Back to Journals » OncoTargets and Therapy » Volume 10
Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma
Authors Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, Dai Z, Cao Y, Fan J, Zhou J
Received 20 April 2017
Accepted for publication 13 June 2017
Published 1 August 2017 Volume 2017:10 Pages 3843—3851
DOI https://doi.org/10.2147/OTT.S140062
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Weifeng Liu,1–3,* Jie Hu,1,2,* Kaiqian Zhou,1,2,* Feiyu Chen,1,2 Zheng Wang,1,2 Boyi Liao,1,2 Zhi Dai,1,2 Ya Cao,4 Jia Fan,1,2,5 Jian Zhou1,2,5,6
1Liver Cancer Institute, Zhongshan Hospital, Fudan University, Shanghai, China; 2Key Laboratory of Carcinogenesis and Cancer Invasion, Fudan University, Ministry of Education, Shanghai, China; 3Department of Hepatobiliary and Pancreatic Surgery, the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China; 4Cancer Research Institute, Central South University, Key Laboratory of Carcinogenesis and Cancer Invasion, Ministry of Education, Changsha, China; 5Institute of Biomedical Sciences, Fudan University, Shanghai, China; 6Shanghai Key Laboratory of Organ Transplantation, Shanghai, China
*These authors contributed equally to this work
Abstract: Hepatocellular carcinoma (HCC) is the sixth most common cancer worldwide with high mortality. Circulating miRNA has been demonstrated as a novel noninvasive biomarker for many tumors. This study aimed to investigate the potential of circulating miR-125b as a prognostic marker of HCC. Exosomes were extracted from serum samples collected from two independent cohorts: cohort 1: HCC (n=30), chronic hepatitis B (CHB, n=30), liver cirrhosis (LC, n=30); cohort 2: HCC (n=128). We found that miR-125b levels were remarkably increased in exosomes compared to those in serum from patients with CHB, LC, and HCC (P<0.01, respectively). However, miR-125b levels in exosomes and the serum from HCC patients were inferior to that of CHB (P<0.01 and P=0.06) and LC patients (P<0.01 for all). Additionally, miR-125b levels in exosomes were associated with tumor number (P=0.02), encapsulation (P<0.01), and TNM stage (P<0.01). Kaplan–Meier analysis indicated that HCC patients with lower exosomal miR-125b levels showed reduced time to recurrence (TTR) (P<0.01) and overall survival (OS) (P<0.01). Furthermore, multivariate analysis revealed that miR-125b level in exosomes, but not in serum, was an independent predictive factor for TTR (P<0.001) and OS (P=0.011). Exosomal miR-125b levels predicted the recurrence and survival of HCC patients with an area under the ROC curve of 0.739 (83.0% sensitivity and 67.9% specificity) and 0.702 (82.5% sensitivity and 53.4% specificity). In conclusion, exosomal miR-125b could serve as a promising prognostic marker for HCC.
Keywords: exosome, miR-125b, hepatocellular carcinoma, prognosis, serum
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the second leading cause of cancer mortality globally.1 Hepatic resection and orthotopic liver transplantation is still the most effective treatment for HCC. However, the efficacy of current therapeutic regimens is poor due to the high frequency of recurrence and metastasis of HCC.2,3 Therefore, the identification of specific and sensitive biomarkers for the recurrence and prognosis of HCC is urgently needed.
miRNAs are crucially implicated in a diverse range of human diseases, including cancer.4,5 Accumulating evidence indicates that dysregulation of miRNAs, especially within natural carriers and exosomes, contributes to the onset and progression of various human tumors such as breast and liver cancer.6–9 Exosomes are small membrane vesicles (40–120 nm) which play key roles in intercellular communication via delivery of active biological molecules.10,11 Extracellular miRNAs in exosomes are relatively stable and shielded against extracellular RNase degradation.12–14 Therefore, exosomal miRNAs may serve as noninvasive biomarkers for the diagnosis and prognosis of various malignancies.8,12,15
Previous studies reported that miR-125b was downregulated in HCC tissue, and plasma miR-125b may be a diagnostic marker for HCC.5,16–20 However, the potential significance of circulating exosomal miR-125b in HCC has not been explored. In this study, we aimed to determine whether the detection of exosomal miR-125b in HCC patients will provide information for predicting the recurrence and prognosis of HCC.
Materials and methods
Patients and samples
Clinical samples were collected from two independent cohorts of patients recruited from Zhongshan Hospital, Fudan University (Shanghai, China) between January and June 2012. Cohort 1 comprised 30 patients with HCC, 30 with chronic hepatitis B (CHB), and 30 with liver cirrhosis (LC); cohort 2 comprised 128 patients with HCC. HCC patients in both cohorts had undergone curative resection and were histologically diagnosed. Clinical samples were collected from all participants after obtaining informed consent. This study and the informed consent was approved by Ethics Committee of Zhongshan Hospital, Fudan University. The clinical characteristics of all participants were shown in Table 1. None of the HCC patients had received any preoperative treatment. Patients were followed every 3 months for the first year, and then every 6 months until June 22, 2016. The follow-up included serum AFP level detection, abdominal ultrasonography, and chest X-ray. When recurrence was suspected, enhanced computed tomography (CT) scanning or enhanced magnetic resonance imaging (MRI) was performed. Time to recurrence (TTR) was defined as the interval between surgery and the detected recurrence. Overall survival (OS) was calculated as the time from surgery to death or the last observation point.
Blood samples were collected and centrifuged at 3,000 rpm for 10 min to separate serum. Exosomes were isolated from serum samples and treated with ExoQuick Exosome Precipitation Solution (System Biosciences, Palo Alto, CA, USA) according to the manufacturer’s protocol. The exosome samples were stored at −80°C for later analysis.
Electron microscopy and nanoparticle tracking analysis (NTA)
The morphology of exosomes was examined by transmission electron microscopy (TEM) as described previously.21 Briefly, exosomes were fixed in 1% glutaraldehyde, and then loaded onto formvar/carbon coated copper grids. The samples were negatively stained with 1% uranyl acetate and examined under a Tesla BS242 transmission electron microscope (Tescan, Brno, Czech Republic) operated at 80 kV. The exosomes were analyzed using NanoSight LM10 system (NanoSight, Malvern, Worcestershire, UK) equipped with fast video capture and NTA (NanoSight).
Western blot analysis
RIPA buffer was used to lyse the exosome pellet and BCA method was performed to determine protein concentration. An amount of 20 μg of protein fraction was run on 10% SDS-PAGE and transferred to nitrocellulose membranes and blocked in tris-buffered saline containing 5% non-fat dry milk and 0.1% tween-20 (TBST) for 1 h. The membranes were incubated with CD9, CD63 or GAPDH antibody (Sigma-Aldrich Co., St Louis, MO, USA) at 4°C overnight. The membranes were washed three times with TBST and incubated with HRP-conjugated secondary antibodies (Santa Cruz Biotechnology Inc., Dallas, TX, USA) for 1 h at room temperature. The bands were visualized using a Clarity™ Western ECL Substrate Kit (Bio-Rad Laboratories Inc., Hercules, CA, USA) and analyzed with an LAS-4000 Luminescent Image Analyzer (Fujifilm, Tokyo, Japan).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the exosomes, whole serum, and exosome-depleted supernatants using MirVana™ miRNA Isolation Kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). TaqMan probe for miR-125b (Applied Biosystems, Thermo Fisher Scientific) was used for qRT-PCR. TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific) was used to synthesize cDNA, and amplification was performed by TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems, Thermo Fisher Scientific). ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Thermo Fisher Scientific) was used to analyze miR-125b relative expression normalized to Caenorhabditis elegans miRNA (Cel-miR-39) (Applied Biosystems, Thermo Fisher Scientific) as previously described.22,23
Statistical analysis
Statistical analysis was performed using SPSS for Windows version 16.0. Categorical data were analyzed by χ2 or Fisher’s exact test. OS and TTR were calculated by the Kaplan–Meier method and the differences were analyzed by the log-rank test. Univariate and multivariate analyses were performed using the Cox proportional hazards regression model. The accuracy of predicting prognosis was evaluated by the ROC curves, and predicting performance was investigated by the area under the ROC curve (AUC). ROC and regression analysis was performed with MedCalc software (version 10.4.7.0; MedCalc, Mariakerke, Belgium). P<0.05 was considered statistically significant.
Results
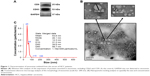
Identification of the isolated serum exosomes
To characterize the isolated serum exosomes, first we detected the expression of CD63 and CD9 in the exosomes isolated from HCC patients (Figure 1A). Western blot analysis showed that exosomal markers CD63 and CD9 were both enriched in the isolated exosome pellets. Next, we employed TEM and NTA to identify the shape and size of the exosomes. TEM revealed that exosomes had a spherical shape and a size of 32.38–70.88 nm (determined from 20 exosomes) (Figure 1B). NTA assay demonstrated that exosomes had a characteristic size of 53.6±12.3 nm and a concentration of 1.16×109 particles/mL (n=3) (Figure 1C). These results indicated that the collected exosomes had been purified adequately.
Differential expression profile of miR-125b
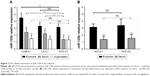
qRT-PCR was performed to detect miR-125b levels in the exosomes, the whole serum, and exosome-depleted supernatants from the participants. As shown in Figure 2A, miR-125b levels in exosomes were significantly increased compared with that in the whole serum or exosome-depleted supernatant (P<0.01, respectively) in patients of cohort 1, including CHB (n=30), LC (n=30), and HCC (n=30). Furthermore, serum miR-125b levels were remarkably decreased in HCC group compared with CHB and LC groups (P<0.01, respectively). Interestingly, exosomal miR-125b level in HCC group was inferior to that in CHB (P<0.01) and LC patients (P=0.06), but only the difference with CHB group was statistically significant (P<0.01). The difference of miR-125b levels in exosome-depleted supernatant among HCC, CHB, and LC groups was not statistically significant. Therefore, miR-125b levels in exosomes and whole serum were validated in HCC patients of cohort 2. The results demonstrated that miR-125b level in whole serum was significantly decreased compared with that in exosomes (P<0.01, respectively) both in HCC patients of cohort 1 (n=30) and 2 (n=128), but the difference in miR-125b level in whole serum or exosomes between the two cohorts was not statistically significant (P>0.05, respectively, Figure 2B).
Association of miR-125b levels with clinicopathological features of HCC patients
The median follow-up for cohort 2 (128 HCC patients) was 29.1 months (range 2.9–52.4 months). OS rate at 3 years after operation was 75.8%. To further explore whether miR-125b levels in serum and exosomes could be associated with clinicopathological parameters of HCC, 128 patients of cohort 2 were divided into two groups based on the median level of miR-125b expression. As shown in Table 2, low expression of miR-125b in serum was significantly correlated with tumor encapsulation (P=0.047), and low expression of miR-125b in exosomes was significantly correlated with tumor number, differentiation, and TNM stage (P<0.01, P<0.01, and P=0.011, respectively). Furthermore, the Kaplan–Meier curves for TTR and OS were plotted according to the levels of exosomal and serum miR-125b (Figure 3). The patients with low levels of exosomal miR-125b had shorter TTR and OS than those with high levels of miR-125b (P<0.01, respectively). However, the patients with low serum miR-125b levels had poorer TTR than those with high levels (P<0.01). Additionally, univariate and multivariate analysis revealed that microvascular invasion (MVI) and miR-125b levels in exosomes, but not serum miR-125b levels, were independent prognostic factors for TTR and OS in patients with HCC (P<0.01, respectively) (Table 3).
ROC and regression analysis
To further explore whether MVI or exosomal miR-125b level could be used as potential predictive prognostic markers of HCC, we used ROC curves to analyze the sensitivity, specificity, and AUC value of MVI and exosomal miR-125b level (Figure 4). The results demonstrated that exosomal miR-125b level showed high accuracy in predicting the recurrence (AUC =0.739) and survival (AUC =0.702) in HCC patients after liver resection. A combination of exosomal miR-125b and MVI had better power of discrimination, with an AUC of 0.807 (recurrence) and 0.765 (survival) (P<0.05, respectively) (Table 4).
Discussion
Despite recent progress in the diagnosis and treatment of HCC, the prognostic outcome of HCC patients is still poor due to the high frequency of recurrence and metastasis after surgery.2,3 In this study, our results showed that serum exosomal miR-125b was an independent predictive factor associated with TTR and OS in HCC patients, thus offering timely and comprehensive treatment alternatives. Therefore, we speculated that exosomal miR-125b is an important prognostic marker to improve the efficacy of current treatment regimens and the survival of HCC patients.
Exosomes are small membrane vesicles secreted by various types of cells.10,11 Exosomes play pivotal roles in intercellular communication via delivery of biological cargoes, including proteins, mRNAs, and miRNAs.10 Recently, miRNAs in exosomes have been investigated as valuable biomarkers of malignancy. First, exosomes can specifically reflect their original cell types and conditions, and they may harbor circulating biomarkers of particular accuracy, even at early stages of cancer which are difficult or impossible to detect.24,25 Second, miRNAs are enriched and relatively stable in exosomes due to the protection of the lipid bilayer and shelter against extracellular RNase degradation.14 In the present study, we measured miR-125b levels in the whole serum, exosome-depleted supernatant, and exosomes in patients with CHB, LC, and HCC. Our results demonstrated that exosomal miR-125b levels were significantly increased compared with miR-125b levels in the serum or exosome-depleted supernatant in all patients. These findings indicate that miR-125b in the exosome is more enriched compared with that in circulating serum.
Dysregulated miRNAs and their target genes are involved in HCC initiation and progression.26,27 miR-125b has been reported as a tumor suppressor for HCC and could suppress epithelial-mesenchymal transition, tumor growth, migration, and invasion of hepatoma cells by directly regulating oncogenes such as SMAD2/4, Sirtuin7, SUV39H1, LIN28B, and PIGF.16,17,28–30 However, the expression pattern of circulating exosomal miR-125b and its clinicopathological significance in HCC remain unknown. Our present study revealed that exosomal miR-125b expression in HCC patients was significantly downregulated compared to CHB patients, consistent with previously reported results.18,31 Furthermore, our results showed that the lower expression of exosomal miR-125b was associated with tumor number, differentiation, and TNM stage of HCC. These clinicopathologic features represent a more aggressive HCC subtype. In addition, HCC patients with lower exosomal miR-125b levels had shorter TTR and OS than those with high levels of miR-125b, indicating that miR-125b in exosomes may suppress HCC growth, migration, and invasion.32
Previous studies have reported that plasma miR-125b levels were downregulated in HCC patients and could be used as diagnostic marker for hepatitis B virus-induced HCC.18,31 In agreement with these results, we found that serum levels of miR-125b in patients with HCC were remarkably downregulated compared to those with CHB and LC. Although low serum level of miR-125b was significantly correlated with tumor encapsulation, it was not an independent prognostic factor for TTR and OS in patients with HCC, which was consistent with the previous study.33 miR-125b in circulating serum, compared with miR-125b in exosomes, may function through a different mechanism in HCC.
There are several limitations to our study. First, our sample size is small and long-term follow-up is required to confirm the relationship between exosomal miR-125b levels and patient outcome. A retrospective study with a large cohort should be considered to confirm the clinical significance of our findings. Second, the study did not include healthy participants. HCC often arises in patients with several risk factors, such as aflatoxin exposure, alcohol abuse, chronic HBV or HCV infection, and LC. However, it has been shown that HCC rarely develops in healthy people without these risks.34 Third, the potential mechanisms of exosomal miR-125b in HCC were not elucidated.
In conclusion, our findings suggest that exosomal miR-125b may serve as a novel serological biomarker with significant accuracy in predicting postoperative recurrence and survival of HCC patients. Exosomal miR-125b may help discriminate HCC patients with high risk of recurrence and poor prognosis, and guide timely comprehensive therapy for these patients.
Acknowledgments
This work was jointly supported by the grants from the National Science Foundation for Distinguished Young Scholars of China (no 81225019), the National Natural Science Foundation of China (no 81572823, 81401929, 81372650, and 81172277), National Key Research and Development Program (no 2016YFC0902400), the National Key Sci-Tech Project (no 2012ZX10002-016, 2013ZX10002007-005), Foundation of Shanghai Science and Technology Commission (no 13ZR1406900), Shanghai Rising-Star Follow-up Program Funding (no 16QA1401000), and Shanghai Hospital Development Center (no SHDC12015104). We thank Xin Zhang, Yong Zhang, and Wenshuai Liu at Zhongshan Hospital for help with the experiments.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150(4):835–853. | ||
Bruix J, Llovet JM. Major achievements in hepatocellular carcinoma. Lancet. 2009;373(9664):614–616. | ||
Roy S, Benz F, Luedde T, Roderburg C. The role of miRNAs in the regulation of inflammatory processes during hepatofibrogenesis. Hepatobiliary Surg Nutr. 2015;4(1):24–33. | ||
Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related micrornas in hepatocellular carcinoma. Hepatology. 2008;47(3):897–907. | ||
Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481(7380):190–194. | ||
Melo SA, Sugimoto H, O’Connell JT, et al. Cancer exosomes perform cell-independent microrna biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. | ||
Zhou W, Fong MY, Min Y, et al. Cancer-secreted mir-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. | ||
Wei JX, Lv LH, Wan YL, et al. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal micrornas in human hepatoma cells. Hepatology. 2015;61(4):1284–1294. | ||
Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. | ||
Zha QB, Yao YF, Ren ZJ, Li XJ, Tang JH. Extracellular vesicles: an overview of biogenesis, function, and role in breast cancer. Tumour Biol. 2017;39(2):1010428317691182. | ||
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. | ||
Munagala R, Aqil F, Gupta RC. Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumour Biol. 2016;37(8):10703–10714. | ||
Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. | ||
Qu Z, Jiang C, Wu J, Ding Y. Exosomes as potent regulators of HCC malignancy and potential bio-tools in clinical application. Int J Clin Exp Med. 2015;8(10):17088–17095. | ||
Liang L, Wong CM, Ying Q, et al. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology. 2010;52(5):1731–1740. | ||
Zhou JN, Zeng Q, Wang HY, et al. MicroRNA-125b attenuates epithelial-mesenchymal transitions and targets stem-like liver cancer cells through small mothers against decapentaplegic 2 and 4. Hepatology. 2015;62(3):801–815. | ||
Chen S, Chen H, Gao S, et al. Differential expression of plasma microRNA-125b in hepatitis B virus-related liver diseases and diagnostic potential for hepatitis B virus-induced hepatocellular carcinoma. Hepatol Res. 2017;47(4):312–320. | ||
Zhao L, Wang W. miR-125b suppresses the proliferation of hepatocellular carcinoma cells by targeting Sirtuin7. Int J Clin Exp Med. 2015;8(10):18469–18475. | ||
Wong CM, Wong CC, Lee JM, Fan DN, Au SL, Ng IO. Sequential alterations of microRNA expression in hepatocellular carcinoma development and venous metastasis. Hepatology. 2012;55(5):1453–1461. | ||
Ge M, Ke R, Cai T, Yang J, Mu X. Identification and proteomic analysis of osteoblast-derived exosomes. Biochem Biophys Res Commun. 2015;467(1):27–32. | ||
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating micrornas as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. | ||
Sohn W, Kim J, Kang SH, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184. | ||
Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. | ||
Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83(11):1484–1494. | ||
Zhou J, Yu L, Gao X, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29(36):4781–4788. | ||
Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of mirna signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56(6):1371–1383. | ||
Kim JK, Noh JH, Jung KH, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57(3):1055–1067. | ||
Fan DN, Tsang FH, Tam AH, et al. Histone lysine methyltransferase, suppressor of variegation 3–9 homolog 1, promotes hepatocellular carcinoma progression and is negatively regulated by microRNA-125b. Hepatology. 2013;57(2):637–647. | ||
Alpini G, Glaser SS, Zhang JP, et al. Regulation of placenta growth factor by microRNA-125b in hepatocellular cancer. J Hepatol. 2011;55(6):1339–1345. | ||
Giray BG, Emekdas G, Tezcan S, et al. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol Biol Rep. 2014;41(7):4513–4519. | ||
Li W, Xie L, He X, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123(7):1616–1622. | ||
Wang L, Liu M, Zhu H, et al. Identification of recurrence-related serum microRNAs in hepatocellular carcinoma following hepatectomy. Cancer Biol Ther. 2015;16(10):1445–1452. | ||
Lutwick LI. Relation between aflatoxin, hepatitis-B virus, and hepatocellular carcinoma. Lancet. 1979;1(8119):755–757. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.