Back to Journals » Journal of Inflammation Research » Volume 15
Single-Cell RNA Sequencing Reveals Transcriptional Changes in the Cartilage of Subchondral Insufficiency Fracture of the Knee
Authors Tang W, Li ZW, Miao GQ, Li ZP, Gui T, Wu CJ, Li ZY, Yang J , Zhao XD, Liu N, Zha ZG, Yao LT, Zhang HT
Received 9 August 2022
Accepted for publication 1 November 2022
Published 5 November 2022 Volume 2022:15 Pages 6105—6112
DOI https://doi.org/10.2147/JIR.S385648
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Wang Tang1 *, Zhen-Wei Li1 *, Gui-Qiang Miao2 *, Zhi-Peng Li,1 Tao Gui,1 Chong-Jie Wu,1 Zhen-Yan Li,1 Jie Yang,1 Xiao-Dong Zhao,2 Ning Liu,1 Zhen-Gang Zha,1 Lu-Tian Yao,3 Huan-Tian Zhang1
1Department of Bone and Joint Surgery, The First Affiliated Hospital of Jinan University, Guangzhou, 510630, People’s Republic of China; 2Department of Orthopedics, Foshan Fosun Chancheng Hospital, Foshan, 528010, People’s Republic of China; 3Department of Orthopedics, The First Hospital of China Medical University, Shenyang, 110001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lu-Tian Yao; Huan-Tian Zhang, Email [email protected]; [email protected]
Purpose: Subchondral insufficiency fracture of the knee (SIFK) is a common cause of knee joint pain that mainly afflicts the elderly. Until now, how a sudden insufficiency fracture of subchondral bone affects the transcriptomic profiles of cartilage in SIFK and OA patients are largely unknown.
Methods: Single-cell RNA sequencing (scRNA-seq) was used to identify various cell subsets and evaluate transcriptomic differences in cartilage of SIFK and OA patients. In addition, the above findings were confirmed by histological evaluation and immunohistochemical (IHC) staining.
Results: We found that the transcriptomic profiles of cartilage in the SIFK patient was completely different from those of normal and OA patients. Accordingly, several novel cell clusters with activation of hypoxia and endochondral ossification signaling were identified in the SIFK cartilage. Chondrocyte trajectories analysis and IHC staining revealed that transcription factors including TCF4 were found to be highly up-regulated during the occurrence of SIFK, which might drive the reactive formation of cartilage and fibrous tissue and the activation of endochondral ossification.
Conclusion: This is the first report to elucidate the transcriptomic alterations and distinct cell type subpopulations in the cartilage of SIFK and OA by the use of scRNA-seq, which provides a new insight in the understanding of the initiation and progression of SIFK.
Keywords: SIFK, scRNA, TCF4, cartilage, hypoxia
Introduction
Spontaneous osteonecrosis of the knee (SONK) is a common disease that causes a sudden onset of knee pain. It was first described in the literature by Ahlback in 1968.1 Later, SONK has been associated with SIFK once the subchondral fragment undergoes true necrosis; therefore, some authors believe the term “SONK” is a misnomer. Until now, the etiology of SIFK has been largely unknown. Low bone mineral density, varus deformity at the proximal tibia, and meniscal dysfunction were reported to be highly associated with the occurrence of SIFK.2 For instance, medial meniscus posterior root tears (MMPRTs) and/or medial meniscus extrusion have been found to be highly correlated with SIFK.3 Recent studies have postulated that subchondral insufficiency fractures resulting from mechanical overloading concentrated on the medial part of the knee could cause SIFK.2 This is supported by the histological finding that a fracture line occurs within the subchondral bone.4 Therefore, a more appropriate term for the condition may be subchondral insufficiency fracture of the knee (SIFK), rather than SONK.
Abnormalities of subchondral bone have been demonstrated to play a fundamental role in mediating cartilage degeneration in osteoarthritis (OA), which can be secondary to SIFK.5 In addition, changes in the medial meniscus (tear and/or extrusion) and/or subchondral bone insufficiency fracture might result in massive stress loading on the cartilage. Therefore, a previous study emphasized the need for assessment of the cartilaginous coating by means of arthroscopy before instituting treatment for SIFK.6 In fact, several studies have reported that the proliferative and redifferentiation abilities of chondrocytes from patients with SIFK are largely compromised compared with those of normal cells.7,8 In agreement with this, levels of chondroitin 6-sulfate (C6S), which originates mainly from the articular cartilage, are found to be markedly elevated in the synovial fluid of SIFK patients compared to patients with medial OA, indicating cartilage breakdown.9 However, how a sudden insufficiency fracture of subchondral bone may contribute to cartilage metabolism alteration and/or ultimately lead to secondary OA is largely unclear. In this work, we were able to identify a subset of cell populations that were highly enriched with hypoxia and endochondral ossification molecules in SIFK cartilage compared with normal and OA cartilage. Furthermore, chondrocyte trajectory analysis and immunohistochemistry (IHC) staining demonstrated that transcription factors, including TCF4, were elevated in SIFK.
Materials and Methods
Clinical Specimen Collection
The diagnosis of SIFK was confirmed on MRI images when the T1-weighted image showed a low signal in the weight-bearing area and the T2-weighted image showed a high signal area surrounding the same site. The cartilage specimens were harvested from a 62-year-old female who underwent unicompartmental knee arthroplasty (UKA) surgery due to SIFK (Koshino classification: stage 2) and they were immediately subjected to chondrocyte isolation for scRNA-seq.
Chondrocyte Isolation and scRNA-Seq
The whole layers of SIFK cartilage specimens were subjected to collagenase type I (LS004196, 2mg/mL, 4h, 37°C) digestion to isolate the chondrocytes. Then, approximately 20,000 cells were used to acquire a single library by a Chromium controller (V3 chemistry version, 10X Genomics Inc, San Francisco, USA); the cells were barcoded and purified as we previously described,10 and then sequenced using a 2×150 paired-end configurations on the Illumina HiSeq platform at a sequencing depth of ~ 400 million reads. Cell Ranger (Version 6.0.2) was used to demultiplex the reads, which was followed by extraction of the cell barcodes and unique molecular identifiers (UMIs). The cDNA insert sequence was aligned to the human reference genome (GRCh38-2020).
Raw scRNA-seq files of normal and OA cartilage tissues were acquired from GEO: GSE169454.11 Seurat package V312 was used for individual or integrated analysis of the datasets. Standard Seurat pipeline was used for filtering, variable gene selection, dimensionality reduction analysis and clustering. Doublets or cells with poor quality (genes >6000, genes <500, or >25% genes mapping to mitochondrial genome) were excluded. Expression was natural log transformed and normalized for scaling the sequencing depth to a total of 1×104 molecules per cell. For the integrated dataset, batch integration was performed using Harmony (version 1.0).13 Briefly, the Seurat objects were first randomly downsampled to get comparable cell number between different group, then the objects were passed on to harmony integration by “RunHarmony” function, followed by downstream dimensional reduction by using the corrected Harmony embeddings. Only chondrocyte subpopulation remained for further analysis. Label transfer was done with Seurat “TransferData” function to classify the query SIFK chondrocyte cells based on reference OA chondrocyte (GSE104782) data,14 since the chondrocyte subtype of GSE169454 was also based on the clustering information of GSE104782. Gene set variation analysis (GSVA) was performed using the GSVA package for analyzing Hallmark and KEGG pathway signaling.15 Gene ontology (GO) analysis was performed using the clusterProfiler package.16 The cell metabolism state was inferred with the scMetabolism package,17 and a list of metabolic gene sets was acquired from the same package. Differentially expressed genes between normal and radiated groups were identified using FindMarkers within Seurat and p values between them were calculated by “test.use=bimod”.
Trajectory analysis was performed using Slingshot18 to computationally delineate the differential progression from normal cartilage to SIFK or OA cartilage. Briefly, Seurat objects were transformed into SingleCellExperiment objects. Slingshot trajectory analysis was conducted using Seurat clustering information and dimensionality reduction produced by UMAP. Cluster C0 was selected as start cluster, since C0 was mostly composed with normal chondrocytes, with specific parameters setting of “extend=‘n’, dist.method =“slingshot”. TradeSeq was used to discover transcription factors that were differentially expressed along trajectory lineages.19
Histology Analysis
Fresh human cartilage specimens from patients who underwent total knee arthroplasty were fixed in 4% paraformaldehyde overnight and decalcified in ethylenediaminetetraacetic acid (EDTA, G1105, Servicebio) for 3 weeks before embedding in paraffin. Then, 6 μm-thick sagittal paraffin sections were cut through the SIFK lesion, and the sections were stained with hematoxylin and eosin (H&E, Leagene), Safranin O/Fast Green (S2255-100 g, Sigma Aldrich) and Masson solutions (Servicebio) according to the manufacturer’s instructions. IHC staining was carried out using rabbit anti-TCF4 (1:50, C48H11, CST) as we described previously.20
Statistical Analysis
The analysis was carried out in the R environment for statistical computing and visualization. For differential proportion, differential expression, and enrichment analyses, P values were adjusted for multiple hypothesis testing using the Benjamini–Hochberg method.
Results
Identification of Novel Chondrocyte Subpopulation in the SIFK Cartilage
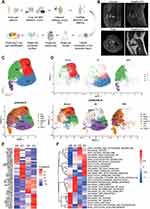
Medial meniscus extrusion and insufficiency fracture of subchondral bone are believed to be the prime factors that contribute to abnormal stress loading on the medial cartilage and the development of SIFK.8,21,22 Ultimately, this can lead to subchondral collapse and secondary OA. Until now, it has been unclear whether the transcriptional profiling in the cartilage of SIFK differ from those of OA. Herein, we sought to uncover the transcriptomic characteristics of cartilage obtained from SIFK using scRNA-seq, as illustrated in Figure 1A. The radiological changes and diagnosis were examined by weight-bearing X-ray and MRI (Figure 1B and Supplementary Figure S1). Of note, a typical subchondral bone lesion and medial meniscus extrusion were revealed by T2/T1-weighted MRI (Figure 1B, Koshino stage 2). Although conservative treatment has been suggested in the early stage of SIFK, in this case, UKA was chosen to rapidly alleviate the patient’s knee pain (Supplementary Figure S1). During the operation, cartilage specimens that covered the lesioned subchondral bone were dissected and subjected to chondrocyte isolation for scRNA-seq. The SIFK cartilage dataset was analyzed with a combination of normal and OA cartilage datasets deposited in GSE169454. After removing the batch effect by the Harmony package, the chondrocytes converged together but still exhibited a differential clustering pattern (Figure 1C and D, Supplementary Figures S2 and S3, Table S1). To further elaborate the cell subtype within the chondrocytes, we transferred the labeling of different stage OA chondrocytes identified by Ji et al14 onto the present integrated datasets. Several subpopulations, including effector chondrocytes (ECs), regulatory chondrocytes (RegCs), proliferative chondrocytes (ProCs) and hypertrophic chondrocytes (HTCs) were found in normal cartilage defined as C0, while ECs were hardly seen in both OA and SIFK group (Figure 1C and D). The C1 cluster was substantially composed of the OA cartilage with ProCs and prehypertrophic chondrocytes (preHTCs) as predicted cell subtype. Of significance, the overwhelming majority of C2 and C3 clusters were composed of the SIFK cartilage, C2 mainly contains the predicted homeostatic chondrocytes (HomCs), preHTCs and HTCs. C3 were predicted to be the fibrocartilage chondrocytes (FCs) and HTCs (Figure 1C and D). The differentially expressed genes (DEGs) in C0-3 clusters were further plotted and presented using a heatmap (Figure 1E). Hallmark, KEGG signaling pathway and GO term analyses of each cluster found that the activation of inflammatory signaling, including TNF-α signaling via NF-κB, IL-6/STAT3 signaling, and cytokine‒cytokine receptor interactions, was found in the OA cartilage, indicating a tendency of breaking down the extracellular matrix and collagens via chronic inflammation (Figure 1F and Supplementary Figure S4). However, inflammatory signaling was reduced in the SIFK cartilage; instead, robust activation of hallmarks of hypoxia and Wnt/β-catenin signaling as well as TGF-β signaling was found in clusters 2 (C2) and 3 (C3), respectively (Figure 1F). Furthermore, we used scMetabolism to investigate cartilage metabolism under different conditions. Unsupervised clustering revealed that normal, OA and SIFK cartilage cells had differential expression patterns of metabolism-associated genes and metabolic pathway scores (Supplementary Figure S5). C2 and C3 SIFK cartilage showed potential upregulation of glycolysis/gluconeogenesis and butanoate metabolism, which are highly relevant to hypoxia, sulfur and glycosaminoglycan metabolism, which are associated with chondrocyte differentiation or endochondral ossification activation. Collectively, these data suggest that a novel subpopulation of cells characterized by hypoxia and ossification activation was found in SIFK.
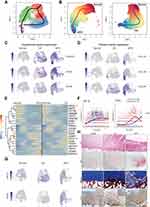
Trajectile of Cartilage in SIFK Differs from That of OA
It is generally accepted that SIFK might lead to the late stage of OA. Therefore, it is presumed that SIFK cartilage may undergo a degenerative process similar to that of OA cartilage. Given the abovementioned novel subpopulation, we then performed trajectory analysis of these cells. Surprisingly, we observed that the trajectory of cartilage from normal conditions to OA was completely different from that of normal conditions to SIFK (Figure 2A and B). In addition, increased expression of biomarkers (COL2A1, ACAN, and SOX9) in hyaline cartilage was found in C2 and C3, along with elevated expression of fibrocartilage markers, including COL1A1, COL1A2, and COL3A1 (Figure 2C and D, Table S2). These findings are in accordance with previous histopathological observations that the formation of cartilage and fibrous tissue in the advanced stage of SIFK is indicative of delayed union or nonunion of the insufficiency fracture of the subchondral bone.4 Transcription factor analysis consistently revealed a shift to HIF1A-mediated hypoxia from normal to SIFK cartilage (Figure 2E–G). The Wnt/β-catenin pathway plays a key role in regulating chondrocyte hypertrophy, promoting bone formation and stimulating endochondral ossification, largely relying on the activity of LEF/TCF family members, including TCF4.20 Herein, we found that TCF4 was predominantly expressed in the SIFK cartilage (clusters C2 and C3) (Figure 2E–G, Table S2). Furthermore, cartilage was obtained from OA (uninvolved lateral compartment of the knee as a normal control) and SIFK patients to validate the above results (Figure 2H, Supplementary Figure S6). As indicated, HE and S/O staining clearly revealed the reactive formation of cartilage and fibrous tissue and the activation of endochondral ossification, which was consistent with a previous report.23 Accordingly, the expression of TCF4 was found to be increased in the ossification region by IHC staining (Figure 2H, Supplementary Figure S7). These results together suggested that transcription factors, including HIF1A and TCF4, might play a role in regulating hypoxia and endochondral ossification in SIFK.
Discussion
SIFK is characterized by acute onset of knee pain that mainly afflicts the elderly. It is often misdiagnosed in the clinic, especially in the early stage, since no change in appearance is observed on radiographs; however, lesions can become conspicuous as the disease progresses, manifesting as radiolucent areas with sclerotic halos and subtle flattening of the articular surface. In advanced cases, subchondral collapse and secondary OA can occur. Abnormalities of subchondral bone are strongly associated with cartilage degeneration in OA, which can be secondary to SIFK.5 Therefore, these progressive changes lead to a plausible assumption that cartilage in SIFK and OA undergo a similar degenerative process. In the present study, we reveal for the first time that cell type subpopulations and transcriptomic profiling of the articular cartilage in SIFK patients are completely different from that in normal and OA patients at the single-cell level.
Articular cartilage is a highly organized structure composed of multiple zones, namely, the superficial, middle, deep and calcified zones, which display three chondrocyte phenotypes: persistent, transient and hypertrophic cells. In recent studies, scRNA-seq has been used to identify several cell populations, including ECs, RegCs, HomC, ProCs, preHTCs, HTCs, FCs, senescent cells and cartilage progenitor cells, in OA.11,14,24 In the current study, we further projected these nomenclatures onto normal and SIFK chondrocytes and shown that ECs which were known for association with metabolism reduced greatly in OA and SIFK chondrocytes. Of note, we were able to identify the overwhelming majority of C2 and C3 clusters in the SIFK cartilage, C2 were shown to contain HomCs and HTCs, while C3 were composed of FCs and HTCs, which is also completely different from OA and normal cartilage regarding hypoxia and endochondral ossification signaling (Figure 1). Our findings are in accordance with several previous studies demonstrating that the proliferative and redifferentiation abilities of chondrocytes from SIFK are largely different from those of normal and OA patients.7–9
Substantial evidence has demonstrated the mutual regulation between cartilage and subchondral bone. For instance, during the epiphyseal extension process, chondrocyte-derived osteoprotegerin (OPG) facilitates bone formation by blocking osteoclast maturation through β-catenin signaling, and the senescence-associated secretory phenotype (SASP) of chondrocytes, which is characterized by the secretion of chemokines and proinflammatory cytokines, accelerates OA progression.25 On the other hand, abnormalities in subchondral bone have been demonstrated to play a fundamental role in mediating cartilage degeneration.26 In our study, the HE and S/O staining of SIFK cartilage revealed a reactive formation of cartilage and fibrous tissue and the activation of endochondral ossification (Figure 2), further validated the above scRNA-seq results (Figure 1). Mechanistically, chondrocyte trajectory analysis and IHC staining revealed that transcription factors, including TCF4, were highly upregulated during the occurrence of SIFK, which might drive the reactive formation of cartilage and fibrous tissue and the activation of endochondral ossification. This is supported by several studies demonstrating that TCF4 plays a key role in embryonic skeletal development and endochondral ossification.27 Additionally, we found that several ossification-related genes, such as SOX5, TRPS1 and ZFHX4, were up-regulated in SIFK cartilage. SOX5 is reported to involve in inducing the expression of Sox9 targets and regulating endochondral ossification.28,29 TRPS1 may coordinate the mineralization of the extracellular matrix by osteoblasts, thus regulating the development of bones formed by intramembranous or endochondral ossification.30 Zfhx4 coordinates the transcriptional network of Osterix and thereby coordinates endochondral ossification.31 Therefore, it is likely that these transcription factors collectively promote the endochondral ossification during SIFK, which together supporting the concept of SIFK, rather than SONK.
Conclusion
To the best of our knowledge, this is the first report to elucidate the transcriptional changes in SIFK cartilage by using scRNA-seq. However, our study has several limitations. First, a limited number of samples were analyzed by scRNA-seq since this is a pilot study, which may lead to statistical bias caused by individual characteristics. Second, age- and stage-matched cartilage is required to eliminate any possible confounding factor. Therefore, if conditions permit, the number of samples from SIFK patients can be increased to confirm the precise mechanisms of chondrocyte metabolism in the SIFK population.
Ethics Approval and Informed Consent
This study was approved (protocol number: KY-2021-065) by the Ethics Committee of the First Affiliated Hospital of Jinan University, in compliance with the Declaration of Helsinki Principle. All patients signed a consent form for the use of their anonymized clinical data and images.
Acknowledgments
The authors thank all whose comments improved the paper.
Funding
This work was supported by the National Key R&D Program of China (2021YFA1102600), the National Natural Science Foundation of China (82072470, 81871809, 81602360, 81672224, 82103781), the Natural Science Foundation of Guangdong Province (2021A1515012154, 2019A1515011082, 2017A030313665), the Guangzhou Science and Technology Project (202102010069, 201707010493), and the Macau Foundation for Development of Science and Technology (0029/2019/A).
Disclosure
The authors have no relevant financial or non-financial interests to disclose in this work.
References
1. Ahlback S, Bauer GC, Bohne WH. Spontaneous osteonecrosis of the knee. Arthritis Rheum. 1968;11:705–733. doi:10.1002/art.1780110602
2. Yamamoto T, Bullough PG. Spontaneous osteonecrosis of the knee: the result of subchondral insufficiency fracture. J Bone Joint Surg Am. 2000;82:858–866. doi:10.2106/00004623-200006000-00013
3. Fujisawa T, Choe H, Kusaba Y, et al. Medial meniscus extrusion and stage are related to the size of spontaneous osteonecrosis of the knee in patients who underwent high tibial osteotomy. Knee. 2022;36:72–79. doi:10.1016/j.knee.2022.04.011
4. Takeda M, Higuchi H, Kimura M, et al. Spontaneous osteonecrosis of the knee: histopathological differences between early and progressive cases. J Bone Joint Surg Br. 2008;90:324–329. doi:10.1302/0301-620X.90B3.18629
5. Husain R, Nesbitt J, Tank D, et al. Spontaneous osteonecrosis of the knee (SONK): the role of MR imaging in predicting clinical outcome. J Orthop. 2020;22:606–611. doi:10.1016/j.jor.2020.11.014
6. Koshino T, Okamoto R, Takamura K, Tsuchiya K. Arthroscopy in spontaneous osteonecrosis of the knee. Orthop Clin North Am. 1979;10:609–618. doi:10.1016/S0030-5898(20)30754-9
7. Sakata K, Furumatsu T, Miyazawa S, et al. Comparison between normal and loose fragment chondrocytes in proliferation and redifferentiation potential. Int Orthop. 2013;37:159–165. doi:10.1007/s00264-012-1728-x
8. Takata N, Furumatsu T, Abe N, Naruse K, Ozaki T. Comparison between loose fragment chondrocytes and condyle fibrochondrocytes in cellular proliferation and redifferentiation. J Orthop Sci. 2011;16:589–597. doi:10.1007/s00776-011-0128-1
9. Berger CE, Kröner A, Kristen KH, et al. Spontaneous osteonecrosis of the knee: biochemical markers of bone turnover and pathohistology. Osteoarthritis Cartilage. 2005;13:716–721. doi:10.1016/j.joca.2005.04.003
10. Mundy C, Yao L, Sinha S, et al. Activin A promotes the development of acquired heterotopic ossification and is an effective target for disease attenuation in mice. Sci Signal. 2021;14(669). doi:10.1126/scisignal.abd0536
11. Fu W, Hettinghouse A, Chen Y, et al. 14-3-3 epsilon is an intracellular component of TNFR2 receptor complex and its activation protects against osteoarthritis. Ann Rheum Dis. 2021;80:1615–1627. doi:10.1136/annrheumdis-2021-220000
12. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902 e1821. doi:10.1016/j.cell.2019.05.031
13. Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296. doi:10.1038/s41592-019-0619-0
14. Ji Q, Zheng Y, Zhang G, et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann Rheum Dis. 2019;78(1):100–110. doi:10.1136/annrheumdis-2017-212863
15. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
16. Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi:10.1016/j.xinn.2021.100141
17. Wu Y, Yang S, Ma J, et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov. 2022;12:134–153. doi:10.1158/2159-8290.CD-21-0316
18. Street K, Risso D, Fletcher RB, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi:10.1186/s12864-018-4772-0
19. Van den Berge K, Roux de Bézieux H, Street K, et al. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun. 2020;11(1):1201. doi:10.1038/s41467-020-14766-3
20. Kitagaki J, Iwamoto M, Liu J-G, et al. Activation of beta-catenin-LEF/TCF signal pathway in chondrocytes stimulates ectopic endochondral ossification. Osteoarthritis Cartilage. 2003;11:36–43. doi:10.1053/joca.2002.0863
21. Hussain ZB, Chahla J, Mandelbaum BR, Gomoll AH, LaPrade RF. The role of meniscal tears in spontaneous osteonecrosis of the knee: a systematic review of suspected etiology and a call to revisit nomenclature. Am J Sports Med. 2019;47:501–507. doi:10.1177/0363546517743734
22. Pareek A, Parkes CW, Bernard C, et al. Spontaneous osteonecrosis/subchondral insufficiency fractures of the knee: high rates of conversion to surgical treatment and arthroplasty. J Bone Joint Surg Am. 2020;102:821–829. doi:10.2106/JBJS.19.00381
23. Tanaka Y, Mima H, Yonetani Y, et al. Histological evaluation of spontaneous osteonecrosis of the medial femoral condyle and short-term clinical results of osteochondral autografting: a case series. Knee. 2009;16(2):130–135. doi:10.1016/j.knee.2008.10.013
24. Wang X, Ning Y, Zhang P, et al. Comparison of the major cell populations among osteoarthritis, Kashin-Beck disease and healthy chondrocytes by single-cell RNA-seq analysis. Cell Death Dis. 2021;12:551. doi:10.1038/s41419-021-03832-3
25. Hu Y, Chen X, Wang S, Jing Y, Su J. Subchondral bone microenvironment in osteoarthritis and pain. Bone Res. 2021;9:20. doi:10.1038/s41413-021-00147-z
26. Hu W, Chen Y, Dou C, Dong S. Microenvironment in subchondral bone: predominant regulator for the treatment of osteoarthritis. Ann Rheum Dis. 2021;80:413–422. doi:10.1136/annrheumdis-2020-218089
27. Li Z, Xu Z, Duan C, et al. Role of TCF/LEF transcription factors in bone development and osteogenesis. Int J Med Sci. 2018;15:1415–1422. doi:10.7150/ijms.26741
28. Hata K, Takahata Y, Murakami T, Nishimura R. Transcriptional network controlling endochondral ossification. J Bone Metab. 2017;24:75–82. doi:10.11005/jbm.2017.24.2.75
29. Ono K, Hata K, Nakamura E, et al. Dmrt2 promotes transition of endochondral bone formation by linking Sox9 and Runx2. Commun Biol. 2021;4(1):326. doi:10.1038/s42003-021-01848-1
30. Piscopo DM, Johansen EB, Derynck R. Identification of the GATA factor TRPS1 as a repressor of the osteocalcin promoter. J Biol Chem. 2009;284:31690–31703. doi:10.1074/jbc.M109.052316
31. Nakamura E, Hata K, Takahata Y, et al. Zfhx4 regulates endochondral ossification as the transcriptional platform of Osterix in mice. Commun Biol. 2021;4:1258. doi:10.1038/s42003-021-02793-9
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.