Back to Journals » International Journal of Nanomedicine » Volume 18
Serum Exosome-Derived piRNAs Could Be Promising Biomarkers for HCC Diagnosis
Authors Rui T, Wang K, Xiang A, Guo J, Tang N, Jin X, Lin Y, Liu J, Zhang X
Received 25 November 2022
Accepted for publication 29 March 2023
Published 13 April 2023 Volume 2023:18 Pages 1989—2001
DOI https://doi.org/10.2147/IJN.S398462
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Phong A Tran
Tao Rui,1,2 Kai Wang,1,2 Aizhai Xiang,1 Jufeng Guo,1 Ning Tang,1 Xin Jin,1 Yimou Lin,3 Jian Liu,1 Xiaobing Zhang1,2
1Department of Surgery, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, 310003, People’s Republic of China; 2Key Laboratory of Integrated Oncology and Intelligent Medicine of ZheJiang Province, Zhejiang University School of Medicine, Hangzhou, 310003, People’s Republic of China; 3Department of Surgery, Collaborative Innovation Center for the Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang University Hangzhou, Hangzhou, 310003, People’s Republic of China
Correspondence: Tao Rui; Xiaobing Zhang, Email [email protected]; [email protected]
Background: Serum exosome-based liquid biopsy has significant advantages for screening and diagnosing hepatocellular carcinoma (HCC). P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs) are novel small silencing RNAs that have been identified to function in cancer-related signalling pathways. However, studies on the presence of piRNAs in serum exosomes from HCC patients and their diagnostic values in HCC are not well reported. Our aim is to validate serum exosome-derived piRNAs as the valuable component of liquid biopsy for diagnosing HCC.
Methods: We used small RNA (sRNA) sequencing to profile piRNAs from serum exosomes and describe the base distribution characteristics of serum exosome-derived piRNAs. Serum exosomes from 125 HCC patients and 44 nontumor donors were included in this study.
Results: We found that piRNAs were components of serum exosomes from HCC patients. A total of 253 differentially expressed serum exosome-derived piRNAs were screened from HCC compared with the piRNAs from nontumor donors. Serum exosome-derived piRNAs from HCC displayed a distinctive base distribution. To further confirm the potential diagnostic value of serum exosome-derived piRNAs in HCC, we detected the levels of the top 5 upregulated piRNAs in our Chinese cohort. The training set and validation set both showed that all 5 piRNAs were dramatically increased in the serum exosomes from HCC compared with the piRNAs from non-tumour donors. The piRNAs could strongly identify HCC patients from non-tumour donors according to the area under the receiver operating characteristic (AUROC) model. Additionally, the piRNAs could also present significant values for the diagnosis of HCC with low tumour burden.
Conclusion: piRNAs enriched the components of serum exosomes from HCC and could serve as promising biomarkers for HCC diagnosis.
Keywords: biomarker, exosome, sRNA-seq, HCC, piRNA
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide. Although great progress has been made in therapeutic strategies, HCC remains the third most common cause of cancer-related death.1 The subtle nature of symptoms of early-stage HCC patients makes in-clinic HCC screening challenging. When patients are diagnosed with HCC, most of them have reached an advanced stage. Serum alpha-fetoprotein (AFP) is a broad marker for the screening of HCC. However, the sensitivity of AFP for identifying HCC only ranges from 41% to 65%.2 Thus, discovering optimal biomarkers for screening and diagnosing HCC is urgent for improving the prognosis of HCC in the clinic.
Exosomes are characterized as nanoscale particles with a diameter of 30–150 nm.3,4 The contents of exosomes are abundant, such as many types of small RNAs, messenger (m)RNAs, proteins, phosphatides, and metabolites. Under transmission electron microscopy (TEM), exosomes are covered by phospholipid bilayer membranes, which prevent degradation of the contents.5 Thus, exosomes can act as optimal cargos for transferring information. This result indicated that the information from cancer cells could be detected from the serum exosomes. During the decade, several studies have reported that exosome-derived contents, especially micro (mi)RNAs, may serve as biomarkers for HCC diagnosis.6–8 However, until now, few have been used in clinical practice. Therefore, we aimed to explore the novel content of exosomes, which may have great potential for use in HCC diagnosis.
P-element-induced wimpy testis (PIWI) -interacting RNA (piRNA) is an animal-specific class of small silencing RNAs with a length of 25–32 nucleotides. piRNAs are spliced from single-stranded precursor transcripts.9,10 The ancestral function of piRNAs is the silencing transposons in the germ lines by binding to the PIWI proteins.11 The newly identified piRNAs also existed in other human tissues and were abnormally expressed in various cancers.12 piRNAs serve as novel therapeutic targets and participate in cancer-related networks and signalling pathways. This finding indicated that piRNAs may have value for cancer diagnosis. Liu et al reported that serum piRNAs can act as potential biomarkers for the diagnosis of cancers.13 To date, several piRNAs may have significant value for the screening of gastric cancer,14 colorectal cancer,15 etc. However, the issue that piRNAs are present in the serum exosomes of HCC has not been confirmed. This study aimed to identify piRNAs as significant components of serum exosomes from HCC. Additionally, we examined the characteristics of the base distribution of exosome-derived piRNAs from HCC and explored their utility in HCC diagnosis.
Materials and Methods
Patients and Clinical Specimen Preparation
All enrolled HCC patients and non-tumour donors aged 18–75 were from the Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine. None of the non-tumour donors were diagnosed with cancer within 1 year. We emphasize that the collection of the serum from the patients occurred before the invasive procedures. The serum was isolated at 1000 × g for 10 min at 4°C. To protect the exosomes in the serum, all the serum was aliquoted and stored at −80°C. All the samples were limited to 1 freeze-thaw cycle. The sample was enrolled in this study after the patient was histologically diagnosed with HCC. We also collected the general information and clinicopathological features of HCC patients. The histological stage of HCC patients was according to the 8th edition of the American Joint Committee on Cancer; tumour, node, metastasis (AJCC-TNM) classification.16,17 This research protocol was approved by the Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine. The participants provided written informed consent to participate in this study.
Serum Exosome Purification, Exosome Identification, and Exosome-RNA Extraction
As previously described, the collected serum from HCC patients and non-tumour donors was centrifuged at 3000 × g at 4°C for 5 min and then filtered with a Millex GV filter unit (0.22 μm, Merck Millipore, Darmstadt, Germany). One millilitre of serum from each sample was used for exosome purification and exosome-RNA extraction according to the manufacturer’s instructions of the exoRNeasy Midi Kit (No. 77144, Qiagen, Venlo, The Netherlands). The extracted exosome-RNA was used for reverse transcription or stored at −80°C.
We identified exosome after the exosome purification. Transmission electron microscopy (TEM) was performed as previously described,18 the purified exosomes were seeded on a copper grid and negatively stained with phosphotungstic acid (2%) for 1.5 min. After the samples were dried for 15 min, the exosomes were observed with a transmission electron microscope (Tecnai G2 Spirit 120 kV, Thermo Fisher Scientific, St. Louis, MO, USA). The hydrodynamic diameter distribution of the exosomes was measured by a Nano-ZS90 instrument (Malvern, UK) according to the manufacturer’s instructions.
Serum Exosome-Derived piRNA Sequencing and Prediction
piRNA sequencing was performed by Novegene (Beijing, China). The small RNAs (sRNAs) of serum exosomes from 4 HCC patients and 4 non-tumour donors were randomly enrolled for piRNA sequencing by using the Multiplex Small RNA Library Prep Set for Illumina® (San Diego, CA, USA) according to the manufacturer’s instructions. After quantification and quality control (Agilent 2100 pic600, Santa Clara, CA, USA), the sRNAs were added to 3′ and 5′ adapters, and a quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed. The products were gel-purified and length filtered according to the range of length. The sRNA tags were matched to genome version GRCh38.
Then, we mapped the reads (mismatch=1) with piRNABank (http://pirnabank.ibab.ac.in/)19 to obtain the known piRNA profiles in serum exosomes. The unmapped reads were used to predict the novel piRNAs according to a k-mer scheme designed by Zhang et al.20 Then, we obtained the reads of 4 bases in each position of the piRNAs. The base distribution of piRNAs is shown. The differentially expressed serum exosome-derived piRNAs between HCC patients and non-tumour donors were screened by using the DESeq R package at the threshold of log2 fold change > 1 and adjusted p value < 0.05.
Quantitative Reverse-Transcription Polymerase Chain Reaction
piR-1029, piR-15254, novel-piR-35395, novel-piR-32132, and novel-piR-43597 were the top 5 upregulated piRNAs with a length of 25–29 nt. Thus, we performed a stem-loop qRT-PCR test to detect the expression of 5 piRNAs, as previously described.21 The piRNA cDNAs was synthesized with the condition of 42°C for 60 min and 70°C for 10 min, and the PCR amplification was performed with the condition of 40 cycles at 95°Cs for 15s and 60°C for 30s. Stem-loop reverse transcription was performed with a riboSCRIPT Reverse Transcription Kit (C11027, Ribobio, Guangzhou, Guangdong, China). qPCR amplification was performed with FS Essential DNA Green Master Mix (4913914001, Roche, Pleasanton, CA, USA) accompanied by a LightCycler 480 instrument (Roche, USA). Data was normalized using the exogenous control (cel-miR-39-3p, miRB0000010-3-1, Ribobio, Guangzhou, Guangdong, China). The primers used in this study are shown in Additional 1, Table S1.
Western Blot Analysis of Exosome Markers
As previously described,18 the protein from the purified exosomes was extracted and electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (FD341-100, Fudebio, Hangzhou, China). Then, all the proteins were transferred onto equilibrated polyvinylidene difluoride membranes. After blocking with 5% nonfat milk for 1 h at room temperature, the membranes were incubated with primary antibodies overnight at 4 °C and secondary horseradish peroxidase (HRP)-linked antibody. The bands were detected by an enhanced chemiluminescence (ECL) system (Biotanon, Shanghai, China) with FDbio-FemtoECL (FD8030, Fudebio, China). The antibodies used in this study are shown in Additional 1, Table S2.
Statistical Analysis
The description of the quantitative variables is presented as the mean ± standard deviation (SD) or median ± interquartile range (IQR). Student’s t-test or the Mann–Whitney test was performed to compare the quantitative variables between the 2 groups. The analysis of variance (ANOVA) followed by Newman-Keuls individual comparisons were used among more than 2 groups. Categorical measures were compared by the chi-square test or Fisher’s exact test. ΔCt levels were used to describe the relative expression levels of 5 serum exosome-derived piRNAs from HCC and non-tumour donors. The area under the receiver operating curve (AUROC) model was used to evaluate the values of the 5 piRNAs in the diagnosis of HCC. Statistical analysis was performed with Statistical Product and Service Solutions (SPSS) software (Version 19.0, IBM, NewYork, USA). All results were 2-tailed, and a p value< 0.05 was set as the significance level. *,**, ***, and **** were used to represent p value of < 0.05, < 0.01, < 0.001, and < 0.0001, respectively.
Results
Patient Characteristics
A total of 130 HCC and 44 non-tumour donors were designed in this study. Serum samples from HCC patients were collected before the patients had undergone hepatectomy or percutaneous liver biopsy. After the patients were histopathologically diagnosed with HCC, the serum of the patients was accepted in this study. One of the patients did not undergo hepatectomy, which led to a lack of clinicopathological features. Two of the patients showed other tumours. The other 2 patients received neoadjuvant therapy before the liver biopsy. Thus, 5 patients were excluded from this study. The characteristics and tumour features of the patients are shown in Table 1.
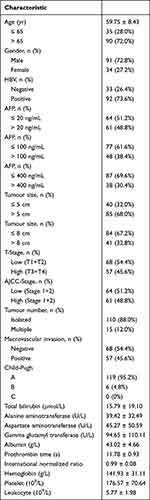 |
Table 1 Patient Characteristics |
Profiling and Predicting the Serum Exosome-Derived piRNAs from HCC Patients
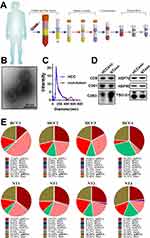
For deep sequencing of piRNAs, total RNA should be extracted from serum exosomes (Figure 1A). We first extracted and identified exosomes from 1 mL of serum. Typical exosomes were observed in the total mesorectal excision (TME). The photographed exosomes had a bilayer membrane structure with a diameter of 100 nm (Figure 1B). The hydrodynamic diameter distribution of the exosomes was also measured. The peak of particle diameter was close to 110 nm (Figure 1C). We extracted total proteins and detected the expression of exosome markers by Western blotting. CD9, CD81, CD63, TSG101, HSP70, and HSP90 were all expressed in the extracted exosomes (Figure 1D).
After the serum exosomes were obtained and identified, we randomly extracted RNAs of serum exosomes from 4 HCC patients and 4 non-tumour donors and performed sRNA sequencing. To obtain the complete piRNA profiles in serum exosomes, we not only matched the sequencing with the known piRNABank (http://pirnabank.ibab.ac.in/) but also predicted the novel piRNAs through the practiced k-mer algorithm (Additional Files 2 and 3, Figure S1). Then, the proportions of various sRNA components were analysed. The results showed that miRNAs were the most important component. However, the composition of piRNAs in exosomes was also abundant. The highest proportion reached 23.72% (Figure 1E). The results showed that piRNAs were the crucial components of the serum exosomes from HCC patients.
The Base Distribution Characteristics of Serum Exosome-Derived piRNAs
Zhang et al have shown the position-specific properties of piRNAs in mice. They noted the biases for base U and A at positions 1 and 10 in piRNAs.22 However, the characteristics of serum exosome-derived piRNAs have never been reported in HCC. The piRNAs have a length of 25–32 nt. Through piRNA sequencing, we acquired the base distribution of each position of piRNAs from the serum exosome. We found that it was specific, both in HCC patients and non-tumor donors (Additional File 3, Figure S2). The proportions of bases U and G accounted for the largest proportion, while base A accounted for the smallest proportion. The density of Base U and G focused on the middle part of piRNAs. Interestingly, the proportion of base C increased dramatically in the last part (26–32 position) of piRNAs. The results showed the specific characteristics of serum exosome-derived piRNAs in HCC patients and non-tumour donors.
The preference of the base U at the first position implies binding to RNase III enzymes or Argonaute-like proteins.23 In this study, we attempted to explore the distribution characteristics of the first base and the last base at different lengths of serum exosome-derived piRNAs. The first base proportion of piRNAs from HCC and non-tumour donors showed significant differences. Compared to bases A and U, bases C and G showed specificity through the analysis of the first base proportion in all lengths of piRNAs (Figure 2A and B). In the serum exosome-derived piRNAs from HCC patients, the proportion of base C decreased significantly compared with those from non-tumor donors (Figure 2C). In contrast, the proportion of base G increased dramatically (Figure 2D). When we analysed the distribution of the last base of piRNAs, we observed an opposite behaviour against the first base (Figure 2E and F). The results showed that the proportion of base C increased and base G decreased (Figure 2G–H). It is particularly obvious in the piRNAs with lengths of 30 and 31 nt. Thus, the results showed that significant differences in the first base and last base distribution between HCC and non-tumour donors could be found in the serum exosome-derived piRNAs. The base distribution characteristics of the serum exosome-derived piRNAs are suggested to be able to be used to identify HCC.
Five Serum Exosome-Derived piRNAs Could Be Biomarkers for the Diagnosis of HCC
We further attempted to discover valuable serum exosome-derived piRNAs as biomarkers for the diagnosis of HCC. At the criteria of log2-fold change > 1 and adjusted p value < 0.05, 253 differentially expressed piRNAs were screened from the results of piRNA sequencing (36 piRNAs were upregulated and 217 piRNAs were downregulated from the serum exosomes of HCC) (Additional File 4). The top 5 significantly upregulated exosome-derived piRNAs (according to the adjusted p value) were set as research candidates (Figure 3A). Of these piRNAs, 2 (piR-1029 and piR-15254) were known piRNAs in the piRNABank, and 3 (novel-piR-35395, novel-piR-32132, and novel-piR-43597) were novel piRNAs.
Then, the expression levels of 5 serum exosome-derived piRNAs from 125 HCC patients and 44 non-tumour donors were detected. All the samples were divided into the training set and validation set. First, from the results of the training set (54 HCC patients and 20 nontumor donors), 5 piRNAs were significantly upregulated in HCC compared with non-tumor donors (Additional File 3. Figure S3A–E). This result indicated that the 5 serum exosome-derived piRNAs were potential biomarkers for the diagnosis of HCC. Then, we calculated the values of 5 piRNAs for distinguishing HCC from non-tumour donors by introducing the 5 piRNAs into the AUROC model. The results showed that all 5 piRNAs had high AUROC values (piR-1029: 0.961; piR-15254: 0.868; novel-piR-35395: 0.898; novel-piR-32132: 0.926; and novel-piR-43597: 0.935). If we combined the 5 piRNAs (5-piRNA signature), the AUROC value would increase to 0.986 (Additional File 3. Figure S3F and G). Second, the results above were confirmed in the validation set (71 HCC patients and 24 nontumor donors). The levels of piR-15254, piR-1029, novel-piR-35395, novel-piR-32132, and novel-piR-43597 in HCC increased 7.46-fold, 1.85-fold, 4.02-fold, 9.08-fold, and 6.57-fold, respectively, compared with those in non-tumour donors (Figure 3B–F). The AUROC values of the 5 piRNAs for diagnosing HCC were higher than 0.85 (piR-1029: 0.861; piR-15254: 0.885; novel-piR-35395: 0.967; novel-piR-32132: 0.885; and novel-piR-43597: 0.981). The value of the 5-piRNA signature was 0.981 (Figure 3G). Therefore, the results of the validation set were consistent with the values of the training set. The 5 serum exosome-derived piRNAs could be optimal biomarkers for diagnosing HCC.
Interestingly, the diagnostic value of AFP was 0.687 in our set (Additional File 3, Figure S4), which was approximately similar to the results reported by Feng et al.24 The 5 piRNAs were observed to have better values than AFP. The 5 piRNAs showed high diagnostic values both in AFP-positive and AFP-negative patients (Additional File 3, Figure S5).
Five Serum Exosome-Derived piRNAs Could Also Serve for the Diagnosis of HCC with Low Tumour Burden
After determining that 5 piRNAs could be used for accurately diagnosing HCC, we explored their values in the diagnosis of HCC with low tumour burden. American Joint Committee on Cancer (AJCC) stage, tumour number, and tumour size were 3 crucial tumour features for describing the tumour burden of HCC. The HCC patients were divided according to the 3 tumour features. The levels of the 5 piRNAs in HCC patients with low AJCC stage, isolated tumour, or small tumour size were all significantly higher than the levels of the 5 piRNAs in non-tumour donors (Table 2 and Additional File 3, Figure S6). There were no significant differences in the expression levels of the 5 piRNAs between the patients with low and advanced tumour burden (low AJCC stage vs high AJCC stage; isolated tumour vs multiple tumours; ≤5 cm vs >5 cm). Thus, the results showed that HCC patients with low tumour burden also had high levels of 5 piRNAs, which indicated that the 5 serum exosome-derived piRNAs could be used for the diagnosis of HCC with low tumour burden. Then, we introduced the levels of 5 piRNAs into the receiver operating characteristic (ROC) model from HCC patients with low and advanced tumour burdens. The AUROC values of 5 piRNAs for diagnosing HCC with low tumour burden were significantly high enough (piR-1029: 0.878; piR-15254: 0.927; novel-piR-35395: 0.986; novel-piR-32132: 0.887; novel-piR-43597: 0.957) (Additional File 3, Figure S7). All the results indicated that the 5 serum exosome-derived piRNAs had the potential to diagnose HCC with a low tumour burden.
 |
Table 2 The Levels of 5 Serum Exosome-Derived piRNAs with Different Statuses of HCC (vs Non-Tumour Donors) |
Discussion
HCC is a well-known lethal tumour. China is one of the countries with a high incidence of HCC due to the high infection rate of Hepatitis B virus (HBV).25 Effective screening and accurate diagnosis are crucial approaches for improving the prognosis of HCC patients. In this study, we proposed that abundant piRNAs existed in the serum exosomes of HCC patients. We showed the base distribution of the serum exosome-derived piRNAs from HCC. Of these piRNAs, 5 serum exosome-derived piRNAs could be optimal biomarkers for the diagnosis of HCC.
The exploration of exosomes on the diagnostic value of HCC has been a hot issue. Exosomes act as cargos for transferring proteins, miRNAs, lncRNAs, mRNAs, tsRNAs, and liquids, which could be potential biomarkers for the diagnosis of HCC.26 Exosome-derived miRNAs could be the most famous content in the research field of diagnosing biomarkers for HCC. Wang et al proposed that serum exosome-derived miR-122, miR-148a, and miR-1246 are significantly higher in HCC.27 Fornari et al revealed that exosome-derived miR-519d could be a better biomarker for HCC than AFP.28 Kogure et al identified a subset of miRNAs that were highly enriched within exosomes of HCC.29 Wang et al showed that serum exosomal miR-21 was significantly higher in HCC than in non-tumour volunteers.30 Our results indeed showed that miRNAs were the most important components compared with others. However, other contents of exosomes also have potential value for diagnosing HCC. Sun et al showed that serum exosome-derived LINC00161 was significantly upregulated in HCC patients and had a high AUROC level for diagnosing HCC.31 Xu et al suggested that serum exosomal hnRNPH1 mRNA levels were significantly higher in HCC, and the AUROC value was 0.865.32 Sanchez et al used lipidomics to identify that lipid species were enriched in the exosomes of HCC and served as biomarkers for the early detection of HCC.33 Zhu et al first proposed that 4 tRNA-derived small RNAs (tRNA-ValTAC-3, tRNA-GlyTCC-5, tRNA-ValAAC-5 and tRNA-GluCTC-5) were potential biomarkers for diagnosing HCC.34 This result indicated that using other content of HCC serum exosomes for the diagnosis of HCC was essential.
piRNAs are small RNAs that are first found in germ lines. Several reports have proposed that other cell types also express functional piRNAs. Gao XQ confirmed that piR-141981 (CHAPIR) regulated cardiac hypertrophy by targeting METTL3-mediated N6-methyladenosine (m6A) methylation of Parp10 mRNA transcripts.35 In the liver, piR-823 increased hepatic stellate cell activation by binding EIF3B, which promoted liver fibrosis and HCC tumorigenesis.36,37 This finding indicated that piRNAs might have potential value in the regulation of HCC. The serum exosomes of cancer patients have been identified with piRNAs. Li et al showed that serum exosomal piRNAs (piR-26925 and piR-5444) could be potential biomarkers for the diagnosis of lung adenocarcinoma.38 However, research on serum exosome-derived piRNAs for diagnosing HCC is lacking. Through sRNA sequencing, we observed that serum exosomes of HCC existed with abundant piRNAs. We also proposed some unknown piRNAs that were not enrolled in the piRNA database and confirmed that the piRNA was the content of serum exosomes from HCC. However, the mechanisms of 5 piRNAs in the regulation of HCC were not reported in this study or the literature.
To our knowledge, we are the first to propose that serum exosome-derived piRNAs could be used for the diagnosis of HCC in the clinic. From the training set and validation set, we found that the 5 piRNAs were significantly upregulated in serum exosomes of HCC. The AUROC model also showed that the 5 piRNAs had high AUROC values for diagnosing HCC. We also showed that the 5 piRNAs were also suitable for the diagnosis of HCC with low tumour burden. Compared with the reported biomarkers in the literature described above and AFP, the 5 piRNAs also showed higher values. Serum exosome-derived piRNAs could serve as valuable biomarkers for HCC diagnosis.
The base distribution of piRNAs has its characteristics. Zhang et al noted the biases for base U and A at positions 1 and 10 in piRNAs.22 The preference of base U at the first position implies binding to RNase III enzymes or Argonaute-like proteins. In this study, we also found that the base distribution of piRNAs in serum exosomes was peculiar. The characteristics of the base distribution of piRNAs could be used to distinguish HCC patients from non-tumour donors. We showed that base C was specifically preferred in the last part of serum exosome-derived piRNAs. In all lengths (25–32 nt) of piRNAs from HCC, base G increased and base C decreased significantly in the first base. On the lower base, we observed the opposite results. However, the mechanisms have not been revealed. The factors that influence miRNA and tRNA characteristics are what we can learn from. Note that the Dicer RNase IIIA domain tends to generate miRNAs with Base U at the 5′,39 resulting in a strong preference for base U at the 5′ end of the miRNA. The specific sequences of 5′ of tsRNA are generated by RNaseZ, and the 3′ of tsRNA results from the transcription process by RNA polymerase III.40 It is interesting that serum exosome-derived piRNAs from HCC showed specificity. The specificity may occur at the stage of piRNA generation from HCC. Alternatively, such characteristic piRNAs are more likely to be released into serum exosomes by HCC.
There were limitations in this study. First, only the Chinese cohort was enrolled. Second, the mechanisms by which piRNAs are packaged into exosomes and transferred into serum have not been elucidated. Third, the reason why serum exosome-derived piRNAs from HCC exhibited specific base distribution should be further investigated.
Conclusions
Our work revealed that piRNAs were components of serum exosomes from HCC patients. Moreover, significant specificity existed when we analysed the base distribution of the serum exosome-derived piRNAs. Notably, from the Chinese HCC cohort, 5 serum exosome-derived piRNAs showed significant values for distinguishing HCC patients from non-tumour donors. The piRNAs could also be used for diagnosing HCC with a low tumour burden. Taken together, the piRNAs from serum exosomes could be promising biomarkers for HCC diagnosis (Figure 4).
 |
Figure 4 piRNAs, as the newly identified components of serum exosomes from HCC patients, could be promising biomarkers for the diagnosis of HCC. |
Abbreviations
HCC, hepatocellular carcinoma; piRNAs, P-element-induced wimpy testis (PIWI) -interacting RNAs; sRNA, small RNA; TEM, transmission electron microscopy; AFP, serum alpha-fetoprotein; AUROC, area under the receiver operating characteristic; AJCC-TNM, American Joint Committee on Cancer; tumour, node, metastasis classification.
Data Sharing Statement
The data used in this study can be acquired from the corresponding author in a reasonable request.
Ethics Approval and Consent to Participate
This research protocol was approved by the Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, according to the Declaration of Helsinki.
Funding
This study was supported by the Zhejiang Provincial Natural Science Foundation of China (Q23H160130) and Medical Health Science and Technology Project of Zhejiang Provincial Health Commission, China (2023RC224).
Disclosure
All authors declare no conflict of interest.
References
1. Forner A, Hessheimer AJ, Isabel Real M, Bruix J. Treatment of hepatocellular carcinoma. Crit Rev Oncol Hematol. 2006;60(2):89–98. doi:10.1016/j.critrevonc.2006.06.001
2. Gupta S, Bent S, Kohlwes J. Test characteristics of alpha-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis. Ann Intern Med. 2003;139(1):46–50. doi:10.7326/0003-4819-139-1-200307010-00012
3. Whiteside TL. The emerging role of plasma exosomes in diagnosis, prognosis and therapies of patients with cancer. Contemp Oncol. 2018;22(1A):38–40.
4. Alcântara-Neto AS, Fernandez-Rufete M, Corbin E, et al. Oviduct fluid extracellular vesicles regulate polyspermy during porcine in vitro fertilisation. Reprod Fertil Dev. 2020;32(4):409–418. doi:10.1071/RD19058
5. Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res. 2019;60(1):9–18. doi:10.1194/jlr.R084343
6. Ghosh S, Bhowmik S, Majumdar S, et al. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int J Cancer. 2020;147(10):2934–2947. doi:10.1002/ijc.33111
7. Alzahrani FA, El-Magd MA, Abdelfattah-Hassan A, et al. Potential effect of exosomes derived from cancer stem cells and MSCs on progression of DEN-induced HCC in rats. Stem Cells Int. 2018;2018:8058979. doi:10.1155/2018/8058979
8. Pan J-H, Zhou H, Zhao -X-X, et al. Role of exosomes and exosomal microRNAs in hepatocellular carcinoma: potential in diagnosis and antitumour treatments (Review). Int J Mol Med. 2018;41(4):1809–1816. doi:10.3892/ijmm.2018.3383
9. Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–324. doi:10.1126/science.1129333
10. Houwing S, Kamminga LM, Berezikov E, et al. A role for piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129(1):69–82. doi:10.1016/j.cell.2007.03.026
11. Lewis SH, Quarles KA, Yang Y, et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol. 2018;2(1):174–181. doi:10.1038/s41559-017-0403-4
12. Chalbatani GM, Dana H, Memari F, et al. Biological function and molecular mechanism of piRNA in cancer. Pract Lab Med. 2019;13:e00113. doi:10.1016/j.plabm.2018.e00113
13. Liu Y, Dou M, Song X, et al. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer. 2019;18(1):123. doi:10.1186/s12943-019-1052-9
14. Cui L, Lou Y, Zhang X, et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem. 2011;44(13):1050–1057. doi:10.1016/j.clinbiochem.2011.06.004
15. Vychytilova-Faltejskova P, Stitkovcova K, Radova L, et al. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol Biomarkers Prev. 2018;27(9):1019–1028. doi:10.1158/1055-9965.EPI-18-0318
16. Chun YS, Pawlik TM, Vauthey JN. 8th edition of the AJCC cancer staging manual: pancreas and hepatobiliary cancers. Ann Surg Oncol. 2018;25(4):845–847. doi:10.1245/s10434-017-6025-x
17. Liao X, Zhang D. The 8th edition American joint committee on cancer staging for hepato-pancreato-biliary ssCancer: a review and update. Arch Pathol Lab Med. 2021;145(5):543–553. doi:10.5858/arpa.2020-0032-RA
18. Rui T, Zhang X, Feng S, et al. MiR-516a-3p is a novel mediator of hepatocellular carcinoma oncogenic activity and cellular metabolism. Engineering. 2021;16:162. doi:10.1016/j.eng.2021.07.020
19. Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered piwi-interacting RNAs. Nucleic Acids Res. 2008;36(Database issue):D173–D177. doi:10.1093/nar/gkm696
20. Zhang Y, Wang X, Kang L. A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics. 2011;27(6):771–776. doi:10.1093/bioinformatics/btr016
21. Rui T, Zhang X, Feng S, et al. The similar effects of miR-512-3p and miR-519a-2-5p on the promotion of hepatocellular carcinoma: different tunes sung with equal skill. Front Oncol. 2020;10:1244. doi:10.3389/fonc.2020.01244
22. Zhang P, Kang JY, Gou LT, et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015;25(2):193–207. doi:10.1038/cr.2015.4
23. Betel D, Sheridan R, Marks DS, Sander C, Kim N. Computational analysis of mouse piRNA sequence and biogenesis. PLoS Comput Biol. 2007;3(11):e222. doi:10.1371/journal.pcbi.0030222
24. Feng H, Li B, Li Z, Wei Q, Ren L. PIVKA-II serves as a potential biomarker that complements AFP for the diagnosis of hepatocellular carcinoma. BMC Cancer. 2021;21(1):401. doi:10.1186/s12885-021-08138-3
25. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
26. Sasaki R, Kanda T, Yokosuka O, Kato N, Matsuoka S, Moriyama M. Exosomes and hepatocellular carcinoma: from bench to bedside. Int J Mol Sci. 2019;20(6):1406. doi:10.3390/ijms20061406
27. Wang Y, Zhang C, Zhang P, et al. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018;7(5):1670–1679. doi:10.1002/cam4.1390
28. Fornari F, Ferracin M, Trerè D, et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS One. 2015;10(10):e0141448. doi:10.1371/journal.pone.0141448
29. Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54(4):1237–1248. doi:10.1002/hep.24504
30. Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. 2014;2014:864894. doi:10.1155/2014/864894
31. Sun L, Su Y, Liu X, et al. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J Cancer. 2018;9(15):2631–2639. doi:10.7150/jca.24978
32. Xu H, Dong X, Chen Y, Wang X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med. 2018;56(3):479–484. doi:10.1515/cclm-2017-0327
33. Sanchez JI, Jiao J, Kwan SY, et al. Lipidomic profiles of plasma exosomes identify candidate biomarkers for early detection of hepatocellular carcinoma in patients with cirrhosis. Cancer Prev Res. 2021;14(10):955–962. doi:10.1158/1940-6207.CAPR-20-0612
34. Zhu L, Li J, Gong Y, et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. 2019;18(1):74. doi:10.1186/s12943-019-1000-8
35. Gao XQ, Zhang YH, Liu F, et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N6-methyladenosine methylation of Parp10 mRNA. Nat Cell Biol. 2020;22(11):1319–1331. doi:10.1038/s41556-020-0576-y
36. Tang X, Xie X, Wang X, Wang Y, Jiang X, Jiang H. The combination of piR-823 and Eukaryotic Initiation Factor 3 B (EIF3B) activates hepatic stellate cells via upregulating TGF-β1 in liver fibrogenesis. Med Sci Monit. 2018;24:9151–9165. doi:10.12659/MSM.914222
37. Rizzo F, Rinaldi A, Marchese G, et al. Specific patterns of PIWI-interacting small noncoding RNA expression in dysplastic liver nodules and hepatocellular carcinoma. Oncotarget. 2016;7(34):54650–54661. doi:10.18632/oncotarget.10567
38. Li J, Wang N, Zhang F, et al. PIWI-interacting RNAs are aberrantly expressed and may serve as novel biomarkers for diagnosis of lung adenocarcinoma. Thorac Cancer. 2021;12(18):2468–2477. doi:10.1111/1759-7714.14094
39. Starega-Roslan J, Galka-Marciniak P, Krzyzosiak WJ. Nucleotide sequence of miRNA precursor contributes to cleavage site selection by dicer. Nucleic Acids Res. 2015;43(22):10939–10951. doi:10.1093/nar/gkv968
40. Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673–695. doi:10.1261/rna.2000810
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.