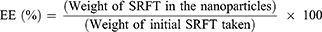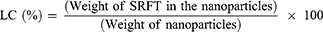Back to Journals » International Journal of Nanomedicine » Volume 17
Sorafenib-Entrapped, Self-Assembled Pullulan–Stearic Acid Biopolymer-Derived Drug Delivery System to PLC/PRF/5 Hepatocellular Carcinoma Model
Authors Chirayil TJ, Kumar GSV
Received 4 June 2022
Accepted for publication 21 August 2022
Published 31 October 2022 Volume 2022:17 Pages 5099—5116
DOI https://doi.org/10.2147/IJN.S377354
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Teena Jacob Chirayil,1,2 G S Vinod Kumar1
1Nano Drug Delivery Systems (NDDS), Cancer Biology Division, Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, Kerala, India; 2Research Scholar, Department of Biotechnology, University of Kerala, Thiruvananthapuram, Kerala, India
Correspondence: G S Vinod Kumar, Tel +91 471 2781217, Email [email protected]
Purpose: This study aimed to design a prototypic drug delivery system (DDS) made of an amphiphilic, pullulan (Pull)-derived biodegradable polymer for targeting the asialoglycoprotein receptor (ASGPR) overexpressed in HCC. Stearic acid (SA) was conjugated to increase the hydrophobicity of pullulan (Pull-SA).
Methods: Pullulan (Pull) was linked to stearic acid (SA) after functional group modifications via EDC/NHS chemistry and characterized. Sorafenib tosylate (SRFT) was entrapped in pullulan–stearic acid nanoparticles (Pull-SA-SRFT) and its particle size, zeta potential, entrapment efficiency (EE), loading capacity (LC), and release efficiency was measured. The competence of Pull-SA-SRFT over SRFT in vitro was assessed using the ASGPR over-expressing PLC/PRF/5 hepatocellular carcinoma (HCC) cell line. This was done by studying cytotoxicity by MTT assay and chromosome condensation assay, early apoptosis by annexin-Pi staining, and late apoptosis by live–dead assay. The cellular uptake study was performed by incorporating coumarin-6 (C6) fluorophore in place of SRFT in Pull-SA conjugates. A biodistribution study was conducted in Swiss-albino mice to assess the biocompatibility and targeting properties of SRFT and Pull-SA-SRFT to the liver and other organs at 1, 6, 24, and 48 h.
Results: The characterization studies of the copolymer confirmed the successful conjugation of Pull-SA. The self-assembled amphiphilic nanocarrier could proficiently entrap the hydrophobic drug SRFT to obtain an entrapment efficiency of 95.6% (Pull-SA-SRFT). Characterization of the synthesized nanoparticles exhibited highly desirable nanoparticle characteristics. In vitro, apoptotic studies urged that Pull-SA-SRFT nanoparticle was delivered more efficiently to HCC than SRFT. The cellular uptake study performed, gave propitious results in 4 hrs. The biodistribution study conducted in immunocompetent mice suggested that Pull-SA-SRFT was delivered more than SRFT to the liver when compared to other organs, and that the system was biocompatible.
Conclusion: Pull-SA-SRFT is a promisingly safe, biodegradable, cell-specific nanocarrier and a potential candidate to target hydrophobic drugs to HCC.
Keywords: pullulan stearic acid nanoparticles, hepatocellular carcinoma, drug delivery system
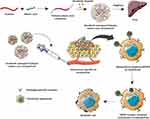
Graphical Abstract:
Introduction
Hepatocellular carcinoma (HCC) is the most prevalent primary liver cancer and the world’s second-biggest cause of cancer-related fatalities.1,2 The incidence of HCC is more prevalent in patients with chronic liver disease, such as chronic hepatitis (B and C), liver cirrhosis, fatty liver disease, exposure to aflatoxin, and excess consumption of alcohol.3–6 Medical treatments such as chemotherapy, chemoembolization, ablation, and proton beam therapy have been ineffective for these patients and the majority of them show disease recurrence, rapid progression, and metastases.7,8 Despite various advancements, pharmacological therapy has had a limited therapeutic impact.9 The majority of anti-cancer medications have high toxicity and low specificity, resulting in systemic toxicity and severe side effects. Due to the lack of effective therapeutic possibilities, the pursuit of alternative pharmaceutical therapies is therefore particularly pressing.
Sorafenib tosylate (Nexavar, BAY-43-9006), the currently FDA-approved drug for the standard treatment of HCC is a multikinase inhibitor that blocks the RAF/MEK/ERK pathway in tumor cells.10 It interferes with cell proliferation and angiogenesis by inhibiting serine/threonine kinases (c-RAF) as well as the receptor tyrosine kinases vascular endothelial growth factor receptor 2 (VEGFR2), VEGFR3, platelet-derived growth factor receptor (PDGFR), FLT3, Ret and c-KIT.11–13 The recommended dose of Nexavar in adults is 400 mg twice daily (a total daily dose of 800 mg)14 leading to severe diarrhea resulting in excretion of high amounts of the drug.13 Regardless of its efficacy, it leads to severe side effects such as nausea, vomiting, high arterial pressure and hand-foot skin reactions.15
Polymeric nanoparticles (NPs) have been used as promising anticancer drug carriers,16 working as long-acting, regulated, and targeted drug delivery systems that boost therapeutic effects while minimising negative effects on normal organs.17–19 Although nanoparticles can accumulate rapidly in the liver, they do not accumulate in the proper intrahepatic cell type. Therefore, specific targeting to cancerous hepatocytes is essential.20 The liver harbours several lectin receptors that can bind to carbohydrates. Among these, the asialoglycoprotein receptor (ASGPR) provides an ideal target for hepatocyte-specific delivery. It is expressed in abundance on hepatocytes (500,000 ASGPR/hepatocyte) and minimally present elsewhere in the body and has the ability to internalise large molecules through clathrin-mediated endocytosis.21,22
Pullulan is a natural, water-soluble, non-mutagenic, biodegradable polymer produced by the yeast like fungus Aureobasidium pullulans.23,24 It is composed of a linear chain consisting of maltotriose units linked by α1,4-glycosidic linkage and the successive maltotriose units are associated by α1,6-glycosidic linkage25,26 Pullulan has a higher affinity to the liver due to the presence of lectin-like receptors on liver cells (hepatocytes and Kupffer cells), which possess biological affinity for sugar residues.27,28 The orientation of hydroxyl groups on pyranose rings in pullulan structure helps in the proper binding of pullulan to specific ASGPRs which further helps in endocytosis.29
Amphiphilic polysaccharides have the ability to self-assemble in an aqueous environment and are employed recently to produce polysaccharide core-shell nanoparticles in which the core is composed of the hydrophobic-modified backbone and the shell is made of the polar carbohydrates.30 This approach is an attractive platform for drug delivery of hydrophobic drugs since they can be retained in the central core of the NPs and their therapeutic efficiency is improved by increasing their solubility, retention time and stability.31
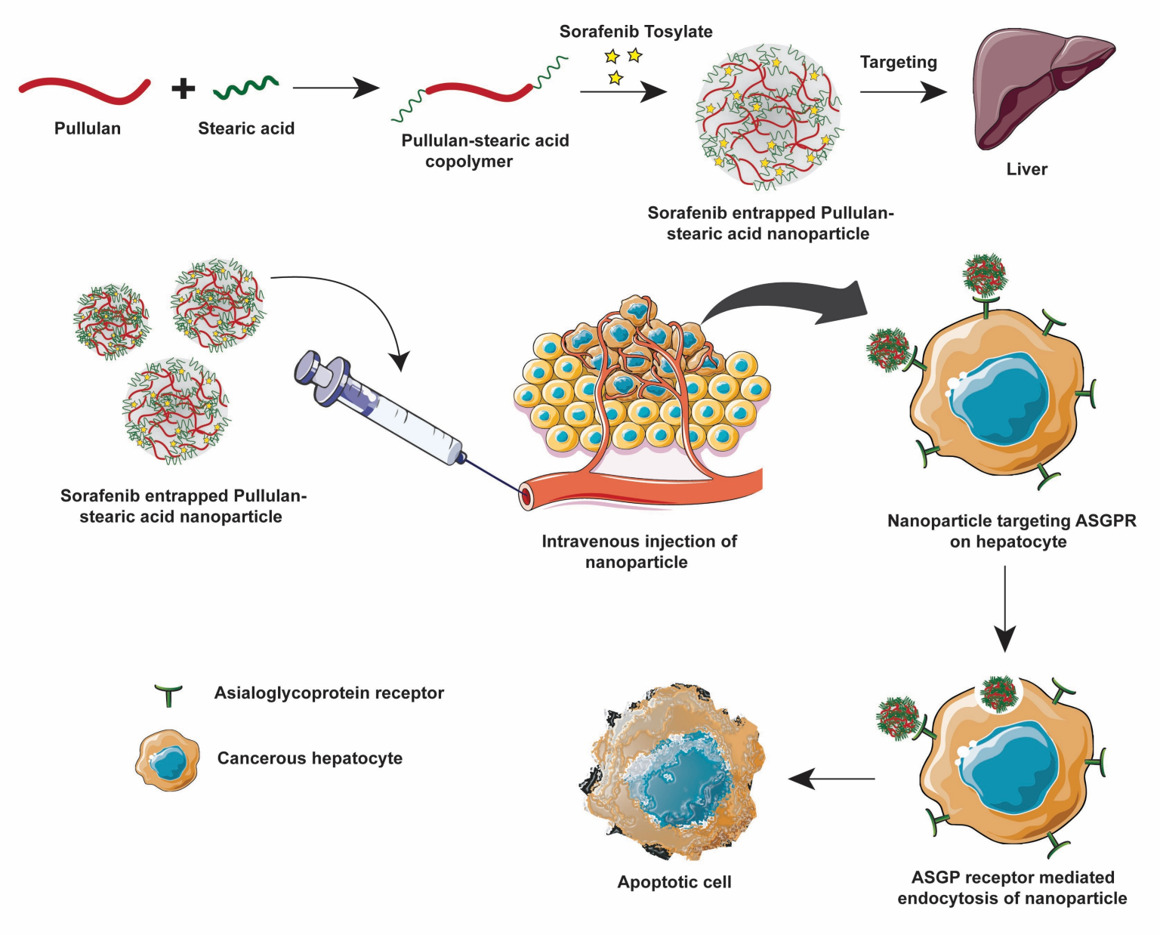
The current study aims to develop a drug delivery system to target ASGPR of HCC cells to induce apoptosis (Figure 1). This was achieved using hydrophobically modified, self-assembled pullulan nanoparticles to entrap sorafenib tosylate (SRFT) that can take advantage of the enhanced permeation and retention (EPR) effect in the tumor environment. Stearic acid, also called octadecanoic acid (18-C), which is an endogenous long-chain saturated fatty acid with low cytotoxicity and high biocompatibility32 was adopted to increase the hydrophobicity of pullulan. Even though various pullulan derivatives with potential anticancer applications have been studied before to improve the hydrophobicity of pullulan, this combination of pullulan–stearic acid biopolymer to entrap the hydrophobic drug Sorafenib approved for the treatment of HCC has not been attempted yet.
Materials and Methods
Cells and Media
Human PLC/PRF/5 (Alexander cells) hepatocellular carcinoma cell line was procured from the cell repository of the National Centre for Cell Science (NCCS), Pune, India. Dulbecco’s Modified Eagle Medium (DMEM) was purchased from Hi-media and fetal bovine serum (FBS) was purchased from Gibco. Penicillin–streptomycin, trypsin, Live/Dead-Viability/Cytotoxicity Assay Kit, Annexin-PI staining kit, Cell MaskTM (plasma membrane staining kit) and Hoechst were purchased from Invitrogen – Thermofisher Scientific.
Chemicals
Pullulan from Aureobasidium pullulans (molecular weight 574.570 Da) and Sorafenib Tosylate was purchased from Santa Cruz Biotechnology, Inc. (California). Stearic acid, dimethyl sulfoxide (DMSO), and HPLC grade solvents used for the biodistribution study were obtained from Merck (USA). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 1-(3-dimethyl aminopropyl)-3-3-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS) and coumarin-6 were procured from Sigma-Aldrich (Steinheim, Germany). Dialysis tubing was purchased from SERVA (Heidelberg, Germany).
Synthesis of Pullulan–Stearic Acid Copolymer
Carboxylation of Pullulan
Four hundred milligrams of pullulan was dissolved in a 10 mL mixture solution of DMSO and pyridine (1/4, v/v). One hundred milligrams of succinic anhydride was then added and the obtained mixture was stirred continuously for 6 h at 50°C. The reaction mixture was then poured into 50 mL of cold dehydrated alcohol, and the produced precipitate was collected by centrifugation. This precipitate was re-dissolved in 20 mL of distilled water and exhaustively dialyzed (Serva, MWCO 12–14 kDa, Germany) against 1L distilled water for 48 h with frequent water changes in the initial hours. Subsequently, the dialyzed solution was then lyophilized to yield the white cotton wool-like carboxylated pullulan product.33
Amino Conversion of Pullulan
The carboxyl functional group-modified Pullulan was allowed to covalently interact with the amino group – (NH2) of Ethylenediamine using EDC and NHS via coupling chemistry to prepare aminated pullulan. Briefly, 400 mg of COOH-converted pullulan was dissolved in 20 mL of 0.1 M MES buffer to keep the solution pH at 6.5. For activation of the carboxyl group of pullulan, EDC (0.574g) and NHS (0.345g) were added to the solution and allowed to stir for 24 h at room temperature. For the preparation of aminated pullulan, 500 µL of EDA was added to the activated pullulan–COOH solution and the reaction proceeded for 48 h at room temperature. The resulting solution was purified by dialysis (MWCO 12–16 kDa) using distilled water and was dried in a freeze drier.34 After the lyophilized product was obtained, the presence of NH2 groups was qualitatively tested for the presence of blue colour by a simple Ninhydrin test and confirmed by doing FTIR and NMR analysis.
Synthesis of Pullulan–Stearic Acid (Pull-SA) Copolymer
Pull-SA copolymer was synthesized by EDC-NHS coupling chemistry in DMSO and dialyzed. Two different ratios of amphiphilic Pull-SA combinations were prepared and characterized, out of which the combination that showed the highest entrapment efficiency and optimum particle size was chosen for further studies (Table 1).
 |
Table 1 Characterization of Sorafenib-Entrapped Pullulan–Stearic Acid Nanoparticles |
Characterisation of the Copolymer
Fourier Transformed Infra-Red Spectroscopy (FT-IR)
The functional group modifications of the pullulan biopolymer and its conjugation to stearic acid were characterized using Fourier transform infrared spectroscopy (FT-IR), (PerkinElmer Spectrum 65FT-IT Spectrometer) by the potassium bromide (KBr) pellet method. The dried polymer samples were mixed with KBr and pressed under mechanical pressure. The sample pellets were placed on the KBr cell and subjected to light within the infrared spectrum. Spectra were recorded between the wavenumber range of 4000 and 600 cm−1.
Proton – Nuclear Magnetic Resonance (1H NMR)
The synthesized copolymer was further characterized by 1H NMR analysis using the Bruker Avance III HD 400 MHz FTNMR Spectrometer. DMSO was used as the solvent for analysis.
Differential Scanning Calorimetry (DSC)
The thermal stability and nature of the synthesized biopolymer compared to free pullulan and stearic acid was analyzed by differential scanning calorimeter (Perkin Elmer 6000, USA). A known amount of sample was sealed in an aluminium pan and was compared to an empty reference pan while heating between 0°C and 160°C at an increment rate of 10°C/min. Nitrogen was purged at 20mL/min to the system.
Preparation of Pullulan–Stearic Acid Nanoparticles
Pull-SA nanoparticles were synthesized by the dialysis method. Briefly, 100 mg of pullulan–stearic acid copolymer (1:10 ratio chosen for further studies from Table 1) was dissolved in 10 mL DMSO by stirring overnight. To this, 5 mg of sorafenib tosylate (SRFT) dissolved in 500 µL DMSO was added drop-wise under stirring conditions. The resultant solution was allowed to react overnight and was dialyzed against distilled water (MWCO 12–16 kDa) for 24 h. Water was replaced every 2 h for the first 6 h and then every 5 h thereafter. Coumarin-6-loaded Pull-SA (Pull-SA-C6) nanoparticles were synthesized by the same protocol, only with SRFT replaced with 10 µg/mL coumarin-6 (10µL from 1mg/mL stock solution in DMSO).
Characterisation of Nanoparticles
Dynamic Light Scattering (DLS) and Zeta-Potential Analysis
The average size, polydispersity index (PDI) and zeta-potential of Pull-SA-SRFT in two different ratios of the conjugate were evaluated by DelsaTM Nanoparticle size analyser (Beckman Coulter, Inc.) A known amount of sample was dispersed in 1mL distilled water, sonicated for 10 mins and analysed.
Morphology Analysis
The morphology characteristics of the developed DDS was studied using Transmission Electron Microscopy (TEM, JEOL 1011, Japan) by coating the Pull-SA-SRFT nanoparticles on a formvar-coated copper grid.
Drug Entrapment Efficiency
The amount of SRFT entrapped in Pull-SA conjugate was determined by calculating the optical density at 265 nm from a standard curve that was plotted beforehand. Measurements were carried out three times for each batch. The entrapment efficiency and loading content were calculated35 as follows:
In vitro Drug Release Kinetics
The drug releasing pattern of SRFT from Pull-SA nanoparticles was assessed by using the dialysis method, under sink conditions, in phosphate-buffered solution (PBS) at pH 7.2 and 6, to simulate the physiological fluid and tumor environment, respectively. Briefly, a known amount of drug-loaded nanoparticles was dispersed in 2 mL PBS, placed in a dialysis tubing (MWCO 12–16 kDa), immersed in 50 mL PBS and was incubated at 37°C under continuous stirring at 100 rpm in an orbital shaker. Aliquots of the external medium (1mL) were withdrawn from the outside of the dialysis tubing in triplicates at fixed time intervals and replaced with equal amounts of fresh medium. The samples were centrifuged and the OD of the supernatant was measured at 265 nm.36
Cell Culture
PLC/PRF/5 (Alexander cells) Hepatocellular carcinoma cells were cultured in DMEM media containing 10% FBS and 1% of penicillin (100 U/mL)/streptomycin (100 mg/mL). Cell culture was done in an atmosphere of 5% CO2 and humidity of 95% at 37°C. Media was replaced all alternate days and cells were sub-cultured after reaching optimal confluence.
Cytotoxicity Assay
Cells were plated in 96-well microtiter plates at a density of 7×103 cells/well in triplicates. After 24 h incubation, the media was removed and replaced with fresh medium containing SRFT, dissolved in DMSO stock solution and then diluted in DMEM at the concentration range of 4–12 µM; equivalent amounts of SRFT-loaded Pull-SA and empty Pull-SA nanoparticles (to check biocompatibility) corresponding to the entrapped SRFT concentration in 4–12 µM. Samples were suspended in DMEM in sterile conditions, sonicated for 10 min and treated the cells for 24 h and 48 h. After treatment, 10 µL of MTT (5mg/mL PBS) solution was added to cells and incubated further for 3 h. After removal of the culture medium, the formazan crystals were dissolved in 200 µL IPA (Isopropyl alcohol), mixed well and the absorbance of each well was measured using ELISA plate reader (Bio-Rad) at a wavelength of 570 nm.37 Untreated cells were taken as control with 100% viability. The drug concentration at which the growth of 50% cells was inhibited (IC50) was calculated by curve fitting of the cell viability data. Percentage of death induced by treatment was calculated as follows:
Chromosome Condensation Assay
PLC/PRF/5 cells were plated in an 8-well chamber at 3000 cells/well. After treatment of cells with the formulations of SRFT and Pull-SA SRFT at 8 µM concentration after sonication for 10 min, the cells were incubated for 48 h. DNA nuclear morphology was examined with Hoechst dye (0.1µg/mL) staining for 10 min. The cells were washed once with PBS and visualized and analyzed using Leica SP 8 WLL Confocal Laser Scanning Microscope.
Cell-Uptake Studies
To visualise the ability of HCC cells to uptake Pull-SA nanoparticles, cells were seeded in an 8-well chamber at a density of 3×103 cells/well followed by incubation for 24 h for cells to attach. Then, coumarin-6-loaded Pull-SA nanoparticles formulated by the dialysis method, were added to cells (at a concentration of 250 ng/mL)38 after sonication for 10 min and incubated for 4 h. After incubation, the cells were carefully rinsed twice with PBS to remove any unbound particles. The plasma membrane was stained red with Cell MaskTM (plasma membrane staining kit) for 10 min and the nucleus was counterstained blue with Hoechst for another 10 min. The cells were then analysed using Leica SP 8 WLL Confocal Laser Scanning Microscope (CLSM).
Early Apoptosis Detection
To detect early apoptosis, cells were seeded in a 6-well plate at a cell density of 1×106 cells/well for FACS analysis and also in an 8-well chamber at a density of 3×103 for CLSM imaging and incubated overnight. The next day, cells were treated with formulations of SRFT and Pull-SA SRFT at 8 µM concentration after sonication for 10 min. Untreated cells served as the control group. After incubation for 12 h, the cells were trypsinised and stained with Annexin V-FITC and propidium iodide (PI) in the dark for FACS analysis39 and the cells were sorted using BD FACS AriaTM III Cell Sorter, USA, and analyzed using BD FACSDiva 7.0 software. In addition, the cells treated for the imaging study were stained according to the kit protocol and visualised using Leica SP 8 WLL Confocal Laser Scanning Microscope (CLSM).
Live–Dead Assay
To investigate the activity of the developed DDS to induce apoptosis in the later stages, cells were treated in a similar manner as mentioned above and incubated for 48 h. The cells were sorted after processing them for analysis using the live–dead apoptosis detection kit protocol and imaging was done using CLSM.
Animal Studies
In vivo Biodistribution Study
In vivo biodistribution study was done for sorafenib and sorafenib-entrapped pullulan–stearic acid nanoparticles. The experiment was performed according to the protocols approved (Protocol number- IAEC/752/GSV/2019) by the IAEC, Rajiv Gandhi Centre for Biotechnology, Kerala, India. Briefly, male Swiss albino mice (4–6 weeks) were divided into two groups (n=3), viz. Group I, sorafenib drug alone and Group II, sorafenib-entrapped pullulan–stearic acid nanoparticles. The animals were weighed and formulations equivalent to 5 mg/kg body weight were administered intravenously to mice. The animals were sacrificed at 1 h, 6 h, 24 h, and 48 h, and blood was collected by cardiac puncture. The organs such as the brain, liver, lungs, kidneys and spleen were isolated. The organs were placed in the mobile phase (acetonitrile:water, 82.5:17.5 v/v) in which the calibration curve of the drug was previously prepared and homogenized using a tissue homogenizer. The homogenate was vortexed for 5 minutes, followed by centrifugation at 10,000 rpm for 10 minutes. The supernatant was injected into the HPLC system (Shimadzu, Japan) coupled with a UV detector (SPD-M20A Diode array detector) and the injector was fitted with a 100µL sample loop. Chromatography was performed on the C18 guard column (Waters, 4.6x300mm) at room temperature under isocratic conditions at a flow rate of 1.5 mL/min with UV detection at a λmax of 265 nm. Sorafenib was analyzed by HPLC and compared with calibration curve against appropriate blank.40
Results and Discussion
Characterization of Amphiphilic Pull-SA Copolymer
Amphiphilic pullulan biopolymer was synthesised to increase the hydrophobicity of pullulan thereby improving its potential to harbour hydrophobic drugs. This was achieved by a three-step reaction as represented by the scheme in Figure 2. First, the carboxylation reaction of pullulan was carried out derived from succinic anhydride. Second, the amino group of ethylenediamine was conjugated to the carboxylated pullulan, Finally, the hydrophobic moiety stearic acid was successfully conjugated to the amino-converted pullulan to yield the final Pull-SA copolymer. The reaction was carried out in DMSO, which is a good solvent for most polysaccharides.41
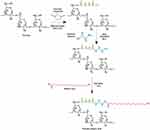 |
Figure 2 Schematic representation of the carboxylic and amino functional group modifications of pullulan and its conjugation to stearic acid. |
FT-IR spectra of free pullulan, carboxylated pullulan, amino-converted pullulan, stearic acid, and pullulan–stearic acid copolymer are presented in Figure 3A. In comparison to the spectrum of free pullulan, the additional signal peak in COOH-modified Pullulan at 1735 cm−1 was attributed to the stretching vibration of the ester bond.33 Also, in aminated pullulan, the spectrum peaks at 1416 cm−1, 1648 cm−1 and 3388 cm−1 indicating the existence of an amino-functional group.34 Furthermore, the characteristic peaks at around 2926 cm−1 ascribed to the symmetric or asymmetric stretching vibrations of CH2 and CH3 groups of stearic acid, were observed in the spectrum of Pull-SA copolymer. These results suggest the successful conjugation of stearic acid to pullulan.
DSC thermogram is commonly used to differentiate synthetic polymers from their physical mixtures. The thermogram of the physicochemical characteristics of Pull, SA and Pull-SA derivatives Figure 3B indicated an endothermic peak characteristic of pullulan in the range of 80–85°C. The thermal analysis of stearic acid designated an endothermic melting peak within the range of 70–80°C. Interestingly, the DSC curve of Pull-SA copolymer exhibited a mild endothermic peak in the range of 45–50°C and a sharp endothermic peak in the range of 85–95°C, which corresponded to pullulan and stearic acid, respectively. Unlike their physical form, there was a shift in the transition temperature of the Pull-SA conjugates. It could be deduced that the crystallinity of pullulan was decreased by the hydrophobic modification which indicates that the Pull-SA copolymer was formed.
In the 1H NMR of Pull-SA copolymer presented in Figure 3C, the proton peaks in the range of 5.57–4.47 ppm correspond to the (1→6) and (1→4) linkage of pullulan. Also, the significant peaks in the range of 4.0–3.0 ppm are mainly associated with the inner methylidene and methylene protons (CH–O and CH2–O) on glucose units of pullulan.42 Additionally, the proton peaks in the range of 1.3 to 0.84 ppm depict the methyl protons (–CH3) and (–CH2) characteristic of stearic acid.43 These results suggest that pullulan–stearic acid copolymer was successfully synthesized.
Preparation and Characterisation of Pull-SA Nanoparticles
In aqueous media, amphiphilic block or graft polymers demonstrate self-aggregation potential.44 The hydrophobicity of pullulan which is a water-soluble, extracellular neutral polysaccharide45 was increased by conjugating it to COOH of stearic acid, which is an 18-C endogenous long-chain saturated fatty acid with low cytotoxicity and high biocompatibility,46 after functional group modifications. Pull-SA conjugates formed self-assembled nanoparticles by the dialysis method. The characteristic parameters of the two different ratios of the drug-entrapped conjugates of Pull-SA (1:1 and 1:10) were measured and are presented in Table 1.
As indicated, the size of nanoparticles decreased from 740 (Figure 4A) to 334 nm by increasing Pull:SA ratio from 1:1 to 1:10. This may have resulted from the enhancement of the inner hydrophobicity of Pull-SA nanoparticles as a consequence of SA increment.46 Enhancement in graft ratio is synchronous with the increase in the percentage of hydrophobic part, which causes the grafted polymers to assemble into significantly smaller micelles in aqueous conditions.47 The morphology study by TEM analysis indicated that the Pull-SA-SRFT nanoparticles were uniformly dispersed. The particle size measurement by TEM analysis revealed that the actual size of nanoparticles is smaller than the size obtained by DLS. This is because, in the light scattering method, the hydrodynamic size is measured and therefore bigger than the actual dry particle size measured by TEM (Figure 4B).
The PDI of 1:10 ratio Pull-SA nanoparticles was very low compared to 1:1 ratio particle indicating that they were monodispersed. The negative surface charge of the particles increased from −5.56 to −11.06 mV when the ratio was increased (Figure 4C), suggesting that the latter was a more stable and safer formulation. Furthermore, the EE% and LC % increased from 67% to 95.6% and 3.2% to 4.83% respectively. This might be due to the more robust and stable association of the hydrophobic drug with the inner hydrophobic core of the nanoparticles, leading to its proper integrity.
Higher entrapment efficiency and desirable nanoparticle characteristics were observed for the 1:10 ratio of Pull-SA conjugate. Henceforth, this ratio of copolymer was chosen to entrap the drug for further experiments in this study.
In vitro release studies presented in Figure 4D indicate that Pull-SA-SRFT nanoparticles have a slow and sustained release and 100% cumulative release was observed over a period of 240 h. This finding was because Pull-SA (1:10) had a higher hydrophobicity by virtue of the higher substitution of stearic acid on its polysaccharide backbone, which led to stronger interaction of SRFT and the inner core of nanoparticles. The surface area of the micelle for mass transfer increases as the size of the micelles decreases, which causes an increase in the release of the encapsulated medication to the medium.48,49 This sustained release is beneficial for longer retention of the drug in the tumor microenvironment. Moreover, the property of delivery materials and the pH of the medium used for drug release from nanoparticles also influenced the drug release profiles.50 It could be noted that the release of SRFT was accelerated in the acidic pH than in the neutral pH, indicating that the Pull-SA nanoparticles were pH-sensitive to some extent. Previous studies have reported that increased aerobic glycolysis, ie, the Warburg effect, is linked to its lower extracellular pH in the tumor microenvironment.51,52 Therefore, the pH-dependent release of SRFT from Pull-SA nanoparticles is beneficial for cancer treatment.
In vitro Cytotoxicity Studies
The biocompatibility of empty Pull-SA nanoparticles and the cytotoxicity induced by free SRFT and Pull-SA-SRFT nanoparticles were studied by MTT assay. As indicated in Figure 5A and B), after 24 h and 48 h incubation at different concentrations of empty Pull-SA particles, more than 90% cells were viable even at the highest concentration of 12 µM. These findings imply that the Pull-SA copolymer exhibited low cytotoxicity, is biocompatible, and can be utilised as a potential drug carrier.
The in vitro anticancer activity of free SRFT and Pull-SA-SRFT nanoparticles displayed a concentration-dependent cytotoxicity against PLC/PRF/5 cells. However, at the same corresponding concentrations, Pull-SA SRFT nanoparticles showed more cytotoxicity than free SRFT. This could be because free SRFT was effluxed out by the transporter proteins expressed by HCC cells. Hepatocytes adopt multiple different ABC transporters to transport organic ions such as bile acids and conjugated bilirubin.53 Perhaps, these pumps and permeability-glycoproteins (p-glycoproteins)54 are more likely to be expressed on the surface of HCC cells, leading to chemotherapy resistance. The Pull-SA-SRFT nanoparticles can bypass the effect of these transporters through an endocytic pathway initiated by the binding of hydroxyl groups of pullulan to the ASGPR expressed on HCC cells. This could maintain the intracellular SRFT at a higher concentration than free SRFT. The drug’s slow and controlled release profile explains the lesser cytotoxicity of Pull-SA-SRFT at 24 h time point than at 48h time point. The IC50 of the drug was observed to be in the range of 6–8 µM. To conclude, a considerable number of Pull-SA-SRFT particles could be delivered to HCC cells due to greater internalisation by receptor-mediated endocytosis.
In addition, the CLSM images of cells subjected to chromosome condensation assay (Figure 5C, depict a greater extent of nuclear condensation and fragmentation by Pull-SA-SRFT treated cells than that of free SRFT.55 These results suggest that the developed drug delivery system could effectively target and kill the HCC cells.
Cellular Uptake and Receptor-Mediated Endocytosis
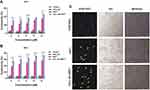
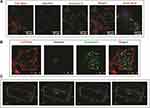
Identifying internalisation pathways for nano-drug delivery carriers leading to the efficient uptake of relevant drugs is significant.56 Therefore, the ability of PLC/PRF/5 cells to uptake the developed drug delivery system was studied by entrapping coumarin-6 fluorophore in Pull-SA conjugate to yield Pull-SA-C6 nanoparticles that were synthesised similarly to Pull-SA-SRFT nanoparticles. Cell nuclei stained with blue Hoechst and the red fluorescence (Cell-mask) on the plasma membrane clearly indicated the localisation of Pull-SA-C6 particles (green). After incubation of cells with the nanoparticles for 4 h, it was observed that cells could proficiently take up the particles (Figure 6A). This result was in line with the higher cytotoxicity of Pull-SA-SRFT, emphasising the importance of nanoparticle binding and internalisation to enhance the cytotoxic effect. Interestingly, the cells exhibited red fluorescence inside the cells after treatment along with the green Pull-SA-C6 particles (Figure 6B Merged). Perhaps, this was the portion of the membrane that was taken inside as result of receptor mediated endocytosis, clearly depicting the internalisation process of the developed drug delivery system. The 3-D images (Figure 6C) proves that the particles have entered inside the cells and are not adsorbed to the surface. These findings of the uptake of Pull-SA-C6 nanoparticles were in accordance with the previously reported studies, that pullulan complexed with the ASGPRs undergo endocytosis and get internalised with high affinity to the hepatocyte cells.57 As discussed earlier, this affinity is because of the orientation of hydroxyl groups on pyranose rings in pullulan structure that promotes proper binding of pullulan to the specific ASGPRs which further aids in endocytosis.
Detection of Early and Late Apoptosis
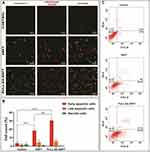
To investigate the cell death mechanism of free SRFT and Pull-SA-SRFT after treatment, early apoptosis was studied by Annexin V-FITC/PI staining after 12 h. Annexin V-FITC (green) can bind to the phosphatidyl serine flipped to the outer leaflet of the plasma membrane of early apoptotic cells. On the other hand, propidium iodide (red) is a membrane impermeant dye that is generally excluded from viable cells. It can rather intercalate with the double-stranded DNA of dead cells. CLSM images (Figure 7A) show that Pull-SA-SRFT could induce more death than free SRFT. This was confirmed by FACS analysis and the obtained results (Figure 7B and C) were in line with the data achieved by CLSM imaging.
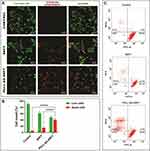
The live–dead assay conveyed the mechanism of late apoptosis after treatment with the formulations for 48h. Analogous results were obtained for this study. Live cells are distinguished by ubiquitous intracellular esterase activity, determined by the enzymatic conversion of the virtually nonfluorescent cell-permeant calcein AM to the intensely fluorescent calcein (green). EthD-1 enters cells with damaged membranes and undergoes a 40-fold enhancement of fluorescence upon binding to nucleic acids, thereby producing a bright red fluorescence in dead cells. The results obtained in Figure 8A–C denote the higher impact of Pull-SA-SRFT particles on cells than free SRFT which is in accordance with early apoptosis. However, it is worth noting that free SRFT and Pull-SA-SRFT could induce apoptosis significantly in the later stages than in the earlier stages and a higher proportion of death was induced by drug-entrapped particles. This is once again ascribed to the efficient cellular uptake of Pull-SA-C6 and the sustained release of SRFT from the conjugate.
Animal Studies
In vivo Biodistribution
To explore the biodistribution and biocompatibility of the developed drug delivery system in vivo, the formulations of free SRFT and Pull-SA-SRFT were injected into immunocompetent Swiss albino mice. The distribution of Pull-SA-SRFT in several organs, as well as its liver targeting property, were evaluated over the period of 1, 6, 24 and 48 h. The findings are shown in Figure 9A. Sorafenib was detected via HPLC analysis in blood, brain, kidneys, liver, lungs and spleen after the administration of free SRFT and Pull-SA-SRFT in mice at varying levels at all time points. However, in the first hour of treatment, it was observed that the concentration of free SRFT accumulated in the kidneys was higher compared to liver. In contrast, a significantly high concentration of Pull-SA-SRFT was observed in the liver from the first hour of treatment. This trend was maintained in the liver throughout the 48-hour study period, indicating the specificity of Pull-SA-SRFT to liver (Figure 9B). In the free SRFT treatment group, the concentration of sorafenib in kidneys at 48 h was higher than in the liver, signifying that the drug is being eliminated from the body, whereas, Pull-SA-SRFT was accumulated in the liver at a higher concentration even at 48 h compared to kidneys. This confirms that Pull-SA-SRFT has more affinity and retention capacity in the liver than compared to free drug. Moreover, no adverse effects were observed in the mice due to the presence of Pull-SA-SRFT for 48 hours. Hence, this proves that Pull-SA-SRFT had not caused any acute toxic effects to the mice and is biocompatible.
Conclusion
This study could develop an efficient drug delivery system to target hydrophobic drugs such as sorafenib tosylate to HCC. An amphiphilic nanocarrier derived from pullulan–stearic biopolymer was successfully synthesized and characterized. The drug-entrapped nanoparticles (Pull-SA-SRFT) had desirable particle size, that effectively entrapped the commercially approved drug with an excellent entrapment efficiency of 95.6%. The nanoparticles also showed a slow and sustained drug release pattern, and an accelerated drug-release in acidic pH. Cytotoxicity assay indicated that blank Pull-SA particles were non-toxic to cells demonstrating the biocompatibility of Pull-SA biopolymer and that Pull-SA-SRFT could induce more death than free SRFT alone in a concentration-dependent manner. This was in accordance with the results obtained by the higher degree of chromosome condensation induced by Pull-SA-SRFT than free SRFT. The cellular uptake study clearly illustrated the efficient uptake of Pull-SA-C6 particles and a thorough understanding of receptor-mediated endocytosis. Early and late apoptosis studies conveyed that SRFT and Pull-SA-SRFT could induce apoptosis significantly in the later stages than in the earlier stages and a higher proportion of death was induced by the drug-entrapped particles. The biodistribution study done in Swiss-albino mice indicates that the system has more affinity and retention capacity to the liver and did not cause any acute toxicity. This is hence a biocompatible, promisingly safe and excellent nanocarrier to deliver hydrophobic drugs to HCC.
Abbreviations
Pull-SA, pullulan–stearic acid copolymer; SRFT, sorafenib tosylate; HCC, hepatocellular carcinoma; DDS, drug delivery system; ASGPR, asialoglycoprotein receptor.
Ethics Approval
We certify that all applicable institutional and governmental regulations concerning the ethical use of animals were followed during the course of this research. The experiments were approved by the Institutional Animal Ethical Committee (IAEC) of Rajiv Gandhi Centre for Biotechnology, Protocol number- IAEC/752/GSV/2019.
Acknowledgments
The authors are thankful to Dr Vishnu Sunil Jaikumar, veterinarian (RGCB); the instrumentation staff of bio-imaging, cell sorting and TEM facility of Rajiv Gandhi Centre for Biotechnology (RGCB), for analyses of experiments conducted; Mr Jibinraj R J, CLIF facility, University of Kerala, for NMR analysis; the Department of Biotechnology, New Delhi, for providing the financial assistance to do this work, and also the Council of Scientific and Industrial Research (CSIR), New Delhi, for the Senior Research Fellowship to Teena Jacob Chirayil.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(2):477–491. doi:10.1053/j.gastro.2018.08.065
3. Forner A, Bruix J. Hepatocellular carcinoma–authors’ reply. Lancet. 2012;380(9840):470–471. doi:10.1016/S0140-6736(12)61286-0
4. Makarova‐Rusher OV, Altekruse SF, McNeel TS, et al. Population attributable fractions of risk factors for hepatocellular carcinoma in the United States. Cancer. 2016;122(11):1757–1765. doi:10.1002/cncr.29971
5. Harkus U, Wankell M, Palamuthusingam P, McFarlane C, Hebbard L. Immune checkpoint inhibitors in HCC: cellular, molecular and systemic data. In: Seminars in Cancer Biology. Elsevier; 2022.
6. Feng S, Zhou J, Li Z, et al. Sorafenib encapsulated in nanocarrier functionalized with glypican-3 specific peptide for targeted therapy of hepatocellular carcinoma. Colloids Surfaces B Biointerfaces. 2019;184:110498. doi:10.1016/j.colsurfb.2019.110498
7. Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(5):S5–S16. doi:10.1053/j.gastro.2004.09.011
8. Bu LL, Yan J, Wang Z, et al. Advances in drug delivery for post-surgical cancer treatment. Biomaterials. 2019;219:119182. doi:10.1016/j.biomaterials.2019.04.027
9. Lopez PM, Villanueva A, Llovet JM. Systematic review: evidence‐based management of hepatocellular carcinoma–an updated analysis of randomized controlled trials. Aliment Pharmacol Ther. 2006;23(11):1535–1547. doi:10.1111/j.1365-2036.2006.02932.x
10. Liu L, Cao Y, Chen C, et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66(24):11851–11858. doi:10.1158/0008-5472.CAN-06-1377
11. Wilhelm SM, Carter C, Tang L, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64(19):7099–7109. doi:10.1158/0008-5472.CAN-04-1443
12. Carlomagno F, Anaganti S, Guida T, et al. BAY 43-9006 inhibition of oncogenic RET mutants. J Natl Cancer Inst. 2006;98(5):326–334. doi:10.1093/jnci/djj069
13. Varshosaz J, Sadri F, Rostami M, Mirian M, Taymouri S. Synthesis of pectin-deoxycholic acid conjugate for targeted delivery of anticancer drugs in hepatocellular carcinoma. Int J Biol Macromol. 2019;139:665–677. doi:10.1016/j.ijbiomac.2019.07.225
14. Guo Y, Zhong T, Duan XC, et al. Improving anti-tumor activity of sorafenib tosylate by lipid-and polymer-coated nanomatrix. Drug Deliv. 2017;24(1):270–277. doi:10.1080/10717544.2016.1245371
15. Granito A, Marinelli S, Negrini G, Menetti S, Benevento F, Bolondi L. Prognostic significance of adverse events in patients with hepatocellular carcinoma treated with sorafenib. Therap Adv Gastroenterol. 2016;9(2):240–249. doi:10.1177/1756283X15618129
16. Wanjale MV, Jaikumar VS, Sivakumar KC, Paul RA, James J, Kumar GSV. Supramolecular hydrogel based post-surgical implant system for hydrophobic drug delivery against glioma recurrence. Int J Nanomedicine. 2022;17:2203. doi:10.2147/IJN.S348559
17. Mahapatro A, Singh DK. Biodegradable nanoparticles are excellent vehicle for site directed in-vivo delivery of drugs and vaccines. J Nanobiotechnol. 2011;9(1):1–11. doi:10.1186/1477-3155-9-55
18. Liu Y, Miyoshi H, Nakamura M. Nanomedicine for drug delivery and imaging: a promising avenue for cancer therapy and diagnosis using targeted functional nanoparticles. Int J Cancer. 2007;120(12):2527–2537. doi:10.1002/ijc.22709
19. Zhang XP, Chen XJ, Li BZ, et al. Active targeted Janus nanoparticles enable anti-angiogenic drug combining chemotherapy agent to prevent postoperative hepatocellular carcinoma recurrence. Biomaterials. 2022;281:121362. doi:10.1016/j.biomaterials.2022.121362
20. Poelstra K, Prakash J, Beljaars L. Drug targeting to the diseased liver. J Control Release. 2012;161(2):188–197. doi:10.1016/j.jconrel.2012.02.011
21. Mcabee DD, Jiang X, Walsh KB. Lactoferrin binding to the rat asialoglycoprotein receptor requires the receptor’s lectin properties. Biochem J. 2000;348(1):113–117. doi:10.1042/bj3480113
22. D’souza AA, Devarajan PV. Asialoglycoprotein receptor mediated hepatocyte targeting—Strategies and applications. J Control Release. 2015;203:126–139. doi:10.1016/j.jconrel.2015.02.022
23. Sugumaran KR, Ponnusami V. Review on production, downstream processing and characterization of microbial pullulan. Carbohydr Polym. 2017;173:573–591. doi:10.1016/j.carbpol.2017.06.022
24. Okada K, Yoneyama M, Mandai T, Aga H, Sakai S, Ichikawa T. Digestion and fermentation of pullulan. Nippon Eiyo Shokuryo Gakkaishi. 1990;43:23–29. doi:10.4327/jsnfs.43.23
25. Catley BJ, Ramsay A, Servis C. Observations on the structure of the fungal extracellular polysaccharide, pullulan. Carbohydr Res. 1986;153(1):79–86. doi:10.1016/S0008-6215(00)90197-6
26. Singh R, Gaur R, Tiwari S, Gaur MK. Production of pullulan by a thermotolerant Aureobasidium pullulans strain in non-stirred fed batch fermentation process. Braz J Microbiol. 2012;43(3):1042–1050. doi:10.1590/S1517-83822012000300030
27. Seymour LW, Duncan R, Chytrý V, Strohalm J, Ulbrich K, Kopeček J. Intraperitoneal and subcutaneous retention of a soluble polymeric drug-carrier bearing galactose. J Control Release. 1991;16(3):255–262. doi:10.1016/0168-3659(91)90001-T
28. Toth CA, Thomas P, Broitman SA, Zamcheck N. Receptor-mediated endocytosis of carcinoembryonic antigen by rat liver Kupffer cells. Cancer Res. 1985;45(1):392–397.
29. Iobst ST, Drickamer K. Selective sugar binding to the carbohydrate recognition domains of the rat hepatic and macrophage asialoglycoprotein receptors (*). J Biol Chem. 1996;271(12):6686–6693. doi:10.1074/jbc.271.12.6686
30. Khutoryanskiy VV. Synthesis and solution properties of hydrophobically modified polysaccharides. Eurasian Chem J. 2005;7(2):99–113. doi:10.18321/ectj621
31. Singh RS, Kaur N, Kennedy JF. Pullulan and pullulan derivatives as promising biomolecules for drug and gene targeting. Carbohydr Polym. 2015;123:190–207. doi:10.1016/j.carbpol.2015.01.032
32. Wang X, Wang J, Bao Y, Wang B, Wang X, Chen L. Novel reduction-sensitive pullulan-based micelles with good hemocompatibility for efficient intracellular doxorubicin delivery. RSC Adv. 2014;4(104):60064–60074. doi:10.1039/C4RA12276C
33. Wang Y, Liu Y, Liu Y, et al. A polymeric prodrug of cisplatin based on pullulan for the targeted therapy against hepatocellular carcinoma. Int J Pharm. 2015;483(1–2):89–100. doi:10.1016/j.ijpharm.2015.02.027
34. Jana P, Mitra T, Selvaraj TKR, Gnanamani A, Kundu PP. Preparation of guar gum scaffold film grafted with ethylenediamine and fish scale collagen, cross-linked with ceftazidime for wound healing application. Carbohydr Polym. 2016;153:573–581. doi:10.1016/j.carbpol.2016.07.053
35. Zhu D, Tao W, Zhang H, et al. Docetaxel (DTX)-loaded polydopamine-modified TPGS-PLA nanoparticles as a targeted drug delivery system for the treatment of liver cancer. Acta Biomater. 2016;30:144–154. doi:10.1016/j.actbio.2015.11.031
36. Cervello M, Pitarresi G, Volpe AB, et al. Nanoparticles of a polyaspartamide-based brush copolymer for modified release of sorafenib: in vitro and in vivo evaluation. J Control Release. 2017;266:47–56. doi:10.1016/j.jconrel.2017.09.014
37. Nanditha CK, Kumar GSV. Bioactive peptides laden nano and micro-sized particles enriched ECM inspired dressing for skin regeneration in diabetic wounds. Mater Today Bio. 2022;14:100235. doi:10.1016/j.mtbio.2022.100235
38. Wang X, Yang B, Li L, et al. Probing the fluorination effect on the self-assembly characteristics, in vivo fate and antitumor efficacy of paclitaxel prodrug nanoassemblies. Theranostics. 2021;11(16):7896. doi:10.7150/thno.61337
39. Su Y, Wang K, Li Y, et al. Sorafenib-loaded polymeric micelles as passive targeting therapeutic agents for hepatocellular carcinoma therapy. Nanomedicine. 2018;13(9):1009–1023. doi:10.2217/nnm-2018-0046
40. Pranatharthiharan S, Patel MD, Malshe VC, et al. Asialoglycoprotein receptor targeted delivery of doxorubicin nanoparticles for hepatocellular carcinoma. Drug Deliv. 2017;24(1):20–29. doi:10.1080/10717544.2016.1225856
41. Urbani R, Cesaro A. Chain conformations of polysaccharides in different solvents. 2001.
42. Huang L, Wang Y, Ling X, et al. Efficient delivery of paclitaxel into ASGPR over-expressed cancer cells using reversibly stabilized multifunctional pullulan nanoparticles. Carbohydr Polym. 2017;159:178–187. doi:10.1016/j.carbpol.2016.11.094
43. Chen L, Wang X, Ji F, et al. New bifunctional-pullulan-based micelles with good biocompatibility for efficient co-delivery of cancer-suppressing p53 gene and doxorubicin to cancer cells. RSC Adv. 2015;5(115):94719–94731. doi:10.1039/c5ra17139c
44. Akiyoshi K, Deguchi S, Tajima H, Nishikawa T, Sunamoto J. Microscopic structure and thermoresponsiveness of a hydrogel nanoparticle by self-assembly of a hydrophobized polysaccharide. Macromolecules. 1997;30(4):857–861. doi:10.1021/ma960786e
45. Teramoto N, Shibata M. Synthesis and properties of pullulan acetate. Thermal properties, biodegradability, and a semi-clear gel formation in organic solvents. Carbohydr Polym. 2006;63(4):476–481. doi:10.1016/j.carbpol.2005.10.008
46. Negahban Z, Shojaosadati SA, Hamedi S. A novel self-assembled micelles based on stearic acid modified schizophyllan for efficient delivery of paclitaxel. Colloids Surfaces B Biointerfaces. 2021;199(September2020):111524. doi:10.1016/j.colsurfb.2020.111524
47. Shaki H, Ganji F, Kempen PJ, Dolatshahi-Pirouz A, Vasheghani-Farahani E. Self-assembled amphiphilic-dextran nanomicelles for delivery of rapamycin. J Drug Deliv Sci Technol. 2018;44:333–341. doi:10.1016/j.jddst.2018.01.010
48. Du YZ, Weng Q, Yuan H, Hu FQ. Synthesis and antitumor activity of stearate-g-dextran micelles for intracellular doxorubicin delivery. ACS Nano. 2010;4(11):6894–6902. doi:10.1021/nn100927t
49. Shaki H, Vasheghani-Farahani E, Shojaosadati SA, Ganji F. Optimizing formulation variables of KCl loaded waxy microspheres. Iran J Pharm Sci. 2014;10(1):37–54.
50. Hsiao MH, Tung TH, Hsiao CS, Liu DM. Nano-hybrid carboxymethyl-hexanoyl chitosan modified with (3-aminopropyl) triethoxysilane for camptothecin delivery. Carbohydr Polym. 2012;89(2):632–639. doi:10.1016/j.carbpol.2012.03.066
51. Vaupel P, Schmidberger H, Mayer A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. Int J Radiat Biol. 2019;95(7):912–919. doi:10.1080/09553002.2019.1589653
52. Bononi G, Masoni S, Di Bussolo V, Tuccinardi T, Granchi C, Minutolo F. Historical perspective of tumor glycolysis: a century with Otto Warburg. In: Seminars in Cancer Biology. Elsevier; 2022.
53. Bonin S, Pascolo L, Crocé LS, Stanta G, Tiribelli C. Gene expression of ABC proteins in hepatocellular carcinoma, perineoplastic tissue, and liver diseases. Mol Med. 2002;8(6):318–325. doi:10.1007/BF03402158
54. Duan B, Huang C, Bai J, et al. Multidrug resistance in hepatocellular carcinoma. Exon Publ. 2019;2019:141–158.
55. Agarwal T, Narayana S, Pal K, Pramanik K, Giri S, Banerjee I. Calcium alginate-carboxymethyl cellulose beads for colon-targeted drug delivery. Int J Biol Macromol. 2015;75:409–417. doi:10.1016/j.ijbiomac.2014.12.052
56. Wang J, Cui S, Bao Y, Xing J, Hao W. Tocopheryl pullulan-based self assembling nanomicelles for anti-cancer drug delivery. Mater Sci Eng C. 2014;43:614–621. doi:10.1016/j.msec.2014.07.066
57. Kaneo Y, Tanaka T, Nakano T, Yamaguchi Y. Evidence for receptor-mediated hepatic uptake of pullulan in rats. J Control Release. 2001;70(3):365–373. doi:10.1016/S0168-3659(00)00368-0
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.