Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 16
Screening of Lymphoma Radiotherapy-Resistant Genes with CRISPR Activation Library
Authors Luo BH, Huang JQ, Huang CY, Tian P, Chen AZ, Wu WH, Ma XM, Yuan YX, Yu L
Received 7 September 2022
Accepted for publication 5 December 2022
Published 30 January 2023 Volume 2023:16 Pages 67—80
DOI https://doi.org/10.2147/PGPM.S386085
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Bi-Hua Luo,* Jian-Qing Huang,* Chun-Yu Huang, Pan Tian, Ai-Zhen Chen, Wei-Hao Wu, Xiao-Mei Ma, Yue-Xing Yuan, Lian Yu
Department of Hematology, Longyan First Hospital Affiliated Fujian Medical University, Fujian, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lian Yu, Department of Hematology, Longyan First Hospital Affiliated Fujian Medical University, No. 105 of Jiuyibei Road, Xinluo District, Fujian, 364000, People’s Republic of China, Tel +86 13859572936, Email [email protected]
Objective: The objective of this study was to screen lymphoma radiotherapy-resistant genes using CRISPR activation (CRISPRa).
Methods: The Human CRISPRa library virus was packaged and then transfected into lymphoma cells to construct an activation library cell line, which was irradiated at the minimum lethal radiation dose to screen radiotherapy-resistant cells. Radiotherapy-resistant cell single-guide RNA (sgRNA) was first amplified by quantitative polymerase chain reaction (qPCR) in the coding region and then subject to next-generation sequencing (NGS) and bioinformatics analysis to screen radiotherapy-resistant genes. Certain radiotherapy-resistant genes were then selected to construct activated cell lines transfected with a single gene so as to further verify the relationship between gene expression and radiotherapy resistance.
Results: A total of 16 radiotherapy-resistant genes, namely, C20orf203, MTFR1, TAF1L, MYADM, NIPSNAP1, ZUP1, RASL11A, PSMB2, PSMA6, OR8H3, TMSB4Y, CD300LF, EEF1A1, ATP6AP1L, TRAF3IP2, and SNRNP35, were screened based on the NGS results and bioinformatics analysis of the radiotherapy-resistant cells. Activated cell lines transfected with a single gene were constructed using 10 radiotherapy-resistant genes. The qPCR findings showed that, when compared with the control group, the experimental group had significantly up-regulated mRNA expression of MTFR1, NIPSNAP1, ZUP1, PSMB2, PSMA6, EEF1A1, TMSB4Y and TAF1L (p < 0.05). No significant difference in the mRNA expression of AKT3 or TRAF3IP2 (p > 0.05) was found between the two groups (p > 0.05).
Conclusion: The 16 genes screened are potential lymphoma radiotherapy-resistant genes. It was initially determined that the high expression of 8 genes was associated with lymphoma radiotherapy resistance, and these genes could serve as the potential biomarkers for predicting lymphoma radiotherapy resistance or as new targets for therapy.
Keywords: lymphoma, radiotherapy resistance, gene, CRISPR/Cas9
Introduction
Lymphoma, a cancer characterized by a malignant tumor of the immune system that originates in the lymph nodes or lymphoid tissue, is one of the most common cancers worldwide. According to GLOBOCAN, the incidence of non-Hodgkin lymphoma (NHL) in both males and females ranked top ten among all cancers in the world in 2020, lymphomas are the most common malignant tumors in China, As most lymphomas are highly invasive, their associated morbidity and mortality are increasing rapidly worldwide. According to statistics.1 Despite significant headway having been achieved in the treatment of lymphoma, some patients suffering from refractory or recurrent conditions are still struggling with poor therapeutic effects and prognosis. Hodgkin lymphoma (HL) has a good prognosis, with a five-year overall survival (OS) rate of 81.3%.2 In contrast, the therapeutic effects of NHL, which accounts for the vast majority of lymphomas, is much worse. Taking subtype diffuse large B-cell lymphoma as an example, it has a five-year OS rate of only 42.9%.3 As lymphoma takes a heavy toll on human health, breakthroughs in pathogenesis and therapy are urgently needed to lower the recurrence and fatality rate.
Radiotherapy is one of the key tools in the treatment of lymphoma as it can effectively improve therapeutic effects and relieve symptoms. Although radiotherapy is very effective against most lymphoma subtypes, some patients still experience relapse or poor outcomes due radiotherapy resistance. As for patients with diffuse histiocytic lymphoma, the local failure rate after radiotherapy remains 21–37% regardless of the dose of radiotherapy.4 Several studies have shown that the level of gene expression is related to the radiosensitivity of tumor cells.5,6 With that being noted, radiotherapy resistance in lymphoma has rarely been reported, meaning the relationship between gene expression and lymphoma radiotherapy resistance as well as the specific mechanism thereof remain unclear. For this reason, we here seek to gain deeper insight into potential radiotherapy-resistant lymphoma genes, which could serve as potential biomarkers to indicate radiotherapy resistance in lymphoma. This can help predict radiotherapy sensitivity and provide guidance in terms of making treatments more accurate. Furthermore, these genes can be used as new treatment targets, providing both an experimental and theoretical basis for the development of lymphoma drugs. This project has great clinical application value and scientific significance.
CRISPR (an acronym for clustered regularly interspaced short palindromic repeats)/Cas (CRISPR-associated proteins) is an adaptive immune system existing in bacteria and archaea that protects bacteria from damage caused by virus invasion by defending against foreign DNA and plasmids.7,8 Built on the CRISPR/Cas system, the newly emerging third-generation genome editing technology CRISPR/Cas9 stands out for its good precision, effectiveness, and practicability.9 CRISPR/Cas9 mainly includes Cas9 protein, which cuts DNA double strands, and single-guide RNA (sgRNA), which guides Cas9 to specific sites in DNA. Currently, CRISPR/Cas9 is used for not only gene editing; additionally, researchers have developed a gene expression regulation technology mediated by such a system. With this technology, two active sites on Cas9 are mutated into dead Cas9 (dCas9), which loses its endonuclease activity yet is still capable of specifically binding to DNA under the guidance of single-guide RNA (sgRNA). CRISPRa (Transcription Activation), as a new member of gene overexpression technology, promotes the transcription of genome specific loci by linking the catalytic inactivated Cas9 (dCas9) to the transcriptional activator. The closer CRISPRa is to the transcription starting site, the stronger the effect of transcriptional activation is. If CRISPRa targets the upstream of the transcription start site (more than 200 bp), the triggered transcription will be more moderate and closer to the physiological level. The vector system can be used to activate the transcription of a single gene or a series of genes, and can also be used for large-scale screening of genomes. The gRNA sequence library is used to generate the gRNA/MS2 expression vector library. CRISPR activation (CRISPRa) connects dCas9 to transcription activators, such as P65 and VP64, to achieve transcriptional activation at specific sites in the genome and promote the expression of specific genes.10 CRISPR systems as a tool to study cancer, has the potential to revolutionize cancer therapy, The impact of CRISPR on cancer biology research The precision medical strategy for cancer relies on the discovery of gene mutations that promote cancer growth. CRISPR gene editing technology can quickly and effectively produce gene knockout, regulate endogenous gene expression and replicate cancer related genomic changes. Cancer cells are constantly changing, resulting in complex genetic and epigenetic maps. CRISPR technology can simulate complex mutation profiles in cancer. Cloned derivatives branch and compete with the evolution of cancer cell populations into different and different mutant entities, while the cellular composition of tumors (cancer cells, stroma and immune cells) is very dynamic, and CRISPR technology can also track the evolution of tumors.11–13
In this study, the genome-wide Human CRISPRa library was used to screen lymphoma radiotherapy-resistant genes by CRISPRa. The library has more than 70,000 sgRNAs, which can target more than 20,000 genes in the human genome with high infection and activation efficiency. The screened genes were then subject to bioinformatics analysis to identify gene pathways that might be related to radiotherapy resistance in lymphoma. This helps to further clarify the mechanism of lymphoma radiotherapy resistance and provide new molecular biological implications for the treatment of lymphoma.
Materials and Methods
Materials
Cell Lines, CRISPRa Library
HANK-1 and NK-92 cell lines (purchased from HYYMed company).
The Human CRISPRa library (SAM-2 plasmid system) was purchased from addgene (http://www.addgene.org/crispr/libraries/).
Reagents
- Modified 1640 Medium (HyClone)
- Dulbecco’s Modified Eagle Medium (DMEM) (HyClone)
- Fetal bovine serum (FBS) (Gibco)
- CCK8 regent (Beyotime)
- Double antibody (penicillin/streptomycin stock) (TBD)
- High pure total RNA fast isolation kit with spin column (BIOTEKE)
- Recombinant DNase I (RNase-free) (TAKARA)
- Recombinant Ribonuclease Inhibitor (TAKARA)
- Oligo (dT)18 Primer (TAKARA)
- Random Primer (hexadeoxyribonucleotide mix: pd(N)6) (TAKARA)
- Reverse Transcriptase M-MLV (RNase H-) (TAKARA)
- Deoxynucleotide triphosphate mixture (TAKARA)
- SYBR Green I (TOYOBO)
- HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme)
- Hieff TMqPCR SYBR® Green Master Mix (LoW Rox Plus) (Yeasen)
- DL 2000 DNA marker (TAKARA)
- Annexin V-APC/7AAD kit (Multi Sciences)
Instruments
- Water-Jacketed CO2 Incubator (Thermo)
- Regular optical microscope (Nikon)
- Multi-function microplate reader (Thermo)
- Fluorescence quantitative PCR instrument (Bioer)
- Tanon-1600 Gel Imaging System (Tanon)
- JY200C Electrophoresis System (Junyi)
- Ultramicro Nucleic Acid Analyzer Nano-200 (AllSheng)
- Flow cytometer (BD)
Primer Design
β-actin primer sequence: F: AGCGAGCATCCCCCAAAGTT, R: GGGCACGAAGGCTCATCATT; product size: 285 bp; sgRNA primer sequence: F: TTTTAGAGCTAGGCCAACATGA, TGGCCAAGTTGATAACGGAC; product size: 86 bp.
Method
Determination of the Minimum Lethal Radiation Dose
Successfully grown HANK-1 and NK-92 cells were subject to radiotherapy for three days (radiation doses: 0, 3, 6, 9, 12, and 15 Gy, respectively). The treated cells were centrifuged, counted, and prepared in a cell suspension (200 µL/well) based on the count and inoculation size (8000 cells/well). They were then inoculated to a 96-well plate and incubated in an incubator (37°C, 5% CO2) for 72 h, with 10 µL of CCK8 solution added to each well. Then, the culture plate was incubated in the incubator for 2 h, after which the absorbance at 450 nm was measured with a microplate reader, and the minimum lethal radiation dose was determined.
Construction of Cell Lines in Activation Library and Verification of Their Activation Effect
Lentivirus Packaging
- One day in advance, 3×106 293T cells were plated in a 10-cm dish, and 10 mL of DMEM medium containing 10% FBS was added.
- The cells were triturated to ensure they had even growth. The cell confluence reached approximately 90% during transfection.
- Plasmid transfection and lentivirus concentration: 9 µg each of Human CRISPRa library, PXD9a, and PXD9a-helper were mixed with viral packaging plasmids (3 µg of S3 + S4 + S5) and 0.6 mL of serum-free DMEM.
- Polyethyleneimine (PEI) reagent (36 µL), mixed well by shaking before use, was added to the plasmid mixture and then triturated evenly or centrifuged spirally.
- The reagent was incubated at room temperature for 15–20 min.
- The transfection reagent–DNA mixture was added to 293T cells. The medium was changed 6 h after the transfection reagent mixture was added.
- The dish was shaken gently to ensure the compounds it contained were evenly distributed.
- The dish was returned to the incubator (37°C, 5% CO2).
- The supernatant was collected after 24 h of transfection. Toxin production usually peaks within 48 hours. Viral fluid (10 mL) was collected in a 15-mL tube and centrifuged at room temperature (3000 rpm, 5 min).
- The virus supernatant was concentrated to 1 mL, transferred to a 1.5 mL EP tube, and stored at –80°C.
Infection of HANK-1 and NK-92 Cells with Lentivirus
- The target HANK-1 and NK-92 cells were plated in a 15-cm dish one day before transfection.
- Large-scale infection was carried out based on a multiplicity of infection of 0.5 to construct activation library group cell lines and negative control (NC) group cell line. The activation library group received 1×107 HANK-1 or NK-92 cells, 20 µL of Human CRISPRa library transfected virus supernatant, 10 µL of PXD9a-helper transfected virus supernatant, 10 µg of Polybrene, and 10 mL of 1640 Medium. The NC group received 2.5×106 HANK-1 or NK-92 cells, 16.4 µL of PXD9a transfected virus supernatant, 4.1 µL PXD9a-helper transfected virus supernatant, 10 µg of Polybrene, and 10 mL of 1640 Medium.
- The cell lines were returned to the incubator (37°C, 5% CO2) for 48 h of incubation.
- Puromycin (2 µg/mL) was added to the culture for positive screening, and cells that survived after 24 hours were considered stably transfected by the CRISPRa library.
- The activation effect of the activation library cell lines was identified by quantitative polymerase chain reaction (qPCR).
Screening of Radiotherapy-Resistant Cells from Cell Lines in Activation Library
HANK-1-NC, HANK-1-activation library, NK-92-NC, and NK-92-activation library cell lines were irradiated with the minimum lethal radiation dose for three days, and the cells that survived were considered radiotherapy-resistant. The cells were digested, inoculated and cultured for three days, and then recorded using a digital camera. All viable cells were collected for subsequent experiments. Another group of non-irradiated cells was used as a blank control group.
Screening of Differential Gene with Next-Generation Sequencing and Bioinformatics Analysis
The screened radiotherapy-resistant cells were subject to an expanding culture, extraction of total RNA, and reverse transcription into copy DNA (cDNA). Afterwards, the coding region of sgRNA was amplified by qPCR, next-generation sequencing (NGS) was performed with a Hiseq2500 System (Illumina Inc., San Diego, CA, USA), and sgRNA reads were counted using Htseq. After organizing the total reads and sgRNA diversity, a Venn diagram was drawn. Differential genes were selected from the intersection, with those with an abundance (50+) of sgRNAs and more than one independent sgRNA considered potential radiotherapy-resistant genes. A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed on the differential genes using the KOBAS online database. By reviewing relevant literature, we selected a number of radiotherapy-resistant genes for subsequent verification as to whether high gene expression could lead to radiotherapy resistance in lymphoma cells.
Construction of Activated Cell Lines Transfected with Single Gene and Verification of Activation Effect
Activated cell lines transfected with single genes were constructed by the transient transfection of cells. Total cell RNA was extracted and reversely transcribed into cDNA, and the activation effect was finally verified with qPCR. The qPCR reaction system included 12 µL of RNA (primer denaturing solution), 4 µL of 5X M-MLV buffer, 1 µL of d(N)TP, 0.5 µL of RRI, and 2 µL of M-MLV (20 µL in total).
Verification of Radiotherapy Resistance of Activated Cell Lines Transfected with Single Genes
Detection of cell apoptosis with flow cytometry: The cell lines in the control group and the activated cell lines transfected with single genes in the experimental group were irradiated at the minimum lethal radiation dose for three days. The cells were then digested, and 100 µL of 1× binding buffer was added to each sample to resuspend the cells (cell quantity in each vial: 1 × 105–1 × 106). Next, 5 µL of Annexin V-APC and 10 µL of 7AAD were added to each sample, followed by mixing in a gentle vortex before 15 min of incubation at room temperature in the dark. Finally, the samples were tested by flow cytometry and then subject to FlowJo analysis (x-coordinates: Annexin V-APC, y-coordinates: 7AAD).
Detection of mRNA Expression of Single-Gene Radiotherapy-Resistant Cell Lines with qPCR
The mRNA expression of single-gene radiotherapy-resistant cell lines was next detected using qPCR.
Statistical Analysis
A statistical analysis and plotting of the data were carried out using SPSS 22.0 and GraphPad Prism 5. All data were expressed as means ± standard deviation, and sample means were compared using an independent-sample t-test or a one-way analysis of variance. All comparisons of the hypothesis test were two-sided, with being p < 0.05 regarded as indicating statistical significance and p < 0.001 as indicating a significant difference.
Results
Minimum Lethal Radiation Dose
After the HANK-1 and NK-92 cells had been subject to 3, 6, 9, 12, or 15 Gy of irradiation for three days, the corresponding optical density value of each group was determined by CCK8. The results revealed that the radiation had a dose-dependent inhibitory effect on cell growth. The HANK-1 cells barely survived after 9 Gy of irradiation, and all died after 12 Gy. Furthermore, all NK-92 cells died after 9 Gy. Therefore, in the subsequent experiments, 9 Gy was selected as the minimum lethal radiation dose for screening radiotherapy-resistant cells.
Successful Construction of Activation Library Cell Lines
The HANK-1 and NK-92 activation library and negative control group cell lines were constructed by lentiviral transfection, including the HANK-1 activation library-pXD9a-helper, NK-92 activation library-pXD9a-helper, HANK −1-pXD9a-pXD9a-helper, and NK-92-pXD9a-pXD9a-helper cell lines. The cells stably transfected with CRISPRa library were obtained after puromycin positive screening, after which the activation effect was verified by qPCR. According to the results on the HANK-1 cells, both the negative control group (pXD9a-helper) (t = 14.44, p = 0.0048) and the activation library group (t = 12.00, p = 0.0069) exhibited a sharp rise in sgRNA expression when compared with the Wild Type (WT) group, indicating that the HANK-1 cell line activation library was positively transfected. As for the NK-92 cells, both the negative control group (pXD9a-helper) (t = 15.07, p = 0.0044) and the activation library group (t = 71.20, p = 0.0002) exhibited a sharp rise in sgRNA expression when compared with the WT group, demonstrating that the NK-92 cell line activation library was positively transfected.
Results of Screening for Radiotherapy-Resistant Cells from Activation Library
The cell lines in the HANK-1 and NK-92 control and activation library groups were irradiated with the minimum lethal radiation dose of 9 Gy, after which the survival rates were investigated using photographic observation. The results are shown in Figure 1. Cells in the HANK-1 activation library group and the NK-92 activation library group survived better at an irradiation dose of 9 Gy when compared with the control group. The cells that survived after 9-Gy radiotherapy were considered radiotherapy-resistant cells and sent for NGS.
Results of NGS and Bioinformatics Analysis
The HANK-1 and NK-92 differential genes were analyzed using a Venn diagram based on the NGS results, wherein there were 129 HANK-1 and NK-92 genes with at least 1 independent sgRNA (see Figure 2A), and 26 with more than 1 independent sgRNA (see Figure 2B). Additionally, there were 47 genes with an abundance (50+) of sgRNA (see Figure 2C), and 16 differential genes with 50+ sgRNA and more than 1 independent sgRNA (see Figure 2D). These included C20orf203, MTFR1, TAF1L, MYADM, NIPSNAP1, ZUP1, RASL11A, PSMB2, PSMA6, OR8H3, TMSB4Y, CD300LF, EEF1A1, ATP6AP1L, TRAF3IP2, and SNRNP35. A total of 16 differential genes were screened, of which 9 (MTFR1, TAF1L, NIPSNAP1, ZUP1, PSMB2, PSMA6, TMSB4Y, EEF1A1, and TRAF3IP2) have been previously reported as associated with tumors. Although the sgRNA abundance of AKT3 was only 1482, it had 3 independent sgRNA. Furthermore, there have been many reports on the correlation between AKT3 and tumors, and the 10 genes above were used in the next part of the experiment to construct activated cell lines transfected with single genes for further verification of the relationship between gene expression and radiotherapy resistance. The sgRNA abundance of these 10 genes is shown in Figure 2E. A KEGG enrichment analysis was performed on 129 genes with at least 1 independent sgRNA, and the results are shown in Figure 2F.
Successful Construction of 10 Activated Cell Lines Transfected with a Single Gene
Activated cell lines transfected with a single gene were constructed using the above 10 screened genes by cell transient transfection, and the activation effect was verified with qPCR. The results are shown in Figure 3. For HANK-1 cells, the mRNA expression of 10 genes was significantly increased after transient transfection when compared with the control group (HANK1-NC) (t = 8.874, p = 0.0009; t = 36.96, p < 0.0001; t = 9.117, p = 0.0008; t = 11.66, p = 0.0003; t = 12.32, p = 0.0002; t = 4.420, p = 0.0115; t = 8.492, p = 0.0136; t = 6.045, p = 0.0038; t = 27.65, p < 0.0001; t = 11.64, p = 0.0003; respectively). This indicated that the cell lines were successfully activated by HANK-1 single-gene transfection. As for the NK-92 cells, all of the 10 genes exhibited a sharp rise in mRNA expression after transient transfection compared with the control group (NK92-NC) (t = 20.84, p < 0.0001; t = 27.86, p < 0.0001; t = 37.11, p < 0.0001; t = 12.23, p = 0.0003; t = 31.20, p < 0.0001; t = 4.906, p = 0.0080; t = 15.05, p = 0.0044; t = 22.45, p = 0.0020; t = 5.470, p = 0.0318; t = 8.912, p = 0.0009; respectively). This indicated that the cell lines were successfully activated by NK-92 single-gene transfection.
Radiotherapy Resistance of Activated Cell Lines Transfected with Single Gene
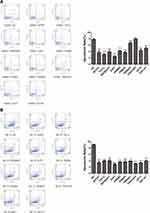
The apoptosis of activated cell lines transfected with a single gene after 9 Gy of irradiation was detected using flow cytometry. As shown in Figure 4, compared with that in the HANK-1-NC group, the apoptosis rates of the activated cells of the MTFR1, TAF1L, NIPSNAP1, ZUP1, PSMB2, PSMA6, TMSB4Y, AKT3, and EEF1A1 genes were significantly lowered (t = 12.75, t = 27.52, t = 11.81, t = 24.15, t = 17.00, t = 18.45, t = 5.286, t = 12.12, and t = 13.01; respectively; all p < 0.01), with significant differences among them. There was no significant difference in the apoptosis rate of the activated cells of the TRAF3IP2 gene (t = 0.5319, p = 0.6229) when compared with the control group. This indicated that the radiotherapy resistance of the TRAF3IP2 gene was uncertain, while the remaining genes were probably radiotherapy-resistant genes. Compared with that of the NK-92-NC group, the apoptosis rates of the activated cells of the MTFR1, TAF1L, NIPSNAP1, ZUP1, PSMB2, PSMA6, TMSB4Y, TRAF3IP2, AKT3, and EEF1A1 genes were significantly decreased (t = 24.07, t = 39.09, t = 20.15, t = 36.65, t = 42.67, t = 18.84, t = 50.73, t = 37.90, t = 42.77, t = 27.34; respectively; all p < 0.01). This difference was significant, indicating that all of the 10 genes could be radiotherapy resistant.
Detection of mRNA Expression of Activated Cell Lines Transfected with Single Gene After Radiotherapy with qPCR
The mRNA expression of the activated cell lines transfected with single genes after radiotherapy was detected using qPCR, and the results are shown in Figure 5. Compared with the control group (NK-92), the mRNA expressions of MTFR1, NIPSNAP1, ZUP1, PSMB2, PSMA6, EEF1A1, TMSB4Y and TAF1L in the experimental group (NK-92, 9 Gy) were significantly up-regulated (t = 120.5, p < 0.0001; t = 120.134, p < 0.0001; t = 48.20, p = 0.0004; t = 26.06, p = 0.0002; t = 45.49, p = 0.0005; t = 27.99, p = 0.0013; t = 4.814, p = 0.0406; t = 16.91, p < 0.0001; respectively). At the same time, the mRNA expression was not statistically different between AKT3 and TRAF3IP2 (t = 4.116, p = 0.0543; t = 2.434, p = 0.1354; respectively). The results showed that the high expression of MTFR1, NIPSNAP1, ZUP1, PSMB2, PSMA6, EEF1A1, TMSB4Y and TAF1L can lead to radiotherapy resistance in cells, while the relationship between AKT3, and TRAF3IP2 and radiotherapy resistance is uncertain.
 |
Figure 5 qPCR relative quantitative standardization results (compared with the control group; ***p < 0.01, *p < 0.05). |
Discussion
In China, lymphoma accounted for 2.9% of 9.6 million cancer deaths globally in 2018.14 For this reason, there is a need to identify new reliable biomarkers and therapies to improve the therapeutic effect of lymphoma. Radiotherapy is one of the integral treatment strategies for lymphoma. However, its clinical applications are limited by toxicity and side effects arising from radiation. Furthermore, lymphoma patients may also grapple with clinical radiotherapy failure and disease recurrence and progression due to radiotherapy resistance, which also occurs in the treatment of other malignant tumors. Therefore, 16 potential radiotherapy-resistant genes were here screened using the human genome-wide CRISPRa technology, and further experiments were carried out to confirm that the high expression of MTFR1, NIPSNAP1, ZUP1, PSMB2, PSMA6, EEF1A1, TMSB4Y and TAF1L can reduce the apoptosis of lymphoma cells and increase the probability of radiotherapy resistance. These findings are of great clinical significance for further elucidating the mechanism of radiotherapy resistance and will assist in seeking new treatment targets and predicting the radiosensitivity of lymphoma.
By causing damage to tumor cells, radiotherapy can result in various types of cell death, such as apoptosis, necrosis, and autophagy, constituting one of the mechanisms through which radiation therapy fights against tumors. Existing studies suggest that the mechanism of tumor radiotherapy resistance involves DNA damage repair, apoptosis, cell hypoxia, cell cycle, autophagy, and other biological processes and also that it is related to tumor stem cells and the tumor microenvironment.15 To determine the optimal radiation dose for subsequent screening of radiotherapy-resistant tumor cells, the present study irradiated radiotherapy-sensitive HANK-1 and NK-92 lymphoma cells with six gradient doses of radiation and then detected the cell survival rates using CCK8. According to the results, 9 Gy was set as the minimum lethal radiation dose for screening radiotherapy-resistant cells.
Current studies suggest that the occurrence and development of tumors as well as drug and radiotherapy resistance are all related to abnormal gene expression. Studies have found that high expression of PAPST in human B lymphoid cell lines reduces radiation-induced apoptosis.16 Changes in gene expression were also found in radiotherapy-resistant lymphoma cells. A high expression of APOBEC3G can improve the repair of ionizing radiation-induced genomic DNA double strand breaks by lymphoma cells and the survival of irradiated cells, thereby increasing the radiotherapy resistance of lymphoma cells.17 All these reveal that high tumor specific gene expression may make tumors resistant to radiotherapy.
In recent years, CRISPR/Cas9 has grown rapidly and become a powerful genetic screening tool for use in combination with high-energy sequencing technology which more used than traditional RNA interference (RNAi) technology18. CRISPR/Cas9 technology not only enables genome-wide gene editing but also activates or inhibits gene expression, thereby screening and identifying key genes that regulate phenotypes.19 Currently, this technology has been widely applied in the systematic comprehensive analysis and screening of genes associated with tumor growth, invasion, and drug resistance, with a view to better revealing functional mechanisms of tumors. By using a genome-scale CRISPR/Cas9 knockout library, studies have screened and identified 9 genes that may be involved in imatinib resistance, providing a reference for the in-depth exploration of its molecular mechanisms.20 Most previous studies have inhibited or silenced gene expression with RNAi or CRISPRi to study gene functions. Until now, no study has been reported on the screening of lymphoma radiotherapy-resistant genes with CRISPRa. The present study thus used human genome-wide CRISPRa to screen genes related to radiotherapy resistance in lymphoma. First, cell lines from the CRISPRa library were constructed using lentiviral transfection. Those stably transfected with the library were obtained through a puromycin drug sieve, after which qPCR was used to verify the successful construction of the activation library cell lines.
To further investigate the genes whose high expression makes lymphoma cells less sensitive to radiotherapy and results in radiotherapy tolerance, and to further analyze which signaling pathways are involved in radiotherapy resistance, this study irradiated activation library cell lines and negative control group cell lines with 9 Gy of irradiation and observed the cell survival rates through photographs. The results showed that the activation library group acquired radiotherapy resistance. To screen differential genes and possible gene pathways, the radiotherapy-resistant cells were subjected to NGS, with 16 differential genes screened as potential lymphoma radiotherapy-resistant genes. Furthermore, a KEGG enrichment analysis was carried out for 129 genes with at least 1 independent sgRNA. The KEGG enrichment signaling pathways mainly include the neuroactive ligand–receptor interaction, the nuclear factor-kappaB signaling pathway, FcγR-mediated phagocytosis, the longevity regulation pathway, and the hypoxia-inducible factor-1 signaling pathway. These pathways involve immune and senescence related gene pathways, and these are key pathways that determine cell life, such as aging is a complex process of accumulation of molecular, cellular, and organ damage, leading to loss of function and increased vulnerability to disease and death. Related transcription factors (FOXO, DAF-16, Gis1, and Msn2/4) transactivate genes involved in resistance to oxidative stress, energy metabolism, DNA damage repair, glucose metabolism, autophagy and protection of proteins by chaperones.These signaling pathways may play a important role in the occurrence of radiotherapy resistance.
To further verify that the high expression of differential genes can indeed lead to radiotherapy resistance in lymphoma cells, this study screened 10 genes for experimental verification, wherein 10 cell lines were activated by single-gene transfection were constructed respectively and subject to 9 Gy of irradiation. Afterwards, cell apoptosis was detected by flow cytometry. The results showed that the apoptosis rate of the cell lines activated by single-gene transfection was much lower than that of the control group, indicating that these genes can promote cell survival after transfection, resulting in resistance to radiotherapy. Finally, the mRNA expression of cell lines activated by single-gene transfection after radiotherapy was detected with qPCR to determine the expression of these genes in radiotherapy-resistant cell lines. The results showed that the mRNA expression of MTFR1, NIPSNAP1, ZUP1, PSMB2, PSMA6, EEF1A1, TMSB4Y and TAF1L in the experimental group was significantly up-regulated when compared with that in the control group. Accordingly, it is believed that the high expression of these 8 genes could lead to cell radiotherapy resistance and that they may be radiotherapy-resistant genes.
Except for C20orf203, MTFR1 exhibited the greatest abundance of sgRNA in this study. Previous studies have found that mitochondria play an important role in the process of apoptosis.21 The MTFR1 mitochondrial protein contains a short polyproline-rich region. It targets the inner mitochondrial membrane and promotes mitochondrial fission.22 Studies have shown that mitochondrial fission can promote tumor progression and that MTFR1 has high expression in various tumors, such as squamous cell carcinoma of head and neck, esophageal cancer, tongue cancer, and squamous cell carcinoma of the lung.23,24 Studies have shown that increased expression of MTFR1 can promote the proliferation, invasion, and migration of lung adenocarcinoma and reduce apoptosis by adjusting the AMPK/mTOR signaling pathway.25 Furthermore, studies have also reported that a lack of MTFR1 can lower the antioxidant activity of cells, resulting in oxidative DNA damage, thus inducing apoptosis.26 In this study, a high expression of MTFR1 was found to reduce the apoptosis of lymphoma cells and lead to radiotherapy resistance, which is consistent with the conclusion that MTFR1 can promote tumor progression, although more experiments are required to verify and explore its specific mechanism. We speculate that the reason why MTFR1 may become a biomarker is that MTFR1 gene involves mitochondrial function. Its basic biological function is antibody to mitochondrial fission regulatory protein 1. In the process of radiation damage, the repair of mitochondrial DNA strand damage plays a key role. Therefore, MTFR1 may play a role in radiation resistance of lymphoma and may become a biomarker.
The ZUP1 enzyme is a type of deubiquitinating enzyme (DUB). Deubiquitinating enzymes can be divided into seven major families and are involved in various physiological activities, such as autophagy, DNA damage response regulation, DNA double-strand break repair, and cell cycle and apoptosis regulation.27,28 Members of the DUB family are involved in the post-translational modification of various cancer-related proteins, thereby regulating the occurrence, development, and prognosis of cancer. Ubiquitin-specific protease (USP) 53 is a member of the DUB family. Furthermore, Nrf2, an important antioxidant protein in cells, can reduce the damage of ionizing radiation to cells as its excessive activation can enhance the antioxidant capacity of cells, thereby reducing the sensitivity of cells to radiotherapy. A high expression of USP53 increases the expression of Nrf2 in esophageal cancer Eca109 cells, thus leading to radiotherapy resistance. ZUFSP/ZUP1, a type of cysteine protease (zinc finger-containing ubiquitin peptidase 1), is one of the seven DUB families. ZUFSP/ZUP1 localizes to damaged DNA and plays a key role in the genome stabilization pathway, acting to prevent spontaneous DNA damage and promote cell survival in response to exogenous DNA damage.29 Therefore, a high expression of ZUP1 can promote cell survival, which is consistent with the findings of this study.
It has been previously confirmed that eEF1A is highly expressed in many malignant tumors, such as leukemia, renal cell carcinoma, ovarian cancer, and lymphoma.30,31 Several studies have shown that eEF1A1 can make tumors more invasive and migratory, meaning it is considered an oncogene. Although the functions of eEF1A1 and its physiological processes have been extensively studied, its specific regulatory mechanisms remain unclear. A team led by Huang Yi demonstrated that the eEF1A1 gene is related to the proliferation and apoptosis of human T-cell acute lymphoblastic leukemia cell lines by knocking it out and restoring its expression. The specific mechanism thereof may be related to the PI3K/Akt/NF-κB and PI3K/Akt/mTOR signaling pathways.32 Another study reported that the survival of B-cell lymphoma and the generation of radiotherapy resistance are built on NF-κB signaling and that the inhibition of NF-κB can enhance the sensitivity of ionizing radiation, resulting in significant anti-tumor effects.33 Cell cycles are regulated by a variety of factors, among which cyclin D1 is a key protein in phase G1 of the cell cycle, and its abnormal expression is closely related to cell carcinogenesis.34,35 Studies have shown that after down-regulating the expression of eEF1A1, the proliferation of hepatoma cells is inhibited, cell cycle is arrested in phase G1, and the expression of cyclin D1 is also significantly reduced. Further studies have shown that eEF1A1 can control the proliferation of tumor cells through STAT1-cyclin D1 axis.36 In this experiment, cell lines highly expressing eEF1A1 obtained stronger radiotherapy resistance with decreased apoptosis, and their KEGG was enriched in the NF-κB signaling pathway, which is consistent with the above research conclusions. The mechanism may be that the highly expressed eEF1A1 affects cell cycles by regulating the expression of cyclin D1, thereby promoting the proliferation of tumor cells, resulting in decreased radiosensitivity, meanwhile, we speculate that this may be related to the fact that the methylation of non histone eEF1A can promote tumorigenesis. The regulation and modification of epigenetics and chromatin biology are involved in growth and development and diseases, especially tumorigenesis, through regulating epigenetics (such as p53, RB, RelA), these also needs to be verified by further experiments.
In conclusion, the study found that the high expression of certain genes contributes to the radiotherapy resistance of lymphoma cells. These findings are of both theoretical and clinical application values. However, when verifying the correlation between a single gene and radiotherapy resistance, the present study only verified the high expression of the genes and not gene knockout. Furthermore, the molecular pathways downstream of radiotherapy-resistant genes were not investigated. For this reason, more in-depth studies are required to explore the relevant mechanisms in the future. Finally, as cells were the object of this study, they need to be further verified at the animal level and even in clinical trials.
Conclusion
In summary, this study reported 16 genes that could act as potential lymphoma radiotherapy-resistant genes, and 8 high expression genes was associated with lymphoma radiotherapy resistance based on experimental result, which will provide a new basis for the mechanism of lymphoma radiotherapy resistance. The 8 genes may become potential biomarkers and targets for predicting lymphoma radiotherapy resistance, and they are worthy of further study.
Abbreviation
CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; qPCR, Quantitative Polymerase Chain Reaction; sgRNA, single-guide RNA; GLOBOCAN, Global Cancer Statistics; HL, Hodgkin Lymphoma; NHL, Non-Hodgkin Lymphoma; OS, Overall Survival; Cas, CRISPR-associated proteins; dCas9, Dead Cas9; CRISPRi, CRISPR interference; CRISPRa, CRISPR activation; CCK8, Cell Counting Kit-8; MOI, Multiplicity of Infection; NC, Numerical Control; KEGG, Kyoto Encyclopedia of Genes and Genomes; OD, Optical Density; WT, Wild Type; RNAi, RNA Interference; MTFR1, Mitochondrial Fission Regulator 1; ZUP1, Zinc finger-containing ubiquitin peptidase 1; DUB, Deubiquitinating enzyme; Nrf2, Nuclear factor-erythroid 2-related factor 2; USP53, Ubiquitin-Specific Protease 53; eEF1A, eukaryotic translation elongation factor 1A.
Ethics Approval
This study is a basic study and does not involve human participants and experimental animals.
Acknowledgments
We are particularly grateful to all the people who have given us help on our article.
Disclosure
The authors declare that they have no competing interests.
References
1. Maomao C, Wanqing C. Interpretation on the global cancer statistics of GLOBOCAN 2020. Chin J Front Med Sci. 2021;13(03):63–69. Chinese.
2. Hu YF, Huang YH, Luo W, Chen MX, Zhang J, Gou F. Clinical characteristics and analysis of prognostic factors of 222 patients diagnosed with Hodgkin’s lymphoma. Zhonghua Yi Xue Za Zhi. 2019;99(48):3792–3796. doi:10.3760/cma.j.issn.0376-2491.2019.48.007
3. Han Y, Qin Y, He X-H, et al. Clinical features and prognostic analysis of 370 cases of advanced diffuse large B-cell lymphoma. Chin J Oncol. 2018;40(06):456–461. Chinese. doi:10.3760/cma.j.issn.0253-3766.2018.06.011
4. Fuks Z, Kaplan HS. Recurrence rates following radiation therapy of nodular and diffuse malignant lymphomas. Radiology. 1973;108(3):675–684. doi:10.1148/108.3.675
5. Lee JY, Kim JH, Bang H, et al. EGR1 as a potential marker of prognosis in extranodal NK/T-cell lymphoma. Sci Rep. 2021;11(1):10342. doi:10.1038/s41598-021-89754-8
6. Troschel FM, Linsenmaier M, Borrmann K, Eich HT, Götte M, Greve B. Heparanase expression is associated with cancer stem cell features and radioresistance in Hodgkin’s lymphoma cells. Anticancer Res. 2021;41(7):3299–3308. doi:10.21873/anticanres.15117
7. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi:10.1126/science.1231143
8. Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36(1):244–246. doi:10.1046/j.1365-2958.2000.01838.x
9. Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–1575. doi:10.1046/j.1365-2958.2002.02839.x
10. Perez-Pinera P, Kocak DD, Vockley CM, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10(10):973–976. doi:10.1038/nmeth.2600
11. Katti A, Diaz BJ, Caragine CM, Sanjana NE, Dow LE. CRISPR in cancer biology and therapy. Nat Rev Cancer. 2022;22(5):259–279. doi:10.1038/s41568-022-00441-w
12. Vaghari-Tabari M, Hassanpour P, Sadeghsoltani F, et al. CRISPR/Cas9 gene editing: a new approach for overcoming drug resistance in cancer. Cell Mol Biol Lett. 2022;27(1):49. doi:10.1186/s11658-022-00348-2
13. Liu D, Zhao X, Tang A, et al. CRISPR screen in mechanism and target discovery for cancer immunotherapy. Biochim Biophys Acta Rev Cancer. 2020;1874(1):188378. doi:10.1016/j.bbcan.2020.188378
14. Liu W, Liu J, Song Y, et al. Burden of lymphoma in China, 2006–2016: an analysis of the Global Burden of Disease Study 2016. J Hematol Oncol. 2019;12(1):115. doi:10.1186/s13045-019-0785-7
15. Kaiwen N, Xin Z, Xinting Z, et al. Research progress on the correlation between hypoxic microenvironment and radiotherapy resistance in head and neck squamous cell carcinoma. Chin J Otorhinolaryngol-Skull Base Surg. 2022;28(01):123–127. Chinese.
16. Nakayama F, Umeda S, Ichimiya T, et al. Sulfation of keratan sulfate proteoglycan reduces radiation-induced apoptosis in human Burkitt’s lymphoma cell lines. FEBS Lett. 2013;587(2):231–237. doi:10.1016/j.febslet.2012.12.002
17. Nowarski R, Wilner OI, Cheshin O, et al. APOBEC3G enhances lymphoma cell radioresistance by promoting cytidine deaminase-dependent DNA repair. Blood. 2012;120(2):366–375. doi:10.1182/blood-2012-01-402123
18. Hai-Ling LIU, Xu-Jun W, Dan LI. Application of CRISPR/Cas9 high-throughput screening technology for identifying new target in tumor therapy. Chin J Biochem Mol Biol. 2016;32(02):133–139. Chinese.
19. Wang G, Ma M, Ye Y, et al. High-throughput functional screening using CRISPR/Cas9 system. Hereditas. 2016;38(05):391–401. Chinese. doi:10.16288/j.yczz.15-329
20. Cao J, Wei J, Yang P, et al. Genome-scale CRISPR-Cas9 knockout screening in gastrointestinal stromal tumor with Imatinib resistance. Mol Cancer. 2018;17(1):121. doi:10.1186/s12943-018-0865-2
21. Chun-sun J, Wei-ming X, Quan C. Mitochondrial fission, fusion and apoptosis. Acta Biophysica Sinica. 2007;4:256–264. Chinese.
22. Tonachini L, Monticone M, Puri C, et al. Chondrocyte protein with a poly-proline region (CHPPR) is a novel mitochondrial protein and promotes mitochondrial fission. J Cell Physiol. 2004;201(3):470–482. doi:10.1002/jcp.20126
23. Jia Y, Chen X, Zhao D, Ma S. SNHG1/miR-194-5p/MTFR1 axis promotes TGFβ1-induced EMT, migration and invasion of tongue squamous cell carcinoma cells. Mol Biotechnol. 2022;64(7):780–790. doi:10.1007/s12033-021-00445-1
24. Weiping F, Xinna D, Wenyan C, et al. Expression and prognostic significance of mitochondrial fission regulator 1 based on TCGA pan-cancer analysis. J Mod Oncol. 2020;28(04):648–651. Chinese.
25. Li Y, Liu Y, Jin K, et al. Negatively regulated by miR-29c-3p, MTFR1 promotes the progression and glycolysis in lung adenocarcinoma via the AMPK/mTOR signalling pathway. Front Cell Dev Biol. 2021;9:771824. doi:10.3389/fcell.2021.771824
26. Monticone M, Tonachini L, Tavella S, et al. Impaired expression of genes coding for reactive oxygen species scavenging enzymes in testes of Mtfr1/Chppr-deficient mice. Reproduction. 2007;134(3):483–492. doi:10.1530/REP-07-0199
27. Jia X, Li Q. Research advances on deubiquitinating enzymes involved in the development of hepatocellular carcinoma. Chin J Clin Oncol. 2020;47(05):260–264. Chinese.
28. Kee Y, Huang TT. Role of deubiquitinating enzymes in DNA repair. Mol Cell Biol. 2015;36(4):524–544. doi:10.1128/MCB.00847-15
29. Kwasna D, Abdul Rehman SA, Natarajan J, et al. Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Mol Cell. 2018;70(1):150–164.e6. doi:10.1016/j.molcel.2018.02.023
30. Bao Y, Zhao TL, Zhang ZQ, et al. High eukaryotic translation elongation factor 1 alpha 1 expression promotes proliferation and predicts poor prognosis in clear cell renal cell carcinoma. Neoplasma. 2020;67(1):78–84. doi:10.4149/neo_2019_190224N158
31. Gong T, Shuang Y. Expression and clinical value of eukaryotic translation elongation factor 1A1 (EEF1A1) in diffuse large B cell lymphoma. Int J Gen Med. 2021;14:7247–7258. doi:10.2147/IJGM.S324645
32. Huang Y, Hu JD, Wu YA, et al. Effects of eEF1A1 re-expression on proliferation and apoptosis of Jurkat cells with knocked down eEF1A1 gene and its mechanisms. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013;21(2):279–284. doi:10.7534/j.issn.1009-2137.2013.02.004
33. Zhang Z, Zhao X, Wang D, et al. Targeted in vivo delivery of NF-κB decoy inhibitor augments sensitivity of B cell lymphoma to therapy. Mol Ther. 2021;29(3):1214–1225. doi:10.1016/j.ymthe.2020.11.026
34. Tchakarska G, Sola B. The double dealing of cyclin D1. Cell Cycle. 2020;19(2):163–178. doi:10.1080/15384101.2019.1706903
35. Montalto FI, De Amicis F. Cyclin D1 in cancer: a molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells. 2020;9(12):2648. doi:10.3390/cells9122648
36. Huang J. Overexpression of eEF1A1 regulates G1-phase progression to promote HCC proliferation through the STAT1-cyclin D1 pathway. Biochem Biophys Res Commun. 2017;494(3–4):542–549. Chinese. doi:10.1016/j.bbrc.2017.10.116
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.