applied to determine the proliferation of the treated glioma cells. In short, the LINC01614 siRNAs-transfected glioma cells were, respectively, collected and placed into 48‐well plates. After being incubated with 50 μM EdU reagents (100 μL per well) for 2.5 h, the cells were treated with paraformaldehyde (4%) and Apollo Dye Solution. DAPI solution was then used to stain the nuclei. After washing with PBS, images were acquired with a fluorescence microscope.
Apoptosis Detection
Annexin V-FITC apoptosis detection kits were purchased from Beyotime (Nanjing, Jiangsu, China) and were used for the determination of cellular apoptosis. In brief, glioma cells after LINC01614 siRNAs transfection were harvested using 0.25% trypsin, and collected in a centrifuge tube. After centrifugation (500×g/min, 5 min, 4°C), the supernatants were discarded, and the plates were resuspended in 500 μL binding buffer. Then, Annexin V-FITC reagents (5 μL) and PI solution (10 μL) were added into the cells. After incubation in the dark for a quarter a quarter, the cells were subjected to flow cytometry assays.
Caspase 3/9 Determination
Caspase 3/9 activity detection kits (Meijie, Xiamen, Fujian, China) were used for caspase 3/9 determination. In short, glioma cells after transfection with LINC01614 siRNAs were collected and Lysis Buffer was placed into the cells. After incubation for 15 min on ice, the mixtures were centrifuged (10,000 ×g, 10 min) and the supernatants were obtained, followed by adding Ac-DEVD-pNA buffer into the supernatants and incubating for 1.5 h. Then, the OD405 nm absorbance was examined by a microplate reader.
Wound-Healing Assays
Glioma cells after LINC01614 siRNAs treatment were grown in 12-well plates and the cell confluence reached about 100%. Then, the cells were scratched by 200 μL pipette tips and the wounds were generated. Floating cells and debris were removed by washing twice with PBS, followed by being cultured for 48 h. Images of the wound closures were taken at 0 h and 48 h by a microscope.
Transwell Assays
Glioma cells were transfected with LINC01614 siRNAs and then collected in a centrifuge tube. The treated cells were washed and resuspended (2.5×105 cells per well) in 250 μL serum-free media. The cell mixtures were added into Corning transwell inserts (pre-coated with Matrigel; Dongsheng, Qingdao, Shandong, China). Then, 650 μL chemo-attractants (RPMI-1640 media with 15% FBS) were added into the bottom chambers of each transwell. Twenty-four hours later, the invading cells were treated with methanol (100%) and crystal violet (0.2%). After washing with PBS three times, the images of the invasive cells were acquired by using a microscope.
Subcellular Fractionation Assays
Life Technologies PARIS kits were applied for the separation of nuclear and cytosolic fractions in accordance with the kits’ protocols. In brief, cells were collected and resuspended in 450 μL ice-cold fractionation buffer, followed by being incubated for 15 min. After the mixtures were centrifuged (500 ×g/min, 5 min, 4°C), the cytoplasmic fraction was attentively aspirated and then used for RNA extraction. Subsequently, the nuclear pellet was lysed, and the samples were then used for RNA isolation. Finally, the RNAs isolated from nuclear and cytosolic fractions were subjected to qRT-PCR assays. U2 was used as the control for cytonuclear expressions and HPRT for cytoplasmic expressions.
ChIP Assays
EpiQuik ChIP assay kits (Aimeijie, Suzhou, Jiangsu, China) were used for ChIP assays. In short, 1×107 LINC01614 siRNA-transfected glioma cells were collected and treated with formaldehyde (final concentration: 1%) for 15 min at 37°C, followed by adding with glycine (final concentration: 125 nM) and incubating at room temperature for 5 min. Subsequently, the cells were washed with PBS and added with lysis buffer, followed by incubation at 4°C for 15 min. Then, the cell lysates were sonicated to generate about 200 to 400 bp DNA fragments. Afterwards, the supernatants were obtained by centrifuging and incubated with anti-SP1 antibody (Abcam, Cambridge, MA, USA) or IgG (as a negative control; BOSTER, Wuhan, Hubei, China). Then, the complexes were precipitated according to the kits’ protocols, and the DNA was eluted and purified, followed by being detected by qPCR analysis as described above.
RNA-Pull Down Assays
Biotin-labeled LINC01614 (LINC01614-biotin) and negative control biotin-labeled probe (NC-biotin) were bought from Fungene Biological company (Changsha, Hunan, China). NC-biotin or LINC01614-biotin was, respectively, mixed with U251 or LN299 cell lysates. Subsequently, magnetic beads (Invitrogen, Pudong, Shanghai, China) were put to the binding reactions for incubation at room temperature. Then, the beads were washed, and the eluted RNAs were measured by qPCR analysis.
Luciferase Reporter Assays
The sequence region containing predicted binding site 2 (S2 WT) of LINC01614 promoter or the corresponding mutant-type (MUT) sequence (S2 MUT) was, respectively, cloned into pGL3 reporter plasmid. LINC01614 sequence was then constructed into pGL3 reporter plasmid (LINC01614 wild-type), and LINC01614 sequence containing the predicted mutant-type binding site between LINC01614 and miR-383 was also cloned into pGL3 reporter plasmid (LINC01614 mutant-type). In addition, the luciferase reporter plasmids of wild-type ADAM12 (ADAM12 wt1, ADAM12 wt2) or matched mutant-type ADAM12 (ADAM12 mut1, ADAM12 mut2) were also constructed. The vector construction was performed by Geno Biological company (Changsha, Hunan, China). Glioma cells were co-transfected with corresponding luciferase reporters plus Renilla luciferase plasmid (pRL-TK) and miR-383 mimics. Then, the cells were harvested after 48 hrs for luciferase detection by using Promega Luciferase Reporter detection kits (Songhong, Qingdao, Shandong, China).
Statistical Analysis
SPSS version 19.0 software (Chicago, Chicago, IL, USA) was employed to do statistical analysis. Kaplan–Meier methods with Log-rank tests were applied to determine the overall survivals. Student’s t-test and a one-way ANOVA were carried out to assess the significant differences. A Cox proportional hazards model was used for the multivariate assays. p values <0.05 were considered as being statistically significant.
Results
Highly Expressed LINC01614 in Glioma Tumor Samples and Cells
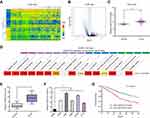
To screen potential functional lncRNAs in glioma, we used “R” statistical software for the assays of microarray data from TCGA datasets. The expression pattern of dysregulated lncRNAs was shown using Heat Map (Figure 1A) and Volcano plots (Figure 1B). Of all these lncRNAs, LINC01614 was distinctly upregulated, with an average increase of 2.9 times (Figure 1C). In addition, we also observed that the upregulation of LINC01614 was a common event in the great majority of tumors (Figure 1D). Then, the levels of LINC01614 were examined in 112 glioma patients using RT-PCR. Data revealed that LINC01614 was distinctly upregulated in tumor specimens compared with corresponding normal brain specimens (p < 0.01, Figure 1E). Moreover, we assessed the expressions of LINC01614 in several glioma cell lines using RT-PCR, finding that LINC01614 was obviously elevated in five glioma cell lines compared with that in NHAs cells (Figure 1F). Overall, our findings suggested that overexpression of LINC01614 may be involved in the progression of glioma.
LINC01614 UpRegulation Associated with Clinical Outcome of Glioma Patients
To study the clinical significance of LINC01614 in glioma patients, the LINC01614 expressions were classified as low or high in relation to the median value. As shown in Table 2, the results of chi-square test revealed that high LINC01614 expressions were associated with higher KPS (p = 0.017) and advanced WHO grade (p = 0.012). However, no significant difference in LINC01614 expression was observed with other clinical factors (p > 0.05). Moreover, we performed Kaplan–Meier analysis and Log-rank test to explore the associations between LINC01614 expression and survival of glioma patients, finding that the patients with higher levels of LINC01614 expression had significantly shorter survival time, compared with those with lower LINC01614 expression (p = 0.0075, Figure 1G). On the other hand, the univariate analysis identified five prognostic factors: KPS, WHO grade and LINC01614 expression (all p > 0.05, Table 2). When it comes to multivariate, we observed that KPS (p = 0.021), WHO grade (p = 0.013), and LINC01614 expression level (HR=2.731, 95% CI: 1.217–4.387, p = 0.024) served as independent prognostic factors for glioma patients (Table 3).
 |
Table 2 Clinical Association Between LINC01614 Expression and Clinicopathological Variables in Glioma Patients |
 |
Table 3 Univariate and Multivariate Analysis of Overall Survival in Glioma Patients |
SP1 Activated LINC01614 Expression Through Binding to Its Promoter
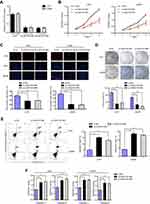
Since LINC01614 was up-regulated in glioma, we next sought to uncover the mechanisms that contributed to LINC01614 high expression. First, we searched LncBook algorithm (https://bigd.big.ac.cn/lncbook/index) and found that the methylation levels of LINC01614 promoter region in glioma tumor specimens were remarkably lower than that of normal samples, which indicated that transcription factors (TFs) might bind to LINC01614 promoter and activate its expression (Figure 2A). Therefore, the Jaspar database was searched, and we found that SP1 might be a potential TF that could bind to LINC01614 promoter and stimulate its high expression (Figure 2B). Next, we synthesized SP1 siRNAs (si-SP1) and constructed SP1 overexpressing plasmid, ov-SP1 (Figure 2C). They were then, respectively, transfected into U251 cells and qPCR analyses were carried out to determine LINC01614 expression. The results showed that repression of SP1 markedly reduced LINC01614 levels while enhancing SP1 levels remarkably increased LINC01614 levels in glioma cells (Figure 2D). Subsequently, ChIP assays validated that there were notable SP1-binding activities in the S2 site of LINC01614 promoter (Figure 2E). In addition, the data from luciferase activity detection assays revealed that enhancing SP1 expression caused obviously increased luciferase activities in GB cells transfected with S2 wild-type (WT) luciferase reporter plasmids but not the S2 mutant-type (MUT) reporters (Figure 2F). These results demonstrated that SP1 could activate LINC01614 aberrantly high expression in glioma cells.
LINC01614 Knockdown Depressed Glioma Cell Proliferation and Induced Cell Apoptosis
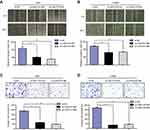
We next aimed to study the functional roles of LINC01614 in glioma, and U251 and LN299 were selected for cell functional experiments. Then, LINC01614 siRNAs were transfected into glioma cells and qPCR was conducted. As presented in Figure 3A, after treatment with si-LINC01614#1 and si-LINC01614#2, the levels of LINC01614 markedly decreased in glioma cells. Then, CCK-8 assays were performed and we observed that the cell viability of glioma cells was significantly reduced after transfection with LINC01614 siRNAs (Figure 3B). Accordingly, data from EdU assays proved that depression of LINC01614 caused remarkably decreased cell number of proliferative cells (Figure 3C). Similarly, cell colony formation abilities were also notably reduced after silencing LINC01614 by si-LINC01614#1 and si-LINC01614#2 (Figure 3D). Thereafter, flow cytometry was carried out to examine the cell apoptosis. The data suggested that transfection with LINC01614 siRNAs contributed to significantly increased cell apoptotic rates (Figure 3E). Mechanically, we detected the caspase 3/9 activities in glioma cells after silencing LINC01614 expression. The results proved that depleting LINC01614 expression caused remarkably increased caspase 3/9 activities in glioma cells (Figure 3F). Hence, these data validated that LINC01614 played a promoting role in glioma cellular growth.
The Metastatic Potentials of GBC Cells Were Regulated by LINC01614
Next, wound-healing and transwell invasion experiments were conducted for evaluating the functional roles of LINC01614 in cellular migration and invasion of glioma cells. As illustrated in Figure 4A and B, the data of wound-healing assays revealed that the wound closures of glioma cells with LINC01614 depletion were remarkably inhibited, which implied that the migratory capabilities of glioma cells were markedly impaired by transfection with LINC01614 siRNAs. Subsequently, transwell assays were conducted to determine the cellular invasion. The data demonstrated that the cellular invasion decreased markedly after repressing LINC01614 expression by si-LINC01614#1 or si-LINC01614#2 when compared with si-NC in glioma cells (Figure 4C and D). Therefore, the data suggested that LINC01614 also served as a promoting role in cellular metastasis of glioma cells.
LINC01614 Directly Bound to miR-383 in Glioma Cells
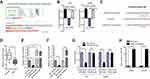
To do further mechanistic studies, we first evaluated the protein-coding abilities of LINC01614 using Coding Protein Calculator program (http://cpc.cbi.pku.edu.cn/). The results revealed that LINC01614 was more likely to be a non-coding RNA (Figure 5A). Therefore, LINC01614 might exert its function by targeting specific miRNAs, particularly when LINC01614 existed in cytoplasm. We thereby further assessed the distribution of LINC01614 in glioma cells using subcellular fractionation experiments, and our group observed that LINC01614 mainly expressed in cytoplasm, indicating LINC01614 might be a miRNA sponge (Figure 5B). Next, we used miRDB algorithm (http://mirdb.org/) to predict the potential targeting of LINC01614, and the results indicated that miR-383, a widely reported anti-tumor factor, was a potential target of LINC01614 (Figure 5C).19,20 In fact, data from qPCR revealed that miR-383 was down-regulated in glioma tumor specimens (Figure 5D). In addition, the miR-383 levels in U251 cells were significantly inhibited by LINC01614 overexpression, while its expression was notably elevated by LINC01614 knockdown (Figure 5E). Vice versa, LINC01614 levels were also suppressed by transfection with miR-383 mimics, while its expression was elevated by miR-383 inhibition (Figure 5F). To further certify that miR-383 was a target of LINC01614, luciferase reporter assays were conducted. Co-transfection of LINC01614 wild-type reporters with miR-383 mimics in glioma cells could result in a significantly decreased luciferase activity compared to the controls (Figure 5G). Furthermore, RNA-pull down assays demonstrated that biotinylated LINC01614 was able to precipitate miR-383 in glioma cells (Figure 5H). Taken together, the data revealed that miR-383 served as a direct target of LINC01614 in glioma.
LINC01614 Modulated ADAM12 Expression via miR-383 in Glioma Cells
Furthermore, to investigate the detailed mechanisms of miR-383 in glioma progression, we intersected the predicted miR-383 target genes using miRDB and TargetScan programs, and 2000 up-regulated genes in glioma tumor samples using GEPIA algorithm (http://gepia.cancer-pku.cn/) analysis, and we found that there were 50 commonly expressed genes (Figure 6A). Then, we employed the GEPIA algorithm to analyze the overall survivals of the 50 commonly expressed genes and found that only six genes (high expression) were significantly associated with poor prognosis of glioma patients (Figure 6B). In addition, cBioPortal algorithm (https://www.cbioportal.org/) was also employed to analyze the genetic alternation of the six genes in glioma specimens (Figure 6C). Among the six genes, we selected ADAM12, an onco-promoter in diverse cancer types for further study.21,22 In fact, ADAM12 was a remarkably high expression in glioma tumor samples (Figure 6D and E). UALCAN algorithm (http://ualcan.path.uab.edu/index.html) analysis using TCGA data also revealed that ADAM12 was up-regulated in multiple cancer types (Figure 6F). Besides, genes positively correlated with ADAM12 in glioma were obtained by UALCAN algorithm, and GO and KEGG pathway analysis revealed that these genes were associated with pathways in cancer, indicating that ADAM12 was involved in tumor function regulation (Figure 6G). Hence, we next sought to investigate whether ADAM12 was involved in LINC01614/miR-383 regulating axis. The two predicted binding sites between miR-383 and 3ʹUTR of ADAM12 mRNA was exhibited in Figure 6H. Subsequently, luciferase reporter plasmids of wild-type ADAM12 (ADAM12 wt1, ADAM12 wt2) and mutant-type ADAM12 (ADAM12 mut1, ADAM12 mut2) were constructed, and luciferase reporter assays were conducted. The results suggested that co-transfection of ADAM12 wt1 or ADAM12 wt2 luciferase reporter with miR-383 mimics in U251 cells resulted in a decreased reporter activity (Figure 6I). Additionally, we found that both LINC01614 and ADAM12 expression were inhibited by miR-383 mimics while elevated by miR-383 inhibitors (Figure 6J). Furthermore, the relative ADAM12 levels in U251 cells were suppressed by miR-383 overexpression, while enhancing LINC01614 expression was able to restore ADAM12 levels (Figure 6K). Similarly, ADAM12 overexpression could also reverse the suppressive effects of miR-383 on LINC01614 (Figure 6L). To sum up, these findings demonstrated that LINC01614 could regulate ADAM12 expression via sponging miR-383 in glioma cells.
Discussion
Growing studies highlighted the dysregulation of lncRNAs and its important functions in tumorigenesis, suggesting the potential of lncRNAs used as novel biomarkers and effective therapeutic targets for tumors.23,24 However, only a few lncRNAs have been functionally characterized. For instance, increased lncRNA DLEU1 was observed in glioma patients with advanced stages and promoted the proliferation and migration in tumor cells via modulating miRNA-421/MEF2D axis.25 LncRNA FOXD2-AS1 was shown to be highly expressed in glioma and its forced expression promoted the metastasis ability of tumor cells via targeting miRNA-185-5p/CCND2 axis.26 Recently, LINC01614 was found to display its oncogenic roles in lung cancer by regulating miR-217, which suggested the potential tumor-related effects of LINC01614 in tumorigenesis.17 Herein, we firstly provided evidence that LINC01614 expression was upregulated in glioma tissues and cell lines. A clinical study indicated that high LINC01614 was associated with high KPS, advanced WHO grade and shorter overall survival of glioma patients. Moreover, multivariate assays demonstrated LINC01614 expression as an unfavorable prognostic indicator for glioma patients, which highlighted the clinical application of LINC01614 as a novel biomarker. These results stimulated us to investigate whether LINC01614 had a functional effect on tumor behaviors. Lost-of-function assays revealed that knockdown of LINC01614 suppressed the proliferation, migration and invasion in glioma cells. Further observation suggested that LINC01614 may protect glioma cells from apoptosis through inhibiting the expression of Caspase 3/9. Our findings indicated that LINC01614 served as an oncogene and drove carcinogenesis via promoting tumor growth and metastasis in glioma.
Transcriptional activation is a crucial mechanism resulting in the overexpression of lncRNAs.27 Recently, growing studies have reported that the dysregulation of lncRNAs was caused by various transcription factors.28 For instance, nuclear transcription factor SP1 induced the distinct upregulation of lncRNA-ZFAS1 in colorectal cancer, resulting in increased abilities of tumor cells in the proliferation, migration and invasion.29 Overexpression of lncRNA LINC00467 was activated by STAT1 in lung adenocarcinoma and further contributed to the proliferation and invasion of cancer cells via epigenetically inhibiting DKK1.30 However, in glioma, only a few transcription factors targeting lncRNAs were identified. In order to explore the mechanism involved in the upregulation of LINC01614, we searched JASPAR tools, finding that SP1 may be a potential regulator for LINC01614 due to several SP1-binding sites in the LINC01614 promoter regions with high scores. Further dual-luciferase reporter analysis and ChIP assays confirmed that the upregulation of LINC01614 in glioma cells may be induced by SP1. Thus, our data firstly reported the possible mechanisms involved in the upregulation of LINC01614.
Inspired by a novel regulatory network (competitive endogenous RNAs) and growing cellular experiments demonstrate that some lncRNAs involved in tumorigenesis may participate in this regulatory meshwork, our group hypothesized that LINC01614 may function as a ceRNA.31,32 The results of subcellular fractionation indicated that a larger proportion of LINC01614 was observed in the nucleus, indicating that LINC01614 may act as a ceRNA in glioma cells by competitively binding miRNAs. Moreover, using bioinformatics assays and luciferase activity reporter assays, LINC01614 was demonstrated to directly bind to miR-383 in glioma cells. MiR-383 was a well-studied tumor-related miRNA. Recently, the dysregulation of miR-383 and its biological function in several tumorigenesis had been frequently reported.19,20 In glioma, the levels of miR-383 were distinctly down-regulated, which was consistent with our experiments.20,33 In addition, previous several studies reported that miR-383 served as a tumor suppressor in the cellular behaviors of glioma via targeting some mRNAs. Thus, our findings revealed that LINC01614 may display its carcinogenic roles in glioma via sponging miR-383.
A disintegrin and metalloproteinase 12 (ADAM12), a kind of type-I transmembrane and soluble glycoproteins, is a newly identified regulatory factor that has been demonstrated to be involved in the biological progress of cellular signal transduction, the hydrolysis of proteins and metastasis.34,35 In tumor research, the distinct upregulation of ADAM12 was observed in several cancer patients, and its levels had been proved to be strongly associated with disease status, stages, and tumor risk.36 In addition, the tumor-promotive roles of ADAM12 and its potential used as a novel diagnostic and prognostic biomarker were also confirmed in several tumors, such as breast cancer and bladder cancer.37,38 Previous studies also provided evidence that overexpression of ADAM12 promoted the proliferation and metastasis of glioma cells, indicating its oncogenic roles in glioma progression.39,40 In this study, we used bioinformatic tools and confirmed the potential miR-383 binding sites in the 3ʹ-UTR of ADAM12. The possible effects of ADAM12 were also analyzed using bioinformatic tools, and the results revealed that high ADAM12 expression was associated with poor clinical outcome of glioma patients. In addition, the data of GO analysis revealed that the levels of ADAM12 were correlated with several tumor-related signaling pathways, highlighting ADAM12 as an important regulator in glioma progression. Then, luciferase assays confirmed that ADAM12 was a direct target of miR-383. Moreover, rescue experiments revealed that overexpression of LINC01614 could increase the mRNA levels of ADAM12 which was decreased by miR-383 mimics. Overall, our data suggested that by binding miR-383, LINC01614 modulates the expression of ADAM12, thereby imposing an additional ADAM12 expression at post-transcriptional regulation level.
In conclusion, we determined that SP1-induced upregulation of LINC01614 promoted glioma progression by acting as a molecular sponge of miR-383 to modulate ADAM12 expressions. Our findings may contribute to enrich the study on the molecular mechanism involved in glioma progression, thereby providing a promising therapeutic target for glioma treatment and a novel diagnostic and prognostic biomarker for glioma patients.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Acknowledgments
This study was supported by grants from the Science and Technology Program of Shenzhen (No.:JCYJ20160422164518800).
Funding
This study was supported by grants from the Science and Technology Program of Shenzhen (No. JCYJ20160422164518800).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Gusyatiner O, Hegi ME. Glioma epigenetics: from subclassification to novel treatment options. Semin Cancer Biol. 2018;51:50–58. doi:10.1016/j.semcancer.2017.11.010
3. Ostrom QT, Gittleman H, Stetson L, Virk SM, Barnholtz-Sloan JS. Epidemiology of gliomas. Cancer Treat Res. 2015;163:1–14.
4. Perry A, Wesseling P. Histologic classification of gliomas. Handb Clin Neurol. 2016;134:71–95.
5. Ferris SP, Hofmann JW, Solomon DA, Perry A. Characterization of gliomas: from morphology to molecules. Virchows Archiv. 2017;471(2):257–269. doi:10.1007/s00428-017-2181-4
6. Rizzo D, Ruggiero A, Martini M, Rizzo V, Maurizi P, Riccardi R. Molecular biology in pediatric high-grade glioma: impact on prognosis and treatment. Biomed Res Int. 2015;2015:215135. doi:10.1155/2015/215135
7. Sim HW, Morgan ER, Mason WP. Contemporary management of high-grade gliomas. CNS Oncol. 2018;7(1):51–65. doi:10.2217/cns-2017-0026
8. Esparragosa I, Diez-Valle R, Tejada S, Gallego Perez-Larraya J. Management of diffuse glioma. Presse Med. 2018;47(11–12Pt 2):e199–e212. doi:10.1016/j.lpm.2018.04.014
9. Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46.
10. Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12(6):433–446. doi:10.1038/nrd4018
11. Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35(9):408–419. doi:10.1016/j.it.2014.07.005
12. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–2509. doi:10.1007/s00018-016-2174-5
13. Luo ML. Methods to study long noncoding RNA biology in cancer. Adv Exp Med Biol. 2016;927:69–107.
14. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi:10.1016/j.gpb.2015.09.006
15. Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126(8):2775–2782. doi:10.1172/JCI84421
16. Wang SJ, Wang H, Zhao CD, Li R. Long noncoding RNA LINC01426 promotes glioma progression through PI3K/AKT signaling pathway and serves as a prognostic biomarker. Eur Rev Med Pharmacol Sci. 2018;22(19):6358–6368. doi:10.26355/eurrev_201810_16047
17. Liu AN, Qu HJ, Yu CY, Sun P. Knockdown of LINC01614 inhibits lung adenocarcinoma cell progression by up-regulating miR-217 and down-regulating FOXP1. J Cell Mol Med. 2018;22(9):4034–4044. doi:10.1111/jcmm.13483
18. Vishnubalaji R, Shaath H, Elkord E, Alajez NM. Long non-coding RNA (lncRNA) transcriptional landscape in breast cancer identifies LINC01614 as non-favorable prognostic biomarker regulated by TGFbeta and focal adhesion kinase (FAK) signaling. Cell Death Discov. 2019;5:109. doi:10.1038/s41420-019-0190-6
19. Han RL, Wang FP, Zhang PA, Zhou XY, Li Y. miR-383 inhibits ovarian cancer cell proliferation, invasion and aerobic glycolysis by targeting LDHA. Neoplasma. 2017;64(2):244–252. doi:10.4149/neo_2017_211
20. Zhao LN, Wang P, Liu YH, et al. MiR-383 inhibits proliferation, migration and angiogenesis of glioma-exposed endothelial cells in vitro via VEGF-mediated FAK and Src signaling pathways. Cell Signal. 2017;30:142–153. doi:10.1016/j.cellsig.2016.09.007
21. Duhachek-Muggy S, Zolkiewska A. ADAM12-L is a direct target of the miR-29 and miR-200 families in breast cancer. BMC Cancer. 2015;15:93. doi:10.1186/s12885-015-1108-1
22. Eckert MA, Santiago-Medina M, Lwin TM, Kim J, Courtneidge SA, Yang J. ADAM12 induction by Twist1 promotes tumor invasion and metastasis via regulation of invadopodia and focal adhesions. J Cell Sci. 2017;130(12):2036–2048. doi:10.1242/jcs.198200
23. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
24. Fan Q, Yang L, Zhang X, et al. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018;414:107–115. doi:10.1016/j.canlet.2017.10.040
25. Feng L, He M, Rao M, Diao J, Zhu Y. Long noncoding RNA DLEU1 aggravates glioma progression via the miR-421/MEF2D axis. Onco Targets Ther. 2019;12:5405–5414. doi:10.2147/OTT.S207542
26. Shen F, Chang H, Gao G, Zhang B, Li X, Jin B. Long noncoding RNA FOXD2-AS1 promotes glioma malignancy and tumorigenesis via targeting miR-185-5p/CCND2 axis. J Cell Biochem. 2019;120(6):9324–9336. doi:10.1002/jcb.28208
27. Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56(1):29–42. doi:10.1016/j.molcel.2014.08.023
28. Huang B, Chang C, Wang BL, Li H. ELK1-induced upregulation of lncRNA TRPM2-AS promotes tumor progression in gastric cancer by regulating miR-195/HMGA1 axis. J Cell Biochem. 2019;120(10):16921–16933. doi:10.1002/jcb.28951
29. Chen X, Zeng K, Xu M, et al. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9(10):982. doi:10.1038/s41419-018-0962-6
30. Yang J, Liu Y, Mai X, Lu S, Jin L, Tai X. STAT1-induced upregulation of LINC00467 promotes the proliferation migration of lung adenocarcinoma cells by epigenetically silencing DKK1 to activate Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun. 2019;514(1):118–126. doi:10.1016/j.bbrc.2019.04.107
31. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
32. Huang B, Jiang C, Zhang R. Epigenetics: the language of the cell? Epigenomics. 2014;6(1):73–88. doi:10.2217/epi.13.72
33. Xu Z, Zeng X, Tian D, et al. MicroRNA-383 inhibits anchorage-independent growth and induces cell cycle arrest of glioma cells by targeting CCND1. Biochem Biophys Res Commun. 2014;453(4):833–838. doi:10.1016/j.bbrc.2014.10.047
34. Jacobsen J, Wewer UM. Targeting ADAM12 in human disease: head, body or tail? Curr Pharm Des. 2009;15(20):2300–2310. doi:10.2174/138161209788682389
35. Nyren-Erickson EK, Jones JM, Srivastava DK, Mallik S. A disintegrin and metalloproteinase-12 (ADAM12): function, roles in disease progression, and clinical implications. Biochim Biophys Acta. 2013;1830(10):4445–4455. doi:10.1016/j.bbagen.2013.05.011
36. Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM. Cellular roles of ADAM12 in health and disease. Int J Biochem Cell Biol. 2008;40(9):1685–1702. doi:10.1016/j.biocel.2008.01.025
37. Duhachek-Muggy S, Qi Y, Wise R, et al. Metalloprotease-disintegrin ADAM12 actively promotes the stem cell-like phenotype in claudin-low breast cancer. Mol Cancer. 2017;16(1):32. doi:10.1186/s12943-017-0599-6
38. Frohlich C, Albrechtsen R, Dyrskjot L, Rudkjaer L, Orntoft TF, Wewer UM. Molecular profiling of ADAM12 in human bladder cancer. Clin Cancer Res. 2006;12(24):7359–7368. doi:10.1158/1078-0432.CCR-06-1066
39. Lulli V, Buccarelli M, Martini M, et al. miR-135b suppresses tumorigenesis in glioblastoma stem-like cells impairing proliferation, migration and self-renewal. Oncotarget. 2015;6(35):37241–37256. doi:10.18632/oncotarget.5925
40. Kodama T, Ikeda E, Okada A, et al. ADAM12 is selectively overexpressed in human glioblastomas and is associated with glioblastoma cell proliferation and shedding of heparin-binding epidermal growth factor. Am J Pathol. 2004;165(5):1743–1753. doi:10.1016/S0002-9440(10)63429-3
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.