Back to Journals » OncoTargets and Therapy » Volume 9
Small interfering RNA targeting S100A4 sensitizes non-small-cell lung cancer cells (A549) to radiation treatment
Authors Qi R, Qaio T , Zhuang X
Received 17 February 2016
Accepted for publication 16 April 2016
Published 23 June 2016 Volume 2016:9 Pages 3753—3762
DOI https://doi.org/10.2147/OTT.S106557
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Ruixue Qi, Tiankui Qiao, Xibing Zhuang
Department of Oncology, Affiliated Jinshan Hospital, Fudan University, Shanghai, People’s Republic of China
Objective: This study aimed to investigate the impact of S100A4-small interfering RNA (S100A4-siRNA) on apoptosis and enhanced radiosensitivity in non-small-cell lung cancer (A549) cells. We also explored the mechanisms of radiosensitization and identified a new target to enhance radiosensitivity and gene therapy for non-small-cell lung cancer.
Methods: RNA interference is a powerful tool for gene silencing. In this study, we constructed an effective siRNA to knock down S100A4. A549 cells were randomly divided into three groups: blank, negative control, and S100A4-siRNA. To investigate the effect of S100A4-siRNA, the expression of S100A4, E-cadherin, and p53 proteins and their messenger RNA (mRNA) was detected by Western blot and quantitative real-time polymerase chain reaction. Transwell chambers were used to assess cell invasion. Cell cycle and apoptosis were analyzed by flow cytometry. Radiosensitivity was determined by colony formation ability.
Results: Our results demonstrate that S100A4-siRNA effectively silenced the S100A4 gene. When siRNA against S100A4 was used, S100A4 protein expression was downregulated, whereas the expressions of E-cadherin and p53 were upregulated. In addition, a clear reduction in S100A4 mRNA levels was noted compared with the blank and negative control groups, whereas E-cadherin and p53 mRNA levels increased. Transfection with S100A4-siRNA significantly reduced the invasiveness of A549 cells. S100A4 silencing induced immediate G2/M arrest in cell cycle studies and increased apoptosis rates in A549 cells. In clonogenic assays, we used a multitarget, single-hit model to detect radiosensitivity after S100A4 knockdown. All parameters (D0, Dq, α, β) indicated that the downregulation of S100A4 enhanced radiosensitivity in A549 cells. Furthermore, S100A4-siRNA upregulated p53 expression, suggesting that S100A4 may promote A549 cell proliferation, invasion, and metastasis by regulating the expression of other proteins. Therefore, siRNA-directed S100A4 knockdown may represent a viable clinical therapy for lung cancer.
Conclusion: S100A4 downregulation potentially enhances the sensitivity of human A549 cells to radiotherapy.
Keywords: lung cancer, S100A4, small interfering RNA, A549 cells
Introduction
Lung cancer is the leading cause of cancer-related deaths in the world, among which non-small-cell lung cancer (NSCLC) accounts for ~85% of the cases.1–3 Radiotherapy coupled with surgery and chemotherapy is the most important method of contemporary NSCLC treatment.4 However, the use of this therapy is also confronted with challenges due to the intrinsic radioresistance of tumor cells. Hence, enhancing the radiosensitivity of resistant tumor cells is a common strategy in the clinical application of radiotherapy.5
Located in the 1q21 human chromosome region, S100A4 (also known as mts1) is a member of the S100 family of transcription factors.6 S100A4 modulates invasion, metastasis, apoptosis, and cell cycle progression of a variety of malignant tumors through different mechanisms. S100A4 promotes tumor cell invasion and metastasis by downregulating intercellular and cell-substratum adhesion, remodeling the extracellular matrix, altering cytoskeletal dynamics, and promoting angiogenesis.7–10 S100A4 has also been implicated in modulating cell cycle progression, likely by suppressing p53.11 S100A4 also decreases proapoptotic gene expression and inhibits apoptosis.12 S100A4 is detectable in numerous cancer types, and its presence is associated with poor prognosis in many malignant tumors, such as breast,13 bladder,14 esophagus,15 and colon.16 Therefore, inhibition of S100A4 may be a good antitumor strategy to reduce cancer cell growth.
Ionizing radiations can result in lethal cell damage, which is correlated with DNA damage induction and repair.17 The tumor suppressor gene p53 encodes a transcriptional regulatory protein that plays a crucial role in controlling cell cycle progression and apoptosis. Grigorian et al18 used several in vitro approaches to demonstrate that S100A4 binds to the extreme end of the p53 C-terminal regulatory domain. Mutations in the p53 gene are noted in almost all types of human cancer with a frequency ranging from 20% to 60%.19 Jiang et al20 demonstrated that β-Elemene increases the radiosensitivity of A549 cells and that the mechanism involved may be related to the upregulation of p53 and induction of cellular apoptosis. When p53 is mutated, cells with DNA damage can escape apoptosis and become cancer cells.21
In this study, we designed a short interfering RNA (siRNA) against S100A4 and evaluated the inhibitory effect of siRNA transfection on cell cycle, apoptosis, and invasion in A549 cells. This effort provides insight into the mechanism by which S100A4 promotes motility, invasion, and metastasis in A549 cells to determine whether S100A4-siRNA sensitizes human NSCLC cells to radiotherapy.
Materials and methods
Cell culture
The study was approved by the ethical committee of Jinshan Hospital of Fudan University. The human lung adenocarcinoma cell line A549 was obtained from the Chinese Academy of Science (Shanghai, People’s Republic of China). Cells were maintained at 37°C in a humidified incubator under 5% CO2 and cultured in RPMI-1640 medium (Sigma-Aldrich Co., St Louis, MO, USA) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, and 10% heat-inactivated fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA). The medium was replaced every 2 or 3 days. Cells in the logarithmic growth phase were used to perform the experiments described in the following section.
Transfection of siRNA
S100A4-siRNA and nontargeting negative control (NC) siRNA were purchased from GenePharm (Shanghai, People’s Republic of China). The presence of siRNA sequences was confirmed by DNA sequencing. The following sequences were used: S100A4, sense: 5′-UGUCCACCUUCCACAAGUATT-3′ and antisense: 5′-UACUUGUGGAAGGUGGACATT-3′; NC, sense: 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense: 5′-ACGUGACACGUUCGGAGAATT-3′. Opti-MEM and Lipofectamine 2000 were purchased from the Cell Signaling Technology Corporation (Danvers, MA, USA). A549 cells in logarithmic growth phase were seeded in six-well culture plates. When the cells reached 50% confluence, 20 μM of S100A4-siRNA was transfected with 5 μL of Lipofectamine 2000 and 250 μL of serum-free Opti-MEM medium without antibiotics according to the manufacturer’s instructions. After incubation for 6 hours, the medium was replaced with the standard culture medium as previously described. After an additional 42-hour incubation, cells were used for the experiments described in the following section. The siRNA silencing experiment was repeated thrice.
Western blot analysis
After cell lysis, total protein was collected and run on 12% Bis-Tris gradient gels (Thermo Fisher Scientific) and electrophoretically transferred onto polyacrylamide membranes (Thermo Fisher Scientific). Nonspecific binding sites were blocked by incubating the membranes for 1 hour at 37°C with 5% nonfat dried milk in Tris-buffered saline containing 0.05% Tween-20 (TBST). Membranes were incubated overnight at 4°C with the following primary antibodies: anti-S100A4 (1:1,000; Santa Cruz Biotechnology Inc., CA, USA), anti-p53 (1:1,000; Santa Cruz Biotechnology Inc.), anti-E-cadherin (1:400; Santa Cruz Biotechnology Inc.), and anti-β-actin (1:1,000; Abcam, Cambridge, UK). Then, membranes were additionally washed and incubated with a goat anti-rabbit secondary antibody (1:20,000; Abcam). Bands were visualized using an enhanced chemiluminescence system (ECL, Pierce, Rockford, IL, USA).
Quantitative real-time polymerase chain reaction
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. RNA concentrations were determined by an ultraviolet spectrophotometer (SHIMADZU UV-2450; Shimadzu Corporation, Kyots, Japan). Subsequently, 2 μg of total RNA was reverse transcribed to produce single-stranded DNA using SuperScript II (Thermo Fisher Scientific). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the ABI Prism 7700 Sequence Detection System (PerkinElmer Inc., Waltham, MA, USA). The primer sequences were: 5′-AGCAACTTGGACAGCAACAG-3′ (forward) and 5′-TCTTCCTGGGCTGCTTATCT-3′ (reverse) for S100A4; 5′-AGGCCTTGGAACTCAAGGAT-3′ (forward) and 5′-CCCTTTTTGG ACTTCAGGTG-3′ (reverse) for p53; 5′-CCAAAGCCTCAGGTCATAA ACA-3′ (forward) and 5′-TTCTTGGGTTGGGTCGTTGTAC-3′ (reverse) for E-cadherin. Control PCR was performed using the following primers for GAPDH: 5′-CATCTTCTTTTGCGTCGCCA-3′ (forward) and 5′-TTAAAAGCAGCCCTGGTGACC-3′ (reverse). Quantitative PCR (Q-PCR) assays were performed in triplicate, and the mean values were used to calculate mRNA expression. Gene expression was normalized to GAPDH mRNA. The results were calculated and presented as 2−ΔΔCT.
Cell invasion assay
The transwell device containing microporous 8 μm membranes (Corning Incorporated, Corning, NY, USA) was placed in 24-well plates. Then, 1×104 cells/mL were seeded in the upper chambers and incubated at 37°C with 5% CO2 for 24 hours. Then, the cells were fixed and stained with 0.1% crystal violet. We counted the number of transmembrane cells under an optical microscope, selected three high-power fields at random, and assessed each field of vision to measure the invasion of tumor cells in vitro. Cells were counted and compared among groups. The experiments were repeated thrice.
Colony forming assay
This assay was used to measure the effect of S100A4 silencing on radiation sensitivity of A549 cells. A549 cells were randomly divided into three groups as described earlier: control, NC, and siRNA. All groups were irradiated with X-ray doses of 0, 2, 4, 6, 8, and 10 Gy. The following number of cells were plated for each radiation dose: 500 cells for 0 Gy, 1,000 for 2 Gy, 1,500 for 4 Gy, 2,000 cells for 6 Gy, 3,000 for 8 Gy, and 4,000 for 10 Gy. After 24-hour irradiation, the cells were suspended and incubated for ~14 days in a humidified atmosphere containing 5% CO2 at 37°C with fresh media being replaced every 3 days. Cell colonies were fixed with 80% cold methanol and then stained with 0.4% crystal violet for 1 hour. The colony formation units were evaluated by counting the number of colonies/well in a 5× magnification field. Clones of at least 50 cells were counted as one colony. Survival curves were plotted as the log of survival fraction. The experiment was repeated thrice to obtain the average colony formation rate. A standard radiation survival curve was constructed and the parameters D0, Dq, α (the probability of target cell damage occurs when an ionizing particle breaks through the DNA double strand breaks), and β (the probability of target cell damage occurs when two ionized particles break through the DNA double strand breaks) were calculated with the multitarget, single-hit model. D0 is the dose required to reduce the fraction of surviving cells to 37% of its previous value, and Dq is the repair capacity of the cells after radiation.
Cell cycle analysis
Cell cycle phases were analyzed by measuring the DNA fragment staining with propidium iodide (PI, Sigma-Aldrich Co.) and used according to the manufacturer’s instruction. A549 cells in the exponential growth phase were plated in six-well plates, divided, and treated as noted earlier. The cells were harvested and centrifuged at 24, 48, and 72 hours after exposure to10 Gy irradiation. Cell pellets were washed twice with phosphate-buffered saline. The cells were fixed with 70% ethanol at 4°C overnight and incubated with 200 μL DNAse-free RNAse (Sigma-Aldrich Co.) for 30 minutes at room temperature in the dark. Then, the cells were analyzed immediately by flow cytometry with a FACScanTM system using the CellQuestTM software (Version 3.3) (BD Biosciences, San Jose, CA, USA).
Apoptosis assay
A549 cells grown in six-well plates were divided and treated as mentioned earlier. All the cells in each group were harvested at 24, 48, and 72 hours after irradiation. The experiments were performed with an annexin V apoptosis detection kit (BD Biosciences). The cell pellets were resuspended in 100 mL of binding buffer and stained with 5 mL each of annexin V and PI staining solution for 15 minutes. Then, the samples were analyzed by flow cytometry on a FACScan (Becton Dickinson, Mountain View, CA, USA) using the Cell Quest program.
Statistical analysis
All values were expressed as mean ± standard deviation (SD). The statistics package SPSS 21.0 (SPSS Inc., Chicago, IL, USA) was used for all statistics analyses and mapping. A multitarget, single-hit model was used to fit cells to a survival curve. For all statistical analyses, differences were considered significant if P<0.05.
Results
Effect of S100A4-siRNA on S100A4, E-cadherin, and P53 proteins and their mRNAs in A549 cells
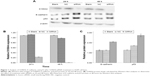
As shown by the Western blot in Figure 1, the expression of S100A4 protein was downregulated when siRNA against S100A4 was used, whereas the expressions of E-cadherin and p53 were upregulated. The nontargeting siRNA did not significantly affect S100A4, E-cadherin, or p53 protein expression, as demonstrated by comparison of the expression of these proteins between the blank and NC groups. S100A4 protein was reduced both at 24 and 48 hours after transfection but was markedly reduced at 48 hours (Figure 1A and B), while E-cadherin and p53 proteins were elevated at 48 hours (Figure 1C). Moreover, after transfection of A549 cells with S100A4-siRNA, mRNA levels were detected in a semiquantitative and quantitative manner using qRT-PCR. A clear reduction in S100A4 mRNA levels was observed compared with the blank and NC group, whereas the E-cadherin and p53 mRNA levels were elevated (Figure 2).
Invasion assay
We investigated the invasive capacity of A549 cells after S100A4 silencing. Transwell assays applying Matrigel-coated filters revealed the number of A549 cells penetrating through the artificial basement membrane. The results suggest that S100A4 knockdown cells exhibited markedly reduced invasion compared with the blank and NC groups. Our data suggest that S100A4 plays an important role in cell invasion (Figure 3).
Effect of S100A4-siRNA on colony formation by A549 cells
To assess the radiosensitivity after knockdown of S100A4, the multitarget, single-hit model was applied to fit the dose-survival curves. We observed that the cells transfected with NC siRNA exhibited radiosensitivity similar to that exhibited by control cells. Specifically, D0, Dq, sensitization enhancement ratio (SER), and survival rate of irradiated 2Gy cells (SF2) were comparable between both experimental groups as shown in Figure 4. In terms of radiobiological parameters, D0 equaled 3.13 Gy, and the SER, calculated by D0 value, equaled 1.34 Gy in the S100A4-siRNA group. All parameters (D0, Dq, α, β) indicated that transfection with S100A4-siRNA significantly (P<0.05) enhanced the radiosensitivity of A549 cells.
A549 cell cycle arrest at the G2/M phase
In the present study, comparison of cell cycle progression after irradiation was conducted by flow cytometry analysis. The effects of S100A4-siRNA on A549 cell cycle progression are presented in Table 1. We observed cell cycle arrest at the G2/M phase in cells that were transfected with siRNA against S100A4. The G2/M cell population was significantly different when compared with cells that were transfected with NC siRNA and blank after 10 Gy radiation at 24, 48, and 72 hours (P<0.05), and G2/M arrest was most evident at 24 hours (Figure 5).
Effect of S100A4-siRNA on apoptosis
After flow cytometry analysis, annexin V-positive cells were used to indicate apoptosis. The apoptotic index for the S100A4-siRNA was significantly increased compared with both NC and blank groups at 24, 48, and 72 hours (Figure 6, P<0.01).
Discussion
S100A4 is a well-established marker and mediator of metastatic disease and tumor growth. We demonstrated that the S100A4 gene is abundantly expressed at both the protein and mRNA levels. It has been previously demonstrated in other cell types that S100A4 downregulation may alter the expression of matrix metalloproteinases (MMPs), E-cadherin, and p53.22,23 The mechanism by which S100A4 protein modulates expression of the E-cadherin and p53 genes in lung cancer cells is not known. Our results suggest that a transcriptional regulation mechanism is involved. In this study, we confirmed the positive correlation between S100A4, p53, and E-cadherin expression in A549 cells. Using an A549 cell line expressing S100A4-siRNA, we demonstrated that knockdown of S100A4 resulted in a decreased expression of p53 and E-cadherin protein and their mRNAs in A549 cells. Our results suggested the possibility that the S100A4 gene could be used as a potential therapeutic target through RNA silencing.
Mahon et al24 demonstrated that knockdown of S100A4 results in an increase in the sensitivity of pancreatic ductal adenocarcinoma cell lines to gemcitabine as well as an increase in the number of apoptotic cells. However, no study has explored the possible correlation between S100A4 expression and lung cancer radiosensitivity to date.
RNA interference (RNAi) reduced mRNA and protein levels of specific genes through posttranscriptional gene silencing mechanisms. This technology is highly efficient, specific, and minimally toxic. This technology is used in functional genomic studies and for therapeutic gene regulation.25,26 In recent years, focus on the impact of S100A4 knockdown on malignant tumors has increased. For example, Ma et al6 reported that siRNA-directed knockdown of S100A4 decreases proliferation and invasiveness of osteosarcoma cells in experiments. Li et al27 demonstrated that S100A4 siRNA inhibits human pancreatic cancer cell invasion in vitro. However, methods to sensitize lung cancer cells to the effects of radiation have remained unclear to date.
Clonogenic survival analysis revealed that S100A4 knockdown markedly potentiated the decrease in NSCLC cell survival induced by radiation. The clonogenic assay acted by modulating the extracellular activity of MMPs.28
Cell cycle regulation is also an important factor in radiosensitivity. Radiosensitivity can vary in different phases of the cell cycle. Cells are most sensitive to radiation during the G2/M phase, less sensitive during the G1 phase, and least sensitive near the end of the S phase.29 Our study demonstrated that S100A4-siRNA altered the cell cycle with S phase reduction and G2/M phase arrest. These results indicate that S100A4-siRNA acts on the G2/M transition checkpoint of the cell cycle. The cells arrested in the G2/M phase tend to undergo radiation-induced apoptosis; therefore, S100A4-siRNA may be a contributing factor to increased radiosensitivity.
Apoptosis is important in mediating radiosensitivity. Resistance to radiotherapy is a major limiting factor in the treatment of cancer, and abnormal regulation of apoptosis is a contributing factor.30,31 In the present study, radiation alone induced minimal cell apoptosis. When radiation was applied in combination with S100A4-siRNA, the apoptotic index significantly increased, suggesting that S100A4-siRNA sensitized A549 cells to radiation, at least in part, by enhancing radiation-induced apoptosis.
Conclusion
This study further substantiates the proposed role of S100A4 knockdown in potentiating the sensitivity of A549 cells to X-rays by assessing the effect of S100A4 knockdown, which resulted in an increase in cells arrested at the G2/M phase, an increase in cell apoptosis, and a decrease in cell survival. Consequently, S100A4 downregulation potentially could serve as an important anticancer therapy.
Acknowledgments
This research was supported by a grant from the Science and Technology Council of the Jinshan District, Shanghai, People’s Republic of China (2013-3-02). We thank Dr Liu Bo for performing FACScan analysis.
Author contributions
All authors read and approved the final manuscript for publication. All authors contributed toward data analysis, drafting, and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Gajra A, Newman N, Gamble GP, Abraham NZ, Kohman LJ, Graziano SL. Impact of tumor size on survival in stage IA non-small cell lung cancer: a case for subdividing stage IA disease. Lung Cancer. 2003;42(1):51–57. | ||
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. | ||
Sechler M, Cizmic AD, Avasarala S, et al. Non-small-cell lung cancer: molecular targeted therapy and personalized medicine – drug resistance, mechanisms, and strategies. Pharmgenomics Pers Med. 2013;6(1):25–36. | ||
Krause S, Debus J, Neuhof D. Radiotherapy. Recent Results Cancer Res. 2011;183(183):285–291. | ||
Terris DJ, Ho EY, Ibrahim HZ, et al. Estimating DNA repair by sequential evaluation of head and neck tumor radiation sensitivity using comet assay. Arch Otolaryngol Head Neck Surg. 2002;128(6):698–702. | ||
Ma X, Yang Y, Wang Y, An G, Lv G. Small interfering RNA-directed knockdown of S100A4 decreases proliferation and invasiveness of osteosarcoma cells. Cancer Lett. 2010;299(2):171–181. | ||
Lakshmi MS, Parker C, Sherbet GV. Expression of the transmembrane glycoprotein CD44 and metastasis associated 18A2/MTS1 gene in B16 murine melanoma cells. Anticancer Res. 1997;17(5A):3451–3455. | ||
Lakshmi MS, Parker C, Sherbet GV. Metastasis-associated MTS1and NM23 genes affect tubulin polymerisation in B16 melanomas: a possible mechanism of their regulation of metastatic behaviour of tumors. Anticancer Res. 1993;13(2):299–303. | ||
Li ZH, Spektor A, Varlamova O, Bresnick AR. Mts1 regulates the assembly of nonmuscle myosin-IIA. Biochemistry. 2003;42(48):14258–14266. | ||
de Silva Rudland S, Martin L, Roshaniall C, et al. Association of S100A4 and osteopontin with specific prognostic factors and survival of patients with minimally invasive breast cancer. Clin Cancer Res. 2006;12(4):1192–1200. | ||
Chen H, Fernig DG, Rudland PS, Sparks A, Wilkinson MC, Barraclough R. Binding to intracellular targets of the metastasis-inducing protein, S100A (p9Ka). Biochem Biophys Res Commun. 2001;286(5):1212–1217. | ||
Schmidt-Hansen B, Klingelhöfer J, Grum-Schwensen B, et al. Functional significance of metastasis-inducing S100A4 (Mts1) in tumor–stroma interplay. J Biol Chem. 2004;279(23):24498–24504. | ||
Cabezón T, Celis JE, Skibshøj I, et al. Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int J Cancer. 2007;121(7):1433–1444. | ||
Davies BR, O’Donnell M, Durkan GC, et al. Expression of S100A4 protein is associated with metastasis and reduced survival in human bladder cancer. J Pathol. 2002;196(3):292–299. | ||
Ninomiya I, Ohta T, Fushida S, et al. Increased expression of S100A4 and its prognostic significance in esophageal squamous cell carcinoma. Int J Oncol. 2001;18(4):715–720. | ||
Taylor S, Herrington S, Prime W, Rudland PS, Barraclough R. S100A4 (P9Ka) protein in colon carcinoma and liver metastases: association with carcinoma cells and T-lymphocytes. Br J Cancer. 2002;86(3):409–416. | ||
Rodemann HP. Molecular radiation biology: perspectives for radiation oncology. Radiother Oncol. 2009;92(3):293–298. | ||
Grigorian M, Andresen S, Tulchinsky E, et al. Tumor suppressor p53 protein is a new target for the metastasis-associated Mts1/S100A4 protein: functional consequences of their interaction. J Biol Chem. 2001;276(25):22699–22708. | ||
Hollstein M, Rice K, Greenblatt MS, et al. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22(17):3551–3555. | ||
Jiang H, Ma S, Feng J. In vitro study of radiosensitization by β-Elemene in A549 cell line from adenocarcinoma of lung. Chinese-German J Clin Oncol. 2009;8(1):12–15. | ||
Lai PB, Chi TY, Chen GG. Different levels of p53 induced either apoptosis or cell cycle arrest in a doxycycline-regulated hepatocellular carcinoma cell line in vitro. Apoptosis. 2007;12(2):387–393. | ||
Boye K, Maelandsmo GM. S100A4 and metastasis: a small actor playing many roles. Am J Pathol. 2010;176(2):528–535. | ||
Berge G, Maelandsmo GM. Evaluation of potential interactions between the metastasis-associated protein S100A4 and the tumor suppressor protein p53. Amino Acids. 2011;41(4):863–873. | ||
Mahon PC, Baril P, Bhakta V, et al. S100A4 contributes to the suppression of BNIP3 expression, chemoresistance, and inhibition of apoptosis in pancreatic cancer. Cancer Res. 2007;67(14):6786–6795. | ||
Dykxhoorn DM, Palliser D, Lieberman J. The silent treatment: siRNAs as small molecule drugs. Gene Ther. 2006;13(6):541–552. | ||
Gazzaniga P, Gradilone A, Giuliani L, et al. Expression and prognostic significance of LIVIN, SURVIVIN and other apoptosis-related genes in the progression of superficial bladder cancer. Ann Oncol. 2003;14(1):85–90. | ||
Li N, Song MM, Chen XH, Liu LH, Li FS. S100A4 siRNA inhibits human pancreatic cancer cell invision in vitro. Biomed Environ Sci. 2012;25(4):465–470. | ||
Kikuchi N, Horiuchi A, Osada R, et al. Nuclear expression of S100A4 is associated with aggressive behavior of epithelial ovarian carcinoma: an important autocrine/paracrine factor in tumor progression. Cancer Sci. 2006;97(10):1061–1069. | ||
Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Phys. 2004;59(4):928–942. | ||
Xie YF, Sheng W, Xiang J, Zhang H, Ye Z, Yang J. Adenovirus-mediated ING4 expression suppresses pancreatic carcinoma cell growth via induction of cell-cycle alteration, apoptosis, and inhibition of tumor angiogenesis. Cancer Biother Radiopharm. 2009;24(2):261–269. | ||
Yi T, Li H, Wang X, Wu Z. Enhancement radiosensitization of breast cancer cells by deguelin. Cancer Biother Radiopharm. 2008;23(3):355–362. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.