Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 13
SLC30A8, CDKAL1, TCF7L2, KCNQ1 and IGF2BP2 are Associated with Type 2 Diabetes Mellitus in Iranian Patients
Authors Vatankhah Yazdi K, Kalantar SM, Houshmand M, Rahmanian M, Manaviat MR, Jahani MR , Kamalidehghan B, Almasi-Hashiani A
Received 4 August 2019
Accepted for publication 18 February 2020
Published 24 March 2020 Volume 2020:13 Pages 897—906
DOI https://doi.org/10.2147/DMSO.S225968
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Antonio Brunetti
Kazem Vatankhah Yazdi,1 Seyed Mehdi Kalantar,1 Massoud Houshmand,2,3 Masoud Rahmanian,4 Masoud Reza Manaviat,5 Mohammad Reza Jahani,6 Behnam Kamalidehghan,2,7 Amir Almasi-Hashiani8
1Department of Genetic, Shahid Sadoughi University of Medical Sciences and Health Services, Yazd, Iran; 2Department of Medical Genetics, National Institute for Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran; 3Research Center, Knowledge University, Erbil, Kurdistan Region, Iraq; 4Yazd Diabetes Research Center, Shahid Sadoughi University of Medical Sciences and Health Services, Yazd, Iran; 5Department of Ophthalmology, Shahid Sadoughi University of Medical Sciences and Health Services, Yazd, Iran; 6Taban Health Care and Diabetes Clinic Lab, Tehran, Iran; 7Medical Genetics Department, Jiroft University of Medical Sciences and Health Services, Jiroft, Iran; 8Department of Epidemiology, School of Health, Arak University of Medical Sciences, Arak, Iran
Correspondence: Seyed Mehdi Kalantar
Reproductive & Genetic Unit, Abortion Research Centre Reproductive Sciences Institute, Shahid Sadoughi University of Medical Sciences and Health Services, Bau Ali Ave. Safaeyeh, Yazd, Iran
Tel +9838247086
Fax +9838247087 Email [email protected]
Background: Type 2 diabetes mellitus (T2DM) is a serious public health issue with significantly increasing rates across the world. The genome-wide association studies (GWAS) have previously manifested involved genes that remarkably enhance the risk of T2DM. In this study, the association of common variants with T2DM risk has been identified among Iranian population from Tehran province of Iran.
Methods: Here, the association of refSNPs with T2DM risk was examined on peripheral blood samples of 268 individuals including control group and patients with T2DM using the tetra amplification refractory mutation system (ARMS) methods and direct genomic DNA sequencing.
Results: Our study demonstrated that SLC30A8 rs13266634 (T/C), CDKAL1 rs10946398 (A/C), TCF7L2 rs7903146 (C/T), KCNQ1 rs2237892 (T/C), and IGF2BP2 rs1470579 (A/C) polymorphisms are significantly associated with type 2 diabetes, but no significant association was identified for FTO rs8050136 and MTNR1B rs10830963 polymorphisms.
Conclusion: The prediction of refSNPs is remarkably needed for pharmacogenetics and pharmacogenomic approaches, in which the information would be useful for clinicians to optimize therapeutic strategies and adverse drug reactions in patients with T2DM.
Keywords: type 2 diabetes mellitus, T2DM, Iranian populations, the tetra amplification refractory mutation system, ARMS, the genome-wide association studies, GWAS
Introduction
Type 2 diabetes mellitus (T2DM) is a multifactorial and complex metabolic disorder, characterized by chronic hyperglycemia due to impairment in insulin secretion and sensitivity.1 The frequency of T2DM is enhancing gradually due to environmental factors and interplay of different variation in multiple genes.2 The World Human Organization (WHO) estimated that the total number of individuals with T2DM will reach to 366 million throughout the world by 2030.3 The hallmark factors, influencing the prevalence of diabetes, include age, gender, ethnicity, lifestyle, and obesity,4 in which the prevalence of T2DM in Iranian population is approximately 7.7%.5 There are several important genetic factors in T2DM including high incidence of diabetes among monozygotic twins compare to dizygotic twins, familial history, ethnicity and migration studies.6,7
IGF2BP2 is extremely expressed in pancreatic islets and binds to IGF-2 (insulin-like growth factor 2) that plays remarkable roles in localization, stability and translation of RNA.8–10 KCNQ1 (potassium voltage-gated channel KQT-like subfamily, member 1) gene is expressed in pancreatic islets11,12 and plays a central role for repolarization of the cardiac action potential and transportation of water and salt in epithelial tissues.13 Mutations in KCNQ1 gene are involved in deafness and long QT syndrome.14 Blockade of the channel with KCNQ1 inhibitors 293B resulting in insulin secretion,11 indicating that KCNQ1 channels may play functionally significant roles in regulation of insulin secretion. Previous studies demonstrated that TCF7L2 is an important regulator of insulin production and is also expressed in pancreatic islets.15 TCF7L2 plays a key role in expression and subsequent conversion of proinsulin into mature insulin through various TCF7L2-target genes and downstream regulatory signaling pathways.15 Additionally, TCF7L2 may also affect the insulin clearance and insulin sensitivity.15,16
The FTO rs8050136 polymorphism is associated with high risk of T2D.17–23 The FTO is a 2-oxoglutarate (2-OG) Fe(II) dependent demethylase. Previous studies demonstrated a potential role of FTO in nucleic acid repair.24 According to the previous studies, the exact molecular signaling pathways of FTO involved in human adiposity, cancer, metabolic disorders, obesity and T2D remain largely unknown.18,25,26 The MTNR1B is a new susceptibility gene involved in the regulation of glucose homeostasis and T2DM, encoding the melatonin receptor MT2, which is expressed in different tissues including pancreatic islets.27–30 The rs13266634 (C/T) in the SLC30A8 gene is associated with increasing risk of T2D, encoding zinc transporter 8, which is primarily expressed in pancreatic β-cells. Additionally, the major C-allele of rs13266634 is associated with a lower early insulin response to glucose and a high risk of T2D.31,32 Due to the limited number of controlled trials, there is to date no overall strong evidence supporting the theory that zinc supplementation may lower the risk of T2D in humans.33 Previous studies demonstrated an interaction between plasma zinc levels and rs13266634 on T2D risk.34 Cyclin-dependent kinase 5 regulatory subunit-associated protein 1-like (CDKAL1) gene, located in 6p22.3, is associated with T2DM.35 CDKAL1 gene encodes tRNA decoration enzyme, namely methyl transfer enzyme which is involved in 2-methylthio-N6-threonylcarbamoyladenosine synthesis of the 37th base of tRNA Lys(UUU).36,37
Zinc is an essential element for insulin secretion and storage.38–40 Pancreatic beta cells contain the highest level of Zinc compared to other cells in the human body.41 The genome-wide association studies (GWAS) have extended the progress and distribution of different genetic components in type 2 diabetes.9 Today, there are at least 20 loci that are associated with T2DM risk, in which the SLC30A8 (rs13266634), CDKN2A/2B (rs10811661), HHEX (rs1111875) and TCF7L2 (rs7903146) play important roles in the risk of T2DM in European Caucasians.9,31,42,43 In this study, the association of different refSNPs with T2DM was investigated for the prediction of T2DM risk among the Iranian population.
Materials and Methods
Specimen Collection and Ethical Statement
In this study, 268 peripheral blood samples, including 106 healthy and unrelated donors and 162 patients with T2DM, were obtained from Tehran Taban Health Care and Diabetes Clinic (TTHCDC) and Aramesh Genetic and Pathobiology Lab from Tehran. The whole peripheral blood samples collected in tubes containing ethylenediamine tetraacetic acid (EDTA) in a final volume of 2 mL. The written informed consent for participating in the study and allowing the publishing of information for genetic analysis were obtained from individuals. Approval to conduct this study was granted by the medical ethics committee of Shahid Sadoughi University of Medical Sciences and Health Services (approval number: IR.SSU.MEDICINE.REC.1395.90) in accordance with the Declaration of Helsinki. The inclusion criteria were patients older than 40 years who had lived with type 2 diabetes for more than 10 years. The exclusion Criteria were: having chronic diseases such as heart failure, chronic kidney disease, chronic lung disease, diabetic foot or limb amputation, and moderate to severe retinopathy. The exclusion criteria in the control group were chronic disease or fasting blood sugar>100 mg/dl.
DNA Extraction Protocol
The DNA from whole peripheral blood samples was extracted using PrimePrep Genomic DNA extraction kit (GeNet Bio). The quantity and quality of extracted DNA was measured using Nanodrop, and then run on a 1% agarose gel electrophoresis.
Primer Design
The forward and reverse primers for identification of genes were designed using the online Primer 1 program, available from http://primer1.soton.ac.uk/public html/primer1.html, developed by Ye and colleagues in 2001. The details of primers were checked using BLAST through https://www.ncbi.nlm.nih.gov/tools/primer-blast/.
The two special set primers were designed by using the primer1 program (http://primer1.soton.ac.uk/public_html/primer1.html) developed by Ye et al (2001). The specificity of primers and their melting temperatures were checked using BLAST (http://www.ncbi.nlm.nih). The details of the primers are summarized in Table 1.
 |
Table 1 Represents List of Forward and Reverse Primers (Inner and Outer) Applied for Detection of SNPs |
Procedure of Tetra Primer ARMS-PCR
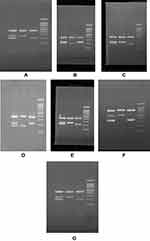
Essential keys for optimization of tetra ARMS are ration determination of outer and inner primers and annealing temperature. The unspecific bands were solved using gradient PCR system and optimization of outer and inner primer concentrations. The PCR reaction was performed in a final volume of 25 ul containing 1 μL Mg, 2.5 μL Buffer, 0.5 μL dNTP, 0.2 μL Taq polymerase, 1μL forward outer primer, 1 μL reverse outer primer, 2 μL forward inner primer, 2 reverse inner primer,1 μL DNA and 13.8 μL H2O. The DNA amplification was carried out using ARMS PCR for SNPs. The details of PCR reaction were as follows: 95°C for 5 min for cycle preparation, 30 cycles of denaturation at (95°C for 30 sec, annealing for 30 sec with different temperatures), polymerization at 72°C for 30 sec, and final extension at 72°C for 5 min with 1 cycle. The amplified fragment was confirmed and analyzed on 1.5% agarose gel (Figure 1).
Single Nucleotide Polymorphisms (SNP) Identification
In this study, the SNPs were sequenced and the results were then identified using the National Center for Biotechnology Information (NCBI) database, available at http://www.ncbi.nlm.nih.gov/SNP.
Sequencing Analysis
The double-stranded DNA of PCR products was examined using an automated ABI sequencing machine (ABI 3130 Genetic Analyzer, Baghiyatallah Hospital, Tehran-Iran). The DNA fragments were confirmed for any nucleotide variation and were then analyzed using Finch TV software (http://www.geospiza.com/finchtv/; PerkinElmer Inc., Waltham, MA, USA) (Figure 2).
Statistical Analysis
The odd ratios (OR) and 95% confidence intervals (CI) are used to determine the significant relationship of SNPs with T2DM. The Chi-square or Fisher exact test is used to analyze the results using STATA (V8) software. The p-values<0.05 is regarded as statistically significant.
Results
The clinical and biomedical characteristic of patients with T2DM were compared with control groups and the results are summarized in Table 2. This study demonstrated that there is a significant correlation (p<0.05) between T2DM patients and control groups with certain clinical parameters, including BMI, SBP, DBP, LDL, HDL, TG, 2hpp, FBS and HbA1c.
 |
Table 2 The Clinical and Biomedical Characteristics of Individuals in This Study |
In our study, genotypic frequency of SNPs variants was analyzed from the blood samples of 162 T2DM patients and 106 non-diabetic individuals from the Iranian population. The genotypic frequencies of the homozygous (C/C), heterozygous (C/T), and homozygous (T/T) variants of the rs13266634 (T/C) significantly (P value<0.001) observed in 16 (9.88%), 64 (39.51%), and 82 (50.62%) of T2DM patients and 5 (4.72%), 23 (21.70%), and 78 (73.58. %) of the control groups, respectively (Table 3). Additionally, the odd ratio of T2DM patients with C/T and C/C genotype was 2.64 and 3.04, respectively. However, patients with C/T (P=0.001) and C/C (P=0.038) genotypes significantly indicated high risk of T2DM in comparison with T/T genotype. The genotypic frequencies of homozygous (A/A), heterozygous (A/C), and homozygous (C/C) variants of the rs10946398 (A/C) significantly (P value<0.001) observed in 31 (19.14%), 104 (64.20%), and 27 (16.67%) of T2DM patients and 46 (43.40%), 50 (47.17%), and 10 (9.43%) of the control groups. Additionally, the odd ratio of T2DM patients with A/C and C/C genotypes were 3.08 and 4.0, respectively. However, patients with C/C and A/C genotypes significantly (P=0.001) indicated high risk of T2DM in comparison with A/A genotype (Tables 3 and 4).
 |
Table 3 The Genotypic Frequency of Polymorphisms Between T2DM and Control Groups |
 |
Table 4 The Genotypic Frequency of Polymorphisms Between T2DM and Control Groups with Odds Ratio |
The genotypic frequencies of homozygous (C/C), heterozygous (C/T), and homozygous (T/T) variants of the rs7903146 (C/T) significantly (P value<0.001) observed in 26 (16.05%), 76 (46.91%), and 60 (37.04%) of T2DM patients and 51 (48.11%), 39 (36.79%), and 16 (15.09%) of control groups. Additionally, the odd ratio of T2DM patients with T/T and C/T genotype were 7.35 and 3.82, respectively. However, patients with T/T and C/T genotypes (P=0.001) indicated high risk in comparison with C/C genotypes (Tables 3 and 4).
The genotypic frequencies of homozygous (A/A), heterozygous (A/C), and homozygous (C/C) variants of the rs1470579 (A/C) significantly (P value<0.001) observed in 67 (41.36%), 40 (24.69%), and 55 (33.95%) of T2DM patients and 55 (51.89%), 40 (37.74%), and 11 (10.38%) of the control groups. Additionally, the A/C genotype was evaluated using odd ratio (OR: 4.1) and it was significantly high (P=0.001) in T2DM patients in comparison with A/A genotype (Tables 3 and 4). The genotypic frequencies of homozygous (T/T), heterozygous (C/T), and homozygous (C/C) variants of the rs2237892 (C/T) significantly (P value=0.002) observed in 1 (0.62%), 9 (5.56%), and 152 (93.83%) of T2DM patients and 6 (5.66%), 15 (14.15%), and 85 (80.19%) of the control groups. Additionally, C/C genotype was evaluated using odd ratio (OR: 10.7) and it was significantly high (P=0.029) in T2DM patients in comparison with the T/T genotype (Tables 3 and 4). Additionally, there is no significant association of rs8050136 FTO (p value = 0.132) and rs10830963 MTNR1B (p value = 0.079) polymorphisms between T2DM patients and control groups (Tables 3 and 4).
Discussion
Type 2 diabetes mellitus (T2DM) is a complicated multi-gene or polygenic disorder involved with unknown contributing genes that increase the risk of T2DM in susceptible individuals. Recently, 16 novel susceptible gene loci for T2DM were identified,44 but the impact of TCF7L2 is much higher in comparison with other confirmed T2D gene candidate.45
The interaction of multiple genes and genetic and environmental factors lead to hyperglycemia due to impaired insulin function or secretion. Therefore, different strategies have been attempted to identify involved genes with T2DM. However, investigation in different ethnicities and geographical region demonstrated different results across the world. SNPs in SLC30A8 (rs13266634), CDKAL1 rs10946398, TCF7L2 rs7903146, KCNQ1 rs2237892, IGF2BP2 rs1470579 and MTNR1B rs10830963 were reported to be associated with T2DM in different studies.46–49 Our study demonstrated that SLC30A8 rs13266634 (T/C), CDKAL1 rs10946398 (A/C), TCF7L2 rs7903146 (C/T),KCNQ1 rs2237892 (T/C), and IGF2BP2 rs1470579 (A/C) polymorphisms are significantly in association with type 2 diabetes, but no significant association was identified for FTO rs8050136 and MTNR1B rs10830963 polymorphisms in present study.
Of 31 provinces in Iran, studies on only two provinces, Gorgan50 and Kurdish,51 revealed that the frequency of the TCF7L2 rs7903146 polymorphism was significant, which is in agreement with present study. In contrast to our study, a lack of remarkable association between TCF7L2 rs7903146 polymorphism and T2D was reported among the Arabian Emirates52 and Saudi Arab population.53 Here, the association of various SNPs with T2DM risk was investigated among Iranian individuals. However, the limitation of our study was to demonstrate the association of mortality and clinical complications with T2DM. The fundamental mechanisms, by which genetic variations within the intron of the TCF7L2 gene confers susceptibility to T2DM, remain to be elucidated. However, a study indicated that genetic variations around 3ʹend of the TCF&L2 gene may affect the function of TCF7L2, due to regulation of alternative splicing.54 The lack of statistically significant differences in the allelic and genotypic frequencies of FTO rs8050136 and MTNR1B rs10830963 polymorphisms between T2DM and control groups may be partially interpreted due to the heterogeneity of the individual’s ethnicity. It is accepted that approximately 14% of the between-studies variances may be attributed to ethnicity differences.55
Conclusion
In conclusion, our study indicated the association of well-established common variants of SLC30A8 rs13266634 (T/C), CDKAL1 rs10946398 (A/C), TCF7L2 rs7903146 (C/T), KCNQ1 rs2237892 (T/C), and IGF2BP2 rs1470579 (A/C) with type 2 diabetes among Iranian population from Tehran. Our study indicated the remarkable presence of the rs13266634, 10946398, 7903146, 2237892 and 1470579 polymorphisms in Iranian population, suggesting susceptibility of individuals to T2DM which may lead to identification of individuals with high risk for developing T2DM. Additionally, this study demonstrated the absence of the common variant FTO rs8050136 (C/A) among Iranian population. It is clinically important to manifest individuals for pharmacogenetics and pharmacogenomics approaches and prevention of harmful complication of this silent disease and adverse drug reactions. Therefore, identification of involved polymorphism analysis improves individual lifestyle and is a helpful tool toward personalized medicine and avoidance of the harmful effects of drugs.
Acknowledgments
The authors would like to appreciate the Shahid Sadoughi University of Medical Sciences and Health Services for providing facilities and financial supports to this study. They also thank Taban Health Care and Diabetes Clinic and Aramesh Genetic and Pathobiology Lab for providing facilities for registration and collection of consent form from diabetic patients and non-diabetic individuals and supporting the laboratory technical equipment. The present article was adopted from research proposal No. 4897 approved by the Shahid Sadoughi University of Medical Sciences and Health Services, Yazd, Iran.
Author Contributions
Conceived the experiments: Seyed Mehdi Kalantar, Massoud Houshmand and Kazem Vatankhah Yazdi; Designed the experiments: Masoud Reza Manaviat and Masoud Rahmanian; Collection of specimens and consent form: Kazem Vatankhah Yazdi, Mohammad Reza Jahani; Performed the experiments: Kazem Vatankhah Yazdi; Analyzed the data: Amir Almasi-Hashiani; Contributed reagents/materials/analysis tools: Kazem Vatankhah Yazdi, Seyed Mehdi Kalantar and Massoud Houshmand; Wrote the paper: Kazem Vatankhah Yazdi and Behnam Kamalidehghan; Revised and approved the manuscript: Kazem Vatankhah Yazdi and Behnam Kamalidehghan. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Nyenwe EA, Jerkins TW, Umpierrez GE, Kitabchi AE. Management of type 2 diabetes: evolving strategies for the treatment of patients with type 2 diabetes. Metabolism. 2011;60(1):1–23. doi:10.1016/j.metabol.2010.09.010
2. Horikawa Y, Oda N, Cox NJ, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26(2):163. doi:10.1038/79876
3. Hu FB. Globalization of diabetes: the role of diet, lifestyle, and genes. Diabetes Care. 2011;34(6):1249–1257. doi:10.2337/dc11-0442
4. Timper K, Donath MY. Diabetes mellitus type 2-the new face of an old lady. Swiss Med Wkly. 2012;142:2930.
5. Esteghamati A, Gouya MM, Abbasi M, et al. Prevalence of diabetes and impaired fasting glucose in the adult population of Iran: national survey of risk factors for non-communicable diseases of Iran. Diabetes Care. 2008;31(1):96–98. doi:10.2337/dc07-0959
6. Ridderstråle M, Groop L. Genetic dissection of type 2 diabetes. Mol Cell Endocrinol. 2009;297(1–2):10–17. doi:10.1016/j.mce.2008.10.002
7. Gloyn AL, McCarthy MI. The genetics of type 2 diabetes. Best Pract Res Clin Endocrinol Metab. 2001;15(3):293–308. doi:10.1053/beem.2001.0147
8. Christiansen J, Kolte AM, Nielsen FC. IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J Mol Endocrinol. 2009;43(5):187–195. doi:10.1677/JME-09-0016
9. Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi:10.1126/science.1142364
10. Nielsen J, Christiansen J, Lykke-andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19(2):1262–1270. doi:10.1128/MCB.19.2.1262
11. Ullrich S, Su J, Ranta F, et al. Effects of I Ks channel inhibitors in insulin-secreting INS-1 cells. Pflugers Arch. 2005;451(3):428–436. doi:10.1007/s00424-005-1479-2
12. Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. K V LQT1 and IsK (minK) proteins associate to form the I Ks cardiac potassium current. Nature. 1996;384(6604):78. doi:10.1038/384078a0
13. Neyroud N, Tesson F, Denjoy I, et al. A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nat Genet. 1997;15(2):186. doi:10.1038/ng0297-186
14. Larsen LA, Fosdal I, Andersen PS, et al. Recessive Romano-Ward syndrome associated with compound heterozygosity for two mutations in the KVLQT1 gene. Eur J Hum Genet. 1999;7(6):724. doi:10.1038/sj.ejhg.5200323
15. Zhou Y, Park S-Y, Su J, et al. TCF7L2 is a master regulator of insulin production and processing. Hum Mol Genet. 2014;23(24):6419–6431. doi:10.1093/hmg/ddu359
16. Elbein S, Chu W, Das S, et al. Transcription factor 7-like 2 polymorphisms and type 2 diabetes, glucose homeostasis traits and gene expression in US participants of European and African descent. Diabetologia. 2007;50(8):1621–1630. doi:10.1007/s00125-007-0717-x
17. McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med. 2010;363(24):2339–2350. doi:10.1056/NEJMra0906948
18. Lindgren CM, Heid IM, Randall JC, et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(6):e1000508. doi:10.1371/journal.pgen.1000508
19. Hotta K, Kitamoto T, Kitamoto A, et al. Association of variations in the FTO, SCG3 and MTMR9 genes with metabolic syndrome in a Japanese population. J Hum Genet. 2011;56(9):647. doi:10.1038/jhg.2011.74
20. Chauhan G, Tabassum R, Mahajan A, et al. Common variants of FTO and the risk of obesity and type 2 diabetes in Indians. J Hum Genet. 2011;56(10):720. doi:10.1038/jhg.2011.87
21. Peng S, Zhu Y, Xu F, Ren X, Li X, Lai M. FTO gene polymorphisms and obesity risk: a meta-analysis. BMC Med. 2011;9(1):71. doi:10.1186/1741-7015-9-71
22. Chang Y-C, Liu P-H, Yu Y-H, et al. Validation of type 2 diabetes risk variants identified by genome-wide association studies in Han Chinese population: a replication study and meta-analysis. PLoS One. 2014;9(4):e95045. doi:10.1371/journal.pone.0095045
23. Xiao S, Zeng X, Quan L, Zhu J. Correlation between polymorphism of FTO gene and type 2 diabetes mellitus in Uygur people from northwest China. Int J Clin Exp Med. 2015;8(6):9744.
24. Loos RJ, Yeo GS. The bigger picture of FTO—the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014;10(1):51. doi:10.1038/nrendo.2013.227
25. Chambers JC, Elliott P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40(6):716. doi:10.1038/ng.156
26. Meyre D, Delplanque J, Chèvre J-C, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41(2):157. doi:10.1038/ng.301
27. Lyssenko V, Nagorny CL, Erdos MR, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41(1):82. doi:10.1038/ng.288
28. Bouatia-naji N, Bonnefond A, Cavalcanti-proença C, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41(1):89. doi:10.1038/ng.277
29. Patel R, Rathwa N, Palit SP, Ramachandran A, Begum R. Association of melatonin & MTNR1B variants with type 2 diabetes in Gujarat population. Biomed Pharmacother. 2018;103:429–434. doi:10.1016/j.biopha.2018.04.058
30. Karamitri A, Jockers R. Melatonin in type 2 diabetes mellitus and obesity. Nat Rev Endocrinol. 2019;15(2):105–125. doi:10.1038/s41574-018-0130-1
31. Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881. doi:10.1038/nature05616
32. Cauchi S, Del Guerra S, Choquet H, et al. Meta-analysis and functional effects of the SLC30A8 rs13266634 polymorphism on isolated human pancreatic islets. Mol Genet Metab. 2010;100(1):77–82. doi:10.1016/j.ymgme.2010.01.001
33. El Dib R, Gameiro OL, Ogata MS, et al. Zinc supplementation for the prevention of type 2 diabetes mellitus in adults with insulin resistance. Cochrane Database Syst Rev. 2015;5:CD005525.
34. Shan Z, Bao W, Zhang Y, et al. Interactions between zinc transporter-8 gene (SLC30A8) and plasma zinc concentrations for impaired glucose regulation and type 2 diabetes. Diabetes. 2014;63(5):1796–1803. doi:10.2337/db13-0606
35. Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39(6):770. doi:10.1038/ng2043
36. Pierrel F, Douki T, Fontecave M, Atta M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J Biol Chem. 2004;279(46):47555–47563. doi:10.1074/jbc.M408562200
37. Wei F-Y, Suzuki T, Watanabe S, et al. Deficit of tRNA Lys modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest. 2011;121:9. doi:10.1172/JCI58056
38. Kang ES, Kim MS, Kim YS, et al. A polymorphism in the zinc transporter gene SLC30A8 confers resistance against posttransplantation diabetes mellitus in renal allograft recipients. Diabetes. 2008;57(4):1043–1047. doi:10.2337/db07-0761
39. Rutter GA. Think zinc: new roles for zinc in the control of insulin secretion. Islets. 2010;2(1):49–50. doi:10.4161/isl.2.1.10259
40. Xiang J, Li X-Y, Xu M, et al. Zinc transporter-8 gene (SLC30A8) is associated with type 2 diabetes in Chinese. J Clin Endocrinol Metab. 2008;93(10):4107–4112. doi:10.1210/jc.2008-0161
41. Chimienti F, Favier A, Seve M. ZnT-8, a pancreatic beta-cell-specific zinc transporter. Biometals. 2005;18(4):313–317. doi:10.1007/s10534-005-3687-9
42. Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. science. 2007;316(5829):1341–1345. doi:10.1126/science.1142382
43. Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi:10.1126/science.1142358
44. Sparsø T, Grarup N, Andreasen C, et al. Combined analysis of 19 common validated type 2 diabetes susceptibility gene variants shows moderate discriminative value and no evidence of gene–gene interaction. Diabetologia. 2009;52(7):1308–1314. doi:10.1007/s00125-009-1362-3
45. Cauchi S, El Achhab Y, Choquet H, et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med. 2007;85(7):777–782. doi:10.1007/s00109-007-0203-4
46. Xu K, Zha M, Wu X, et al. Association between rs13266634 C/T polymorphisms of solute carrier family 30 member 8 (SLC30A8) and type 2 diabetes, impaired glucose tolerance, type 1 diabetes—a meta-analysis. Diabetes Res Clin Pract. 2011;91(2):195–202. doi:10.1016/j.diabres.2010.11.012
47. Jing Y, Sun Q, Bi Y, Shen S, Zhu D. SLC30A8 polymorphism and type 2 diabetes risk: evidence from 27 study groups. Nutr Metab Cardiovasc Dis. 2011;21(6):398–405. doi:10.1016/j.numecd.2009.11.004
48. Chauhan G, Spurgeon CJ, Tabassum R, et al. Impact of common variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 on the risk of type 2 diabetes in 5164 Indians. Diabetes. 2010;59(8):2068–2074. doi:10.2337/db09-1386
49. Han X, Luo Y, Ren Q, et al. Implication of genetic variants near SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, FTO, TCF2, KCNQ1, and WFS1 in type 2 diabetes in a Chinese population. BMC Med Genet. 2010;11(1):81. doi:10.1186/1471-2350-11-81
50. Alami FM, Ahmadi M, Bazrafshan H, et al. Association of the TCF7L2 rs12255372 (G/T) variant with type 2 diabetes mellitus in an Iranian population. Genet Mol Biol. 2012;35(2):413–417. doi:10.1590/S1415-47572012005000029
51. Shokouhi S, Delpisheh A, Haghani K, Mahdizadeh M, Bakhtiyari S. Association of rs7903146, rs12255372, and rs290487 polymorphisms in TCF7L2 gene with type 2 diabetes in an Iranian Kurdish ethnic group. Clin Lab. 2014;60(8):1269–1276. doi:10.7754/Clin.Lab.2013.130809
52. Saadi H, Nagelkerke N, Carruthers SG, et al. Association of TCF7L2 polymorphism with diabetes mellitus, metabolic syndrome, and markers of beta cell function and insulin resistance in a population-based sample of Emirati subjects. Diabetes Res Clin Pract. 2008;80(3):392–398. doi:10.1016/j.diabres.2008.01.008
53. Alsmadi O, Al-rubeaan K, Mohamed G, et al. Weak or no association of TCF7L2 variants with Type 2 diabetes risk in an Arab population. BMC Med Genet. 2008;9(1):72. doi:10.1186/1471-2350-9-72
54. Chang Y-C, Chang T-J, Jiang Y-D, et al. Association study of the genetic polymorphisms of the transcription factor 7-like 2 (TCF7L2) gene and type 2 diabetes in the Chinese population. Diabetes. 2007;56(10):2631–2637. doi:10.2337/db07-0421
55. Yen C-J, Beamer BA, Negri C, et al. Molecular scanning of the human peroxisome proliferator activated receptor γ (hPPARγ) gene in diabetic Caucasians: identification of a Pro12Ala PPARγ2 missense mutation. Biochem Biophys Res Commun. 1997;241(2):270–274. doi:10.1006/bbrc.1997.7798
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.