Back to Journals » Cancer Management and Research » Volume 9
SIRT1 gene polymorphisms and risk of lung cancer
Received 26 May 2017
Accepted for publication 25 July 2017
Published 4 September 2017 Volume 2017:9 Pages 381—386
DOI https://doi.org/10.2147/CMAR.S142677
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Alexandra R. Fernandes
Yanbo Lv, Shuangyan Lin, Fang Peng
Department of Pathology, Zhejiang Hospital, Hangzhou City, Zhejiang, China
Objective: Lung cancer, which is the leading cause of cancer death worldwide, is influenced by a wide variety of environmental and genetic risk factors. The silent information regulator 1 (SIRT1) gene is located on the long arm of chromosome 10 (10q21.3) and has been shown to play crucial roles in lung cancer development in previous studies. In this study, we determined whether variation in the SIRT1 gene is associated with lung cancer in Chinese population.
Methods: The case–control study comprised 246 controls and 257 non-small cell lung cancer patients, comprising 79 squamous cell carcinoma patients and 124 adenocarcinoma patients. All subjects were from Zhejiang, China. Four single-nucleotide polymorphisms of SIRT1 gene were analyzed: rs12778366 (C/T, lies in the 5′ upstream), rs3758391 (C/T, lies in the 5′ upstream), rs2273773 (C/T, lies in the coding) and rs4746720 (C/T, lies in the 3′ untranslated region).
Results: No significant difference of allele and genotype frequencies was observed between the different groups. Haplotype association analysis carried out on the four single-nucleotide polymorphisms within the case–control cohort also did not reveal a significant association with lung cancer (P>0.05).
Conclusion: The results suggest the tested SIRT1 gene polymorphisms may not contribute to lung cancer. Further studies are warranted to demonstrate the functional roles of the SIRT1 polymorphism in lung cancer.
Keywords: SIRT1, SNP, non-small cell lung cancer, adenocarcinoma, squamous cell carcinoma
Introduction
Lung cancer is the leading cause of cancer death worldwide, and non-small cell lung cancer (NSCLC) that consists mainly of squamous cell carcinoma (SCC) and adenocarcinoma (AD) accounts for nearly 80% of all lung cancer cases.1 Thus, understanding of the mechanisms of lung carcinogenesis in the NSCLC subtypes is urgently needed.
The silent information regulator 2 (Sir2) family is a highly conserved group of genes from bacteria to humans and encodes a group of nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylases.2 In mammalian cells, there are seven homologues (Sirts 1–7) of yeast Sir2, termed sirtuins, which share the catalytic domain with Sir2.3 Sirt1, which is located in the long arm of chromosome 10 (10q21.3) and consists of nine exons and eight introns, is the closest homologue of yeast Sir2.4–6 Many studies have shown that SIRT1 regulates a wide variety of cellular functions, such as mitochondrial biogenesis, autophagy, circadian rhythms, stress resistance, apoptosis, and glucose and lipid metabolism.7–9 SIRT1 gene has been extensively studied, and was shown to be implicated in cancer, inflammation, obesity, diabetes, and cardiovascular and neurodegenerative diseases.10–12 SIRT1 deacetylates various transcription factors that are involved in stress response, cell cycle and apoptosis, such as p53, Ku70, nuclear factor kB, FOXO and HIC1.13–18 Animal studies demonstrated that SIRT1 was essential to the embryonic development.15,19 On the other hand, accumulating evidence showed that SIRT1 was overexpressed in various types of malignant cells, and its inhibitors suppressed the growth of tumor cells.
To date, polymorphisms in SIRT1 have been associated with several disease-related phenotypes, including diabetes, body mass index, obesity, cholesterol metabolism, energy expenditure, glucose tolerance and cardiovascular disease.20 However, very little is known about the association of polymorphism of SIRT1 and lung cancer development in the Chinese Han population. Based on the previous reports, we hypothesized that polymorphisms in SIRT1 might also be associated with susceptibility to lung cancer. Therefore, four functional single-nucleotide polymorphisms (SNPs) were selected for further investigation of their association with lung cancer.
Patients and methods
Subject selection
We conducted a retrospective study using in-patient medical records of patients with NSCLC who were treated at Zhejiang Hospital in China. The case–control study consisted of 257 patients with NSCLC and 246 controls. The records of 257 patients with NSCLC admitted to Zhejiang Hospital between January 2010 and June 2015 were reviewed based on the following inclusion criteria: 1) patient was pathologically diagnosed with NSCLC; 2) patient displayed normal cardiac functions and 3) patient displayed normal serum glucose level. Exclusion criteria were the following: 1) patient with small cell lung cancer and 2) patient with malignant tumors except lung cancer.
All participants were provided written informed consent. Our study protocol was approved by the Ethics Committee of Zhejiang Hospital (No. 2013-k-2), and all experiments were conducted in accordance with the Declaration of Helsinki.
SNP selection and genotyping
The candidate SNPs were selected based on the relevant studies of the SNPs associated with human diseases, combined the result analyses by Haploview 4.2 software r2≥0.8 and the allele frequency in public database of the National Center for Biotechnology Information (a minor allele frequency ≥0.05) (http://www.ncbi.nlm.nih.gov/snp/) in SIRT1 gene region (Gene ID: 23411). We downloaded the China Han population’s SNP data of SIRT1 gene from HapMap database (HapMap Genome Browser [phases 1, 2 and 3 merged genotypes and frequencies]). We then analyzed these data by Haploview 4.2 software. We found four TagSNPs with r2≥0.8 and a minor allele frequency ≥0.05: rs12778366 (C/T, lies in the 5′ upstream), rs3758391 (C/T, lies in the 5′ upstream), rs2273773 (C/T, lies in the coding) and rs4746720 (C/T, lies in the 3′ untranslated region [UTR]). Because that the four SNPs (tagSNPs) could capture all the SNPs with minor allele frequency ≥0.05 across the SIRT1 gene region. We then conducted a further analysis about the four TagSNPs for their predicted functions by the online software FASTSNP (a web server that allows users to efficiently identify and prioritize high-risk SNPs according to their phenotypic risks and putative functional effects; FASTSNP is available at http://fastsnp.ibms.sinica.edu.tw.; Table 1).
Genotyping was carried out by ligase detection reaction (LDR). The target DNA sequences were amplified using a multiplex polymerase chain reaction method (the primer and probe sequence are shown in Table 1). LDR (2 min at 95°C, and 15 s at 94°C, 25s at 50°C, for 40 cycles) was proceeded in a final volume of 10 μL, which contained 1 μL 1× New England Biolabs (NEB) buffer for Taq DNA Ligase (New England Biolabs, Beverly, MA, USA), 1 mL of 2 pmol/μL of each common probe, 4 μL of Multi-polymerase chain reaction product and 0.5 μL of 2 U/μL Taq DNA Ligase. The fluorescent products of LDR were differentiated by ABI 377 sequencer (Perkin-Elmer, Foster City, CA, USA). The result was analyzed by GeneMapper Analysis Software 3.7 (Applied Biosystems, Foster City, CA, USA). The primer sequences are shown in the Table 1.
Statistical analysis
The c2 test was used to compare the distribution of the genotypes between lung cancer patients and controls using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). Haploview 4.2 was used to calculate and display linkage disequilibrium (LD) values between the four SNPs. Hardy–Weinberg equilibrium test, genotype frequencies and LD were calculated using the SNPstats software (a web tool for the analysis of association studies: http://bioinfo.icon cologia.net/SNPstats).21 The odds ratio and 95% CI were calculated to evaluate the risk between genotype frequencies and lung cancer. LD between SNPs was measured by calculating r2 values. All tests were two-sided, and a P value <0.05 was considered statistically significant.
Results
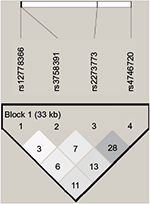
Of the four SNPs genotyped within the lung cancer cases and controls, all achieved a success rate of ≥85%, with an average success rate of 98.2% by LDR detection. A total of 257 NSCLC patients (200 men and 57 women) comprising 79 SCC patients (71 men and 8 women), 124 AD patients (77 men and 47 women) and 246 healthy individuals as controls (186 men and 60 women) were included in the study. The genotype frequencies of SNPs in all the groups did not deviate significantly from the expected values under the Hardy–Weinberg equilibrium (P>0.05). LD of SIRT1 polymorphism analysis using r2 within the case and control samples did not reveal strong LD (Figure 1).
The genotype and allele frequencies for SIRT1 polymorphisms in NSCLC, SCC, AD and control groups are shown in Table 2. We found no significant differences between the NSCLC cases, SCC cases, AD cases and controls for the four SIRT1 polymorphisms when the general distribution of genotypes was compared. Haplotype (TTTC, TTCT, TCTT, CTTT) association analysis carried out on the four SNPs within the case–control cohort also did not reveal a significant association to lung cancer (P>0.05, data not shown).
Discussion
The SIRT1 is located on the long arm of chromosome 10 (10q21.3). It has been shown to play crucial roles in mammalian health and disease and is often associated with the most complex physiologic processes such as metabolism, cancer and aging due to its NAD+-dependent deacetylase activity.22 The mammalian SIRT1 regulates several transcription factors that control the expression of genes involved in the metabolism and endocrine signaling, such as PPARg, PGC1-a, UCP2, NFkB and FOXO1 proteins, and plays important roles in glucose homeostasis and insulin secretion.23–25 In the human SIRT1 gene, multiple variants have been described, several of which are associated with carotid atherosclerosis, body mass index, aging, type 2 diabetes and risk of obesity.26–31 However, very few SIRT1 variants have been reported in lung cancer. Therefore, the main objective of this study was to identify the SNPs in SIRT1 and to estimate the extent of associations between these SNPs and NSCLC.
Overexpression of SIRT1 or treating with SIRT1 activators improves glucose homeostasis and insulin sensitivity in mice.22,32,33 Li et al34 observed a significant association between the C.−274C>G polymorphism and growth traits in Nanyang cattle. They found that the G allele-containing construct displayed a strikingly lower promoter activity compared with the C allele by the luciferase assay system. In addition, they also found g.17379 A>G polymorphism was significantly related to body weight of 6 and 12 months old in NY population.35 Kilic et al36 reported that the oldest people carrying AG genotypes for rs7895833 have the highest SIRT1 level, suggesting an association between rs7895833 SNP and lifespan longevity. The 2827 A>G polymorphism is positioned on exon 2, leading to a histidine-to-arginine conversion. Although Izmirli et al6 found no statistically significant relation between the 2827 A>G at the SIRT1 and cardiovascular disease, Peeters et al37 reported that SIRT1 rs7895833 and rs7069102 polymorphisms were associated with cardiovascular disease. Shimoyama et al38 reported that the different allele frequencies in rs7895833 and rs7069102 between hemodialysis patients and controls could have an impact on the survival of Japanese hemodialysis patients.
Very little is known about the effect of polymorphism of SIRT1 gene on lung cancer until now, despite the fact that many studies had been carried out on the association between the SNPs of SIRT1 and human diseases. In this study, we assessed whether genetic variations in SIRT1 are associated with NSCLC, which consists mainly of SCC and AD and accounts for nearly 80% of all lung cancer cases. According to our previous report,39 the expression of SIRT1 was significantly associated with unfavorable SCC characteristics, but not with AD characteristics; furthermore, we compared the SNP frequencies in SCC patients vs controls and AD patients vs controls. We investigated four TagSNPs: rs12778366 (C/T, lies in the 5′ upstream), rs3758391 (C/T, lies in the 5′ upstream), rs2273773 (C/T, lies in the coding) and rs4746720 (C/T, lies in the 3′ UTR), which could capture all the SNPs with minor allele frequency ≥0.05 across the SIRT1 gene region in 257 NSCLC patients (200 men and 57 women), 79 SCC patients (71 men and 8 women), 124 AD patients (77 men and 47 women) and 246 healthy controls (186 men and 60 women). By LDR detection, we achieved a success rate of 98.2% for the four TagSNPs. However, no association was detected between the SNP rs3758391 and NSCLC in Chinese population, which was similar to the previous study that reported no significant relationship between the SNP rs3758391 and extreme aging in Germans.40,41 Although in another study, rs3758391/C was found more common than rs3758391/T (P=0.026) and rs3758391/CC was more common than rs3758391/CT and rs3758391/TT (P=0.027) in the aging subjects.42 The difference might be due to the different genetic background in different populations. rs4746720, which is located in the 3′ UTR, was also a TagSNP of SIRT1 gene in Chinese Han population. Besides, Zhang et al42 found that rs4746720/C was more common than rs4746720/T (P=0.022) and rs4746720/CC was more common than rs4746720/CT and rs4746720/TT (P=0.049) in the aging subjects. In contrast, our study did not find rs4746720 as a significant marker for lung cancer in the case–control analysis. Figarska et al30 showed an association between the SNP rs12778366 in SIRT1 gene and long-term survival in 1390 subjects of the Vlagtwedde/Vlaardingen cohort. Rai et al43 sequenced the 1.46 kb region upstream of the translation start site of the SIRT1 gene in 1542 samples (692 type 2 diabetes patients and 850 controls) in the North Indian population and observed that rs12778366 (lies in the promoter region of SIRT1) was associated with SIRT1 expression. However, in our case–control study, we did not find any relationship of the rs12778366 polymorphism in the NSCLC and control groups, SCC and control groups and AD and control groups. In addition, Flachsbart et al41 studied the relationship between five SNPs (rs3758391, rs1885472, rs2273773, rs10997870 and rs2234975) in SIRT1 gene in 1026 unrelated German long-lived individuals (mean age: 98.3 years) and found no association between SIRT1 gene polymorphisms and exceptional human longevity. In our study, we also did not find any effect of the rs2273773 polymorphism in the NSCLC and control groups, SCC and control groups, AD and control groups.
The haplotypes (TTTC, TTCT, TCTT, CTTT) observed here are likely to represent some of the variation in the SIRT1 genomic region in the population analyzed. In this study, we found no relationship of the haplotypes of the SIRT1 in the NSCLC and control groups, SCC and control groups, AD and control groups.
To the best of our knowledge, this is the first report about the polymorphism of SIRT1 and lung cancer. In our study, we found no significant association between SIRT1 gene polymorphisms and human NSCLC, since we did not find any significant polymorphism of the SIRT1 gene in the NSCLC and control groups, SCC and control groups, AD and control groups. The findings presented here may be due to the small number of the cases and controls, which is the main limitation of our study, due to the complex multifactorial genetic contribution to lung cancer. Expanding the sample size might be able to find a more meaningful result. Therefore, further studies with more sample size and subtle design are still necessary to delineate the functional roles of SIRT1 polymorphism on lung cancer.
Acknowledgment
This work is supported by the Zhejiang Provincial Health Bureau Foundation (No. 2011KYA024 and No. 2013KYA004) and innovation disciplines of Zhejiang province to Xu-Jiao Chen.
Disclosure
The authors report no conflicts of interest in this work.
References
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.