Back to Journals » Journal of Hepatocellular Carcinoma » Volume 10
Screening and Verification of Key Ubiquitination Genes Related to Immune Infiltration in Stage III/IV Hepatocellular Carcinoma
Authors Tang Y, Cao J, Peng R, Mao X, Su B, Tang H, Tu D, Zhou J, Jiang G , Jin S, Wang Q, Zhang C , Liu R, Zhang C, Bai D
Received 22 February 2023
Accepted for publication 8 May 2023
Published 22 May 2023 Volume 2023:10 Pages 765—781
DOI https://doi.org/10.2147/JHC.S407536
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Imam Waked
Yuhong Tang,1,* Jun Cao,1,* Rui Peng,1 Xingkang Mao,1 Bingbing Su,1 Hao Tang,1 Daoyuan Tu,1 Jie Zhou,1 Guoqing Jiang,1 Shengjie Jin,1 Qian Wang,1 Chen Zhang,2 Renjie Liu,1 Chi Zhang,1,3 Dousheng Bai1,3
1Department of Hepatobiliary Surgery, Clinical Medical College, Yangzhou University, Yangzhou, People’s Republic of China; 2The Second Affiliated Hospital of Xuzhou Medical University, Xuzhou, People’s Republic of China; 3General Surgery Institute of Yangzhou, Yangzhou University, Yangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Dousheng Bai; Chi Zhang, Department of Hepatobiliary Surgery, Clinical Medical College, Yangzhou University, 98 West Nantong Road, Yangzhou, 225000, People’s Republic of China, Email [email protected]; [email protected]
Introduction: Immune checkpoint therapy (ICIs) effectively improves the prognosis of advanced (stage III/IV) hepatocellular carcinoma (HCC) patients. However, its objective response rate (ORR) is below 20%, significantly limiting ICI use in advanced HCC patients. The level of tumour immune infiltration influences ICI response rate. Recent studies have found ubiquitinase to be an important factor that regulates tumour immune infiltration. Therefore, the aim of this study is to explore the key ubiquitination genes that regulate immune infiltration in advanced HCC and further validate them.
Methods: A biotechnological process was performed as a means of classifying 90 advanced HCC patients into three immune subtypes and identifying associations with immune infiltration in the co-expressed modules. Ubiquitination-related genes were then screened with WGCNA. Gene enrichment analysis was performed for the target module and 30 hub genes were screened out by protein–protein interaction network (PPI). ssGSEA, single-gene sequencing and the MCP counter were used for exploring immune infiltration. TIDE score was applied for predicting drug efficacy and GSEA was used for exploring potential pathways. Finally, GRB2 expression in HCC tissue was validated by in vitro experiments.
Results: GRB2 expression was found to have a significant correlation with the pathological stage and prognosis of HCC patients and a positive correlation with immune infiltration and tumour mutation burden (TMB). In addition, significant correlations with the efficacy of ICIs, sorafenib and transarterial chemoembolization (TACE) were identified. GRB2 was found to be most significantly associated with the JAK-STAT signalling pathway and cytosolic DNA sensing pathway. Finally, it was found that GRB2 expression is closely related to the prognosis, tumour size and TMN stage.
Conclusion: A significant association was observed between the ubiquitinated gene GRB2 and the prognosis and immune infiltration of advanced HCC patients and it may potentially be used for predicting therapy efficacy in advanced HCC patients in the future.
Keywords: ubiquitination, growth factor receptor-binding protein 2, liver cancer, tumour-infiltrating immune cells
Introduction
Liver cancer is the third most common cause of cancer death globally with hepatocellular carcinoma (HCC) accounting for the majority of liver cancer.1,2 Early HCC (stage I/II) treatment mainly includes radical surgical resection, radiofrequency ablation and liver transplantation.3,4 However, there is a lack of typical clinical symptoms with early-stage HCC and over 60% of HCC patients are initially diagnosed at an advanced stage, meaning that the best opportunity for surgery is lost and resulting in a low five-year survival rate (below 16%).5 According to the American Gastroenterological Association (AGA) Clinical Practice Guideline on Systemic Therapy for HCC, patients in stage III or IV were only previously able to receive conservative treatment, including molecular targeted drugs (sorafenib, etc.), TACE and radiation therapy.6 However, these therapies demonstrated limited effect in improving advanced HCC patient prognosis. In recent years, immune checkpoint inhibitors (ICIs) that target PD-1, PD-L1 and CTLA-4 pathways have proven to be effective in many advanced cancers.7,8 Recent studies have reported that a combination of camrelizumab and apatinib has outstanding efficacy in the treatment of advanced liver cancer, with tumour ORR of 34.3% (95% CI: 23.3–46.6%) and a one-year survival rate of 74.7% (95% CI, 62.5–83.5%).9,10 Furthermore, a combination of tremelimumab and TACE treatment was found to increase T lymphocyte accumulation in distant untreated lesions.11 Treatment combining oral hydrogel and ICIs can decrease tumour growth by reversing the immunosuppressive tumour microenvironment (TME).12 Therefore, multiple immune cell subsets infiltrate the TME and play an important role in the response rate of ICIs. It has also been found that ubiquitinated proteases play a key regulatory role in tumour immune infiltration, affecting ICI efficacy.12 Therefore, screening the key molecules of ubiquitinated proteases that regulate immune infiltration in advanced HCC may provide new targets for improving ICI efficacy among HCC patients.
Ubiquitinated proteases (E1, E2, E3) are enzymes that catalyse protein degradation by linking ubiquitin as a means of targeting proteins in a three-step cascade and are closely related to antitumour immunity.13,14 Increasing evidence that ubiquitinated proteases play a key role in TME has been identified in recent years. Yu et al reported that ubiquitinase RNF126 affects the TME and promotes nasopharyngeal carcinoma progression through the regulation of PTEN ubiquitination.15 Wang et al discovered that an overexpression of USP24 in M2 macrophages can promote IL-6 transcription and tumour malignancy.16 It was previously predicted that the ubiquitination function gene CDC20 is related to HCC prognosis and its expression has a positive correlation with the infiltration of B cells, CD8 + T cells, CD4 + T cells and DCs by bioinformatics methods.17 The important role of RNF128 in HCC was also identified and it was found that RNF38 promotes TGF-β signalling in HCC by ubiquitinating and degrading AHNAK.18,19 Recently, immune infiltration was used for constructing a patient prognostic model of the ubiquitinates TRIM family as a means of predicting effective prognostic markers and therapeutic targets for HCC.20 These studies suggest that ubiquitinated proteases play a crucial role in tumourigenesis and immune infiltration. Therefore, an exploration of the key molecules of ubiquitinated genes that regulate the immune infiltration of HCC can provide new targets for improving ICI efficacy among HCC patients.
In this study, the immune infiltration-related ubiquitinase GRB2 was screened using various bioinformatics approaches. Subsequently, the high expression of GRB2 was found to be associated with a worse prognosis and higher immune cell infiltration levels. In addition, GRB2 expression was found to be closely related to sorafenib, TACE and ICI treatment efficiency among HCC patients. Generally, GRB2 could be a novel predictive biomarker for the prediction of therapy efficacy among advanced HCC patients in the future.
Materials and Methods
Data and Information
A total of 1597 ubiquitin-protein ligases were obtained from the Integrated Annotations for Ubiquitin and Ubiquitin-like Conjugation Database (IUUCD) (http://iuucd.biocuckoo.org/species.php?spe=Homo_sapiens).21 Gene expression data and the corresponding clinical information of 90 advanced HCC patients were obtained from the International Cancer Genome Consortium (ICGC) database (https://dcc.icgc.org/projects/LIRI-JP) and TCGA database (https://tcga-data.nci.nih.gov/tcga/). In addition, single-cell RNA sequencing data was taken from the “LIHC GSE140228 10X” and “LIHC GSE98638” datasets of the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo).
Patients and Samples
One hundred and forty pairs of HCC tissues were collected from Yangzhou Clinical Medical College. Thirty pairs were used for quantitative real-time polymerase chain reaction (qRT-PCR) and eight pairs were chosen for Western blotting. All patients in the immunohistochemical staining chip received a radical resection of liver cancer in the Department of Hepatobiliary Surgery, Clinical Medical College, Yangzhou University and the clinical data was complete. Two clinical pathologists independently confirmed the HCC diagnosis. Clinicopathological information was obtained for correlation analysis. All HCC subjects gave their written informed consent in accordance with the Declaration of Helsinki and studies were approved by the Institutional Review Board at Clinical Medical College, Yangzhou University.
ESTIMATE and CIBERSORT Analysis, WGCNA, Enrichment Analysis and PPI
Single-sample gene set enrichment analyses (ssGSEA) derived from the gene set variation analysis (GSVA) package was used for scoring immune infiltration in 90 advanced HCC. In addition, the cumulative distribution function (CDF) method was used to determine the optimal grouping according to the CDF delta region. In order to verify differences in the immunity of clusters, the ESTIMATE package for R (https://sourceforge.net/projects/estimateproject) was used for calculating the tumour purity, stromal score, immune score and ESTIMATE score of each HCC tumour sample. The CIBERSORT package was then used for assessing the distribution of 24 immune cell types in each sample. Results with p < 0.05 were then used in further analysis.
The R package WGCNA was used for co-expression analysis. Based on the above grouping, selected genes relating to ubiquitination were constructed in a sample clustering tree. Differential genes were then screened as a means of constructing a matrix of similarity by Pearson correlation analysis. A weighted adjacency matrix was then constructed based on soft threshold β. Finally, the 1597 ubiquitination-related genes were divided into four modules and the module with the highest correlation with immune infiltration was identified. The clusterProfiler package was used for performing enrichment analysis of the selected module, based on Gene Ontology (GO) function annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG).
On the STRING, module genes in the selected module were utilised for the construction of a PPI network. An interaction score >0.9 was taken as the threshold and the degree score for each gene was determined by Network Analyzer in Cytoscape (https://cytoscape.org).
Validation of GRB2 in TME
The correlation between GRB2 expression and immune infiltration cells was verified using ssGSEA and MCP counter was used to score immune infiltration in the TCGA and ICGC databases. The R package heatmap was then used for visualising their correlations according to the coefficient. The data of mutations from the TCGA database were downloaded and visualised using the R maftools package and a p-value of 0.05 was set as the statistical significance threshold.
Efficacy Analysis and Gene Set Enrichment Analysis (GSEA)
The Tumor Immune Dysfunction and Exclusion (TIDE) algorithm22 was used to predict the response efficiency to anti-PD-1. “GSE109211” and “GSE104580” from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) were used for evaluating sorafenib and TACE efficacy. All HCC samples in the TCGA and ICGC databases were divided into a GRB2 low group and GRB2 high group based on the median expressions GRB2. Single-gene GSEA was used for exploring the significant biological pathways between the two groups through GSEA_4.0.1 software (https://www.gsea-msigdb.org/gsea/index.jsp). All results were plotted using the ggplot2 package.
In vitro Experiments
qRT-PCR assay, immunohistochemistry (IHC) and Western blot methods were conducted based on previous literature.22 The following primers were used: GAPDH forward 5′-GT CTCCTCTGACTTCAACAGCG-3′ and reverse 5′-ACCACCCTGTTGCTGTAGCCAA3′; GRB2 forward 5′-TCCTCTGGGTGGTGAAGTTCAATTC-3′ and reverse 5′-GCTGTGGCACCTGTTCTATGTCC3′, and the primary antibodies anti-Tubulin (1:5000) and anti-GRB2 (1:1000) were obtained from Proteintech (Wuhan, China).
Statistical Analysis
All statistical analytical methods were based on IBM SPSS statistical software (version 22.0) and R software (4.0.5). Spearman correlation was used for determining the association between GRB2 expression and immune infiltration cells. In this study, the scatter plot of qRT-PCR results was performed using GraphPad Prism 8 software. A p-value < 0.05 was considered to be statistically significant.
Results
Unsupervised Clustering Grouped Advanced HCC Patients into Three Clusters
Firstly, ssGSEA was used for scoring immune infiltration in 90 advanced HCC patients from the TCGA database. The optimal number of clusters was determined by the consistent cumulative distribution function graph and the delta area plot. The final partitioning result with K = 3 clusters was obtained (Figure 1A and B). The hierarchical clustering showed three clusters with significant interconnectivity (Figure 1C). In order to determine the relationship of immune scores between clusters in the TCGA database, the ESTIMATE package was used for evaluating immune scores for three clusters and cluster 1 was found to be the immune desert type, cluster 2 the intermediate type and cluster 3 the immune enrichment type (Figure 2A).
To validate the above results, it was found that most human leukocyte antigen (HLA) genes exhibited significantly higher expression levels in Immunity_H but lower expression levels in Immunity_L (p < 0.05) (Figure 2B). In addition, the CIBERSORT algorithm quantified the immune cell infiltration level. The results showed the levels of plasma cells, CD8+T cells, memory resting CD4+T cells, and active NK cells to be remarkably increased in cluster 3 (p < 0.001) (Figure 2C). The results suggest that the grouping was representative and advanced HCC patients were further analysed based on these clusters.
WGCNA and PPI Screened Out the Hub Genes
A total of 1597 ubiquitination-related genes were first obtained from the IUUCD database and a WGCNA network was constructed. β = 5 was then chosen as the optimal soft-thresholding parameter (Figure 3A). A module–immune cell correlation heat map was drawn to explore the correlation between the four modules and immune cell infiltration. Four modules with co-expressed genes were found to be clustered and the brown module was significantly correlated with cluster 3 (r=0.46, p=2e-06) (Figure 3B and C). Therefore, the brown module was chosen as the focus module for the research.
The GO pathway showed most of the genes in this module to be related to TME, including protein polyubiquitination, interferon-gamma-mediated signalling pathway and autophagosome (Figure 4A). The KEGG pathways suggested ubiquitin-mediated proteolysis, bacterial invasion of epithelial cells, Kaposi sarcoma-associated herpes virus infection and Chemokine signalling pathway may be activated (Figure 4B).
 |
Figure 4 Enrichment analysis of genes in the brown module. (A) GO enrichment analysis of genes in the brown module. (B) KEGG enrichment analysis of genes in the brown module. |
On the STRING, all genes in the brown module were then used for building a PPI network and an interaction score greater than 0.9 was taken as the threshold (Figure 5A). The degree scores for each gene were determined by the Network Analyzer in Cytoscape. A degree value ≥20 was defined as a hub gene and 30 hub genes were screened in total (Figure 5B).
At the same time, the TCGA and ICGC databases were used for analysing the relationship between hub gene expression and HCC stage (Figure 6A and B). GRB2, SH3KBP1, LCK and UBE2D1 expressions were all found to be correlated with HCC stage in both databases. Furthermore, a high expression of GRB2 was found to have the ability to predict poor prognosis in advanced HCC patients (Figure 6C and D) and higher pathological stage (Figure 6E and F) in both databases. These results suggest that GRB2 expression has a close correlation with HCC patient stage and prognosis. Therefore, the role of GRB2 in HCC requires further analysis.
Association Between GRB2 Expression and TME
TME has been found to play a crucial role in immunotherapy in recent studies.23,24 Therefore, the relationship between GRB2 and immune cells was examined (Figure 7A and B). The results showed the high expression of GRB2 to be positively related to immune cells, including CD8+T cells, central memory CD8+T cells, activated CD4 + T cells, activated CD8 T cells and Myeloid dendritic cells. The single-cell sequencing also found there to be a significant increase in tumour suppressive immune cells, including CD4 Conv T cells, Treg cells, T profile cells, CD8 +T cells and CD8 Tex cells infiltration in the gene with a high expression of GRB2 from GSE140228_10X and GSE98638 (Figure 8A and C). The results suggest that GRB2 may affect the prognosis of advanced HCC patients by altering the immune cell infiltration level.
 |
Figure 8 Single-cell sequencing of GRB2 (A–C). There are various immune cell infiltrations in highly expressed GRB2 such as CD4 conv T cells, Treg cells, prolif T cells, CD8T cells and CD8T cells. |
Relationship Between GRB2 Expression and TMB
Somatic mutations may serve as immunotherapy response predictors,25 so 87 HCC patients were processed with gene mutation data using the maptools R package. The most common type of DNA base mutation in both low and high expression groups was found to be C>T (Figure 9A and B). The differences in somatic mutations between high and low expression groups were also compared. The most commonly mutated genes in the high expression group were TP53 (35%), TTN (26%), CTNNB1 (26%) and MUC16 (23%) and the most commonly mutated genes in the low expression group were CTNNB1 (27%), TTN (20%), APOB (16%) and TP53 (16%). Furthermore, the median mutation number in the low expression group was 70.5 and 64 in the high expression group (Figure 9C and D). Genes with different mutations may contribute to immune infiltration characteristics.
GRB2 Expression Was Found to Be Associated with Sensitivity to ICIs, Sorafenib and TACE Treatment
Several studies have found that TMB and immune infiltration can affect the treatment effect of HCC patients.26–28 Therefore, the relationship between GRB2 expression and HCC treatment was further evaluated. The TIDE algorithm was used for verifying the capacity of GRB2 expression in ICIs. The results found that most HCC patients who had a better response to ICIs had higher GRB2 expressions (p = 0.0065) and higher TIDE scores in comparison to a low expression of GRB2 (90%VS 80%) (Figure 10A). Conversely, patients with low GRB2 expressions had a higher response rate than those with a high GRB2 expression to sorafenib (42% vs 21%) and TACE (70% vs 41%) (Figure 10B and C). The results demonstrate that GRB2 has a high value in predicting ICI, sorafenib and TACE treatment efficacy among advanced HCC patients.
GSEA of GRB2
The potential biological functions of GRB2 in HCC were explored preliminarily using GSEA. From GO, it was found that a high GRB2 expression is often enriched in immune-related pathways, such as B cell proliferation, cytokine activity, cytokine binding and cytokine receptor binding (Figure 11A and B). KEGG suggested that GRB2 is involved in the interferon-related pathways, such as JAK-STAT signalling pathway and cytosolic DNA sensing pathway (Figure 11C and D). These results may further help with the elucidation of the potential mechanism of action of GRB2 in HCC.
 |
Figure 11 GSEA for GRB2 in HCC. (A and C) The data were derived from the TCGA database. (B and D) The data were derived from the ICGC database. |
GRB2 Expression is Upregulated in HCC
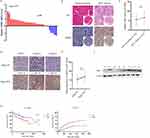
Further confirming the clinical significance of GRB2 expression in HCC, the RNA expression level of GRB2 was found to be upregulated in 24 of the 30 pairs of HCC tissue and adjacent non-cancer tissues (80%) (p < 0.05) (Figure 12A). Similarly, IHC and Western blot were performed as a means of confirming that GRB2 was highly expressed in HCC protein levels (Figure 12B, C and F). In addition, immunohistochemical chip results showed there to be a positive correlation between GRB2 expression and tumour staging (Figure 12D and E). GRB2 expression was found to have a significant correlation with the presence or absence of liver cirrhosis (p = 0.007), tumour size (p = 0.002) and TNM stage (p = 0.049) (Table 1). The overall survival curve and disease-free survival curve can be seen in Figure 12G. The results of this study matched those above.
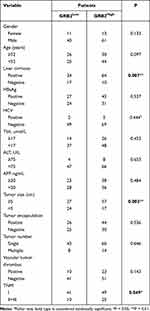 |
Table 1 Correlation Between GRB2 Expression and Clinicopathological Characteristics of HCC Patients |
Discussion
TME is an important factor that affects the efficacy of sorafenib, TACE and ICIs among advanced HCC patients.29,30 Recent studies have shown that ubiquitination-related enzymes play a crucial regulatory role in tumourigenesis and TME.31,32 Therefore, screening out ubiquitinated genes that regulate key molecules of immune infiltration among advanced HCC patients may provide new targets for improving HCC treatment efficacy.
This study used ssGSEA for scoring immune infiltration in 90 advanced HCC patients from the TCGA database and divided them into three clusters using unsupervised cluster analysis. In comparison to the other clusters, cluster 3 was found to be the most immune enrichment type. WGCNA was then established as a means of obtaining the most relevant ubiquitinated gene module for immune infiltrating cells and PPI was performed for identifying hub genes in this module. The GRB2 were screened out, which was correlated with immune cell infiltration, prognosis and HCC stage in both the TCGA and ICGC databases.
Growth factor receptor-bound protein 2 (GRB2) is a scaffold protein that has two SH2 and one SH3 domain. It has been proven that the SH3 domain regulates ubiquitination modifications of proteins by binding to ubiquitin.33 In addition, several studies have reported that GRB2 contributes to the progression or metastasis of several carcinomas, including colorectal, gastric and breast cancer.34–36 With liver cancer, lncRNA AC092171.4 upregulates GRB2 and promotes HCC progression by competitively binding miR-1271.4.37 Yu et al found the inhibition of GRB2 expression significantly inhibits cancer cell proliferation and survival.38 Furthermore, this work provides evidence that the mRNA and protein expression levels of GRB2 are high in HCC tissues and HCC patients with high expressions of GRB2 have worse prognosis. It was also found that the expression of GRB2 has a positive correlation with immune infiltration cells (CD4 Conv T cells, Treg cells, T profile cells, CD8+ T cells and CD8Tex cells), which plays a central role in HCC immunotherapy and improves patient prognosis.39,40 Recent studies have shown that PD1 or CTLA-4 inhibitors have obvious effects on Treg cells mediated immunosuppression and anti-tumour immunity.41 With NSCLC, patients with high expressions of PD1 and CD8+T have a longer progression free survival (PFS) after they receive ICIs than those with low expressions of PD1 or CD8+T.42 Furthermore, GRB2 plays a crucial role in immune infiltration cells. DNaM-1 promotes NK cell activation by coupling GRB2 and GRB2 regulated CD8+T cells and promoting the secretion of IL-2.43,44
Tumour mutation burden (TMB) also affects response to HCC immunotherapy. Mutation analysis has shown that the immune-related mutation rate is significantly higher in patients with high expressions of GRB2 expression than those with low expressions and the types of mutated genes differ widely between the groups. TP53 is the most frequently mutated gene in the high expression group (35%). With breast cancer, TP53 mutation significantly promotes the infiltration of Tregs, T helper cells and M0 macrophages.45 TP53 mutation has an inverse correlation with anti-CTLA-4 efficacy in melanoma.46 Therefore, TMB is an important prognostic factor for HCC and TMB-related genes could be potential HCC therapeutic targets. In the individual patient-level analysis of patients who were treated with ICIs, TERT, CTNNB1, BRD4, MLL or TP53 mutations were found to be associated with a significantly worse survival rate.47 Recent studies have also found that cabozantinib and nivolumab combination therapy improves the prognosis of advanced HCC patients with high TMB significantly.27
Sorafenib, TACE and ICIs are currently the main treatment modalities for advanced HCC patients.48 ICI efficacy is directly proportional to high GRB2 expressions. Conversely, patients with high GRB2 expressions have poor efficacy on sorafenib and TACE. Recent studies have reported that miR-378a-3p targets MAPK1/GRB2 sensitise ovarian cancer cells to cisplatin.49 Lymphocylin inhibits the GRB2/ERK/STAT3 signalling pathway and reduces the resistance of NSCLC cells to EGFR-TKI.50 Another study has reported that patients with high GRB2 expressions have a better response to PD-1/PD-L1 and TP53 frameshift mutation, which is potentially related to the high immune cell infiltration that is caused by GRB2, and this could improve the efficacy of PD-1/PD-L1.51 The above results provide further proof that GRB2 may exert critical function in HCC treatment.
Finally, GSEA was performed as a means of exploring the mechanism of GRB2. The results suggest that GRB2 is involved in the cancer-related and cellular regulation pathways, such as the JAK-STAT3 signalling pathway, cytosolic DNA sensing pathway and regulation of autophagy. Recent studies have shown that the overexpression of STAT3 results in the up-regulation of Mcl-1 expression, which makes the hepatoma cell Huh7 resistant to sorafenib.52 With prostate cancer, JAK2-STAT3 inhibits the cGAS-STING and plays a crucial role in tumour rejection and immune cell infiltration.53 With liver cancer, a combination of sorafenib and STAT3 knockout induces endoplasmic reticulum stress and activates the cGAS-STING axis.54 GRB2 also regulates the genomic stability of (DNA damage repair) DDR by affecting the Rad51 expression.55 Therefore, it is supposed that GRB2 may activate STAT3 and the cGAS-STING axis through DDR and affect anti-tumour immunity. Recent studies have determined that a combination of STAT3 antiagonists and STING agonists could inhibit tumour growth among mice by increasing CD8+ T cells while reducing Tregs and MDSCs.53 Furthermore, STING agonist cannot individually induce IFN expression in prostate cancer cell DU145, but inhibition of the JAK2-STAT3 pathway may restore responsiveness to STING agonist.
Conclusion
To summarise, through a comprehensive evaluation of the molecular, cellular and clinical characteristics of GRB2, it was discovered that there may be important implications for the selection of sorafenib, TACE and ICI strategies for advanced HCC patients. However, there are limitations to the study that require refinement. Further experiments are required exploring the potential biological mechanism of GRB2 in HCC treatment.
Abbreviations
HCC, hepatocellular carcinoma; GRB2, growth factor receptor-bound protein 2; SH3, Src homology 3; TMA, tissue microarray; IHC, immunohistochemistry; qRT-PCR, quantitative real-time polymerase chain reaction; TCGA, The Cancer Genome Atlas; ICGC, International Cancer Genome Consortium; GSEA, gene set enrichment analysis; PPI, protein–protein interaction; TIMER, Tumor Immune Estimation Resource; NK, nature kill cell; ILC, innate lymphoid cell; DC, dendritic cell.
Ethics Approval
This study was approved by the ethics committee of Clinical Medical College, Yangzhou University, China.
Consent for Publication
All of the HCC patients in the study have given their consent to publish their data.
Acknowledgments
YT especially thank these people who gave enough support and encouragement to him.
Author Contributions
(I) Conceptualization: D Bai, C Zhang, Y Tang, J Cao; (II) Study design: D Bai, C Zhang, R Peng; (III) Acquisition of data: Y Tang, J Cao, X MAO, H Tang, D Tu, Y Tang, J Zhou; (IV) Execution: S Jin, R Liu, G Jiang, Q Wang, B Su and Q Wang; (V) Analysis and interpretation Y Tang, C Zhang. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 81871909, 82203716), the “13th five-year Plan” Science and Education strong Health Project Innovation team of Yangzhou (LJRC20181; YZCXTD201801), Provincial-level discipline leader of the NJPH (DTRA202214), Cross-cooperation special projects of the NJPH (YJCHZ-2021-08), Beijing iGanDan Foundation (GDXZ-08-19), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX22_3568) Yangzhou Key Social Development Project (YZ2022093). The National Natural Science Foundation of China (Grant No. 82203716), the “13th five-year Plan” Science and Education strong Health Project Innovation team of Yangzhou (LJRC20181; YZCXTD201801), Provincial-level discipline leader of the NJPH (DTRA202214), Cross-cooperation special projects of the NJPH (YJCHZ-2021-08), Beijing iGanDan Foundation (GDXZ-08-19), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX22_3568).
Disclosure
All the authors declare that they have no competing interests.
References
1. Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462.
2. Lawal G, Xiao Y, Rahnemai-Azar AA, et al. The immunology of hepatocellular carcinoma. Vaccines. 2021;9(10):1184. doi:10.3390/vaccines9101184
3. Clavien PA, Lesurtel M, Bossuyt PM, Gores GJ, Langer B, Perrier A. Recommendations for liver transplantation for hepatocellular carcinoma: an international consensus conference report. Lancet Oncol. 2012;13(1):e11–e22. doi:10.1016/S1470-2045(11)70175-9
4. Vitale A, Peck-Radosavljevic M, Giannini EG, et al. Personalized treatment of patients with very early hepatocellular carcinoma. J Hepatol. 2017;66(2):412–423. doi:10.1016/j.jhep.2016.09.012
5. Xing M, Wang X, Kiken RA, He L, Zhang JY. Immunodiagnostic Biomarkers for Hepatocellular Carcinoma (HCC): the first step in detection and treatment. Int J Mol Sci. 2021;22(11):6139. doi:10.3390/ijms22116139
6. Gordan JD, Kennedy EB, Abou-Alfa GK, et al. Systemic therapy for advanced hepatocellular carcinoma: ASCO guideline. J Clin Oncol. 2020;38(36):4317–4345. doi:10.1200/JCO.20.02672
7. Whiteside TL, Demaria S, Rodriguez-Ruiz ME, Zarour HM, Melero I. Emerging opportunities and challenges in cancer immunotherapy. Clin Cancer Res. 2016;22(8):1845–1855. doi:10.1158/1078-0432.CCR-16-0049
8. Hoos A. Development of immuno-oncology drugs - from CTLA4 to PD1 to the next generations. Nat Rev Drug Discov. 2016;15(4):235–247. doi:10.1038/nrd.2015.35
9. Xu J, Shen J, Gu S, et al. Camrelizumab in combination with apatinib in patients with advanced hepatocellular carcinoma (RESCUE): a nonrandomized, open-label, phase II trial. Clin Cancer Res. 2021;27(4):1003–1011. doi:10.1158/1078-0432.CCR-20-2571
10. Cheng AL, Hsu C, Chan SL, Choo SP, Kudo M. Challenges of combination therapy with immune checkpoint inhibitors for hepatocellular carcinoma. J Hepatol. 2020;72(2):307–319. doi:10.1016/j.jhep.2019.09.025
11. Duffy AG, Ulahannan SV, Makorova-Rusher O, et al. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J Hepatol. 2017;66(3):545–551. doi:10.1016/j.jhep.2016.10.029
12. Hu X, Wang J, Chu M, Liu Y, Wang ZW, Zhu X. Emerging role of ubiquitination in the regulation of PD-1/PD-L1 in cancer immunotherapy. Mol Ther. 2021;29(3):908–919. doi:10.1016/j.ymthe.2020.12.032
13. Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–351. doi:10.1038/35085597
14. Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75(3):495–505. doi:10.1016/0092-8674(93)90384-3
15. Yu C, Xue B, Li J, Zhang Q. Tumor cell-derived exosome RNF126 affects the immune microenvironment and promotes nasopharyngeal carcinoma progression by regulating PTEN ubiquitination. Apoptosis. 2022;27(7–8):590–605. doi:10.1007/s10495-022-01738-9
16. Wang YC, Wu YS, Hung CY, et al. USP24 induces IL-6 in tumor-associated microenvironment by stabilizing p300 and β-TrCP and promotes cancer malignancy. Nat Commun. 2018;9(1):3996. doi:10.1038/s41467-018-06178-1
17. Xiong C, Wang Z, Wang G, et al. Identification of CDC20 as an immune infiltration-correlated prognostic biomarker in hepatocellular carcinoma. Invest New Drugs. 2021;39(5):1439–1453. doi:10.1007/s10637-021-01126-1
18. Peng R, Zhang P-F, Yang X, et al. Overexpression of RNF38 facilitates TGF-β signaling by ubiquitinating and degrading AHNAK in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):113. doi:10.1186/s13046-019-1113-3
19. Bai XS, Zhang C, Peng R, et al. RNF128 promotes malignant behaviors via EGFR/MEK/ERK pathway in hepatocellular carcinoma. Onco Targets Ther. 2020;13:10129–10141. doi:10.2147/OTT.S269606
20. Cao J, Su B, Peng R, et al. Bioinformatics analysis of immune infiltrates and tripartite motif (TRIM) family genes in hepatocellular carcinoma. J Gastrointest Oncol. 2022;13(4):1942–1958. doi:10.21037/jgo-22-619
21. Zhou J, Xu Y, Lin S, et al. iUUCD 2.0: an update with rich annotations for ubiquitin and ubiquitin-like conjugations. Nucleic Acids Res. 2018;46(D1):D447–D453. doi:10.1093/nar/gkx1041
22. Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi:10.1038/s41591-018-0136-1
23. Kim KH, Choi KU, Kim A, et al. PD-L1 expression on stromal tumor-infiltrating lymphocytes is a favorable prognostic factor in ovarian serous carcinoma. J Ovarian Res. 2019;12(1):56. doi:10.1186/s13048-019-0526-0
24. Ravelli A, Roviello G, Cretella D, et al. Tumor-infiltrating lymphocytes and breast cancer: beyond the prognostic and predictive utility. Tumour Biol. 2017;39(4):1010428317695023. doi:10.1177/1010428317695023
25. Gajic ZZ, Deshpande A, Legut M, Imieliński M, Sanjana NE. Recurrent somatic mutations as predictors of immunotherapy response. Nat Commun. 2022;13(1):3938. doi:10.1038/s41467-022-31055-3
26. Zhu Y, Yang J, Xu D, et al. Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade. Gut. 2019;68(9):1653–1666. doi:10.1136/gutjnl-2019-318419
27. Yang X, Shi J, Chen X, Jiang Y, Zhao H. Efficacy of cabozantinib and nivolumab in treating hepatocellular carcinoma with RET amplification, high tumor mutational burden, and PD-L1 expression. The Oncologist. 2020;25(6):470–474. doi:10.1634/theoncologist.2019-0563
28. Xie C, Wu H, Pan T, et al. A novel panel based on immune infiltration and tumor mutational burden for prognostic prediction in hepatocellular carcinoma. Aging. 2021;13(6):8563–8587. doi:10.18632/aging.202670
29. Wang Y, Wang Z, Jia F, et al. CXCR4-guided liposomes regulating hypoxic and immunosuppressive microenvironment for sorafenib-resistant tumor treatment. Bioact Mater. 2022;17:147–161. doi:10.1016/j.bioactmat.2022.01.003
30. He Q, Yang J, Jin Y. Development and validation of TACE refractoriness-related diagnostic and prognostic scores and characterization of tumor microenvironment infiltration in hepatocellular carcinoma. Front Immunol. 2022;13:869993. doi:10.3389/fimmu.2022.869993
31. Jiang JH, Liu YF, Ke AW, et al. Clinical significance of the ubiquitin ligase UBE3C in hepatocellular carcinoma revealed by exome sequencing. Hepatology. 2014;59(6):2216–2227.
32. Peng R, Huang X, Zhang C, Yang X, Xu Y, Bai D. Overexpression of UHRF2 in intrahepatic cholangiocarcinoma and its clinical significance. Onco Targets Ther. 2017;10:5863–5872. doi:10.2147/OTT.S149361
33. Arya R, Dangi RS, Makwana PK, Kumar A, Upadhyay SK, Sundd M. Grb2 carboxyl-terminal SH3 domain can bivalently associate with two ligands, in an SH3 dependent manner. Sci Rep. 2017;7(1):1284. doi:10.1038/s41598-017-01364-5
34. Yu GZ, Chen Y, Wang JJ. Overexpression of Grb2/HER2 signaling in Chinese gastric cancer: their relationship with clinicopathological parameters and prognostic significance. J Cancer Res Clin Oncol. 2009;135(10):1331–1339. doi:10.1007/s00432-009-0574-8
35. Zhang Y, Li Z, Yang M, et al. Identification of GRB2 and GAB1 coexpression as an unfavorable prognostic factor for hepatocellular carcinoma by a combination of expression profile and network analysis. PLoS One. 2013;8(12):e85170. doi:10.1371/journal.pone.0085170
36. Ding C, Tang W, Wu H, et al. The PEAK1-PPP1R12B axis inhibits tumor growth and metastasis by regulating Grb2/PI3K/Akt signalling in colorectal cancer. Cancer Lett. 2019;442:383–395. doi:10.1016/j.canlet.2018.11.014
37. Sun C, Huang S, Hou Y, et al. Long noncoding RNA AC092171.4 promotes hepatocellular carcinoma progression by sponging microRNA-1271 and upregulating GRB2. Aging. 2020;12(14):14141–14156. doi:10.18632/aging.103419
38. Yu GZ, Chen Y, Long YQ, Dong D, Mu XL, Wang JJ. New insight into the key proteins and pathways involved in the metastasis of colorectal carcinoma. Oncol Rep. 2008;19(5):1191–1204.
39. Bozward AG, Warricker F, Oo YH, Khakoo SI. Natural killer cells and regulatory T cells cross talk in hepatocellular carcinoma: exploring therapeutic options for the next decade. Front Immunol. 2021;12:643310. doi:10.3389/fimmu.2021.643310
40. Pham L, Kyritsi K, Zhou T, et al. The functional roles of immune cells in primary liver cancer. Am J Pathol. 2022;192(6):826–836. doi:10.1016/j.ajpath.2022.02.004
41. Mitsuiki N, Schwab C, Grimbacher B. What did we learn from CTLA-4 insufficiency on the human immune system? Immunol Rev. 2019;287(1):33–49. doi:10.1111/imr.12721
42. Shimoda Y, Shibaki R, Yoshida T, et al. Concurrent high PD-L1 expression and CD8(+) immune cell infiltration predict PD-1 blockade efficacy in advanced EGFR-mutant NSCLC patients. Clin Lung Cancer. 2022;23(6):477–486. doi:10.1016/j.ebiom.2018.05.019
43. Zhang Z, Wu N, Lu Y, Davidson D, Colonna M, Veillette A. DNAM-1 controls NK cell activation via an ITT-like motif. J Exp Med. 2015;212(12):2165–2182. doi:10.1084/jem.20150792
44. Ritthipichai K, Haymaker CL, Martinez M, et al. Multifaceted role of BTLA in the control of CD8(+) T-cell fate after antigen encounter. Clin Cancer Res. 2017;23(20):6151–6164. doi:10.1158/1078-0432.CCR-16-1217
45. Zhang Z, Hao R, Guo Q, Zhang S, Wang X. TP53 mutation infers a poor prognosis and is correlated to immunocytes infiltration in breast cancer. Front Cell Dev Biol. 2021;9:759154. doi:10.3389/fcell.2021.759154
46. Xiao W, Du N, Huang T, et al. TP53 mutation as potential negative predictor for response of anti-CTLA-4 therapy in metastatic melanoma. EBioMedicine. 2018;32:119–124.
47. Ou Q, Yu Y, Li A, et al. Association of survival and genomic mutation signature with immunotherapy in patients with hepatocellular carcinoma. Ann Transl Med. 2020;8(5):230. doi:10.21037/atm.2020.01.32
48. Cheu JW, Wong CC. Mechanistic rationales guiding combination hepatocellular carcinoma therapies involving immune checkpoint inhibitors. Hepatology. 2021;74(4):2264–2276. doi:10.1002/hep.31840
49. Xu ZH, Yao TZ, Liu W. miR-378a-3p sensitizes ovarian cancer cells to cisplatin through targeting MAPK1/GRB2. Biomed Pharmacother. 2018;107:1410–1417. doi:10.1016/j.biopha.2018.08.132
50. Chen Y, Wu J, Yan H, et al. Lymecycline reverses acquired EGFR-TKI resistance in non-small-cell lung cancer by targeting GRB2. Pharmacol Res. 2020;159:105007. doi:10.1016/j.phrs.2020.105007
51. Dong ZY, Zhong WZ, Zhang XC, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23(12):3012–3024. doi:10.1158/1078-0432.CCR-16-2554
52. Xie L, Zeng Y, Dai Z, et al. Chemical and genetic inhibition of STAT3 sensitizes hepatocellular carcinoma cells to sorafenib induced cell death. Int J Biol Sci. 2018;14(5):577–585. doi:10.7150/ijbs.22220
53. Suter MA, Tan NY, Thiam CH, et al. cGAS-STING cytosolic DNA sensing pathway is suppressed by JAK2-STAT3 in tumor cells. Sci Rep. 2021;11(1):7243. doi:10.1038/s41598-021-86644-x
54. Wang X, Hu R, Song Z, et al. Sorafenib combined with STAT3 knockdown triggers ER stress-induced HCC apoptosis and cGAS-STING-mediated anti-tumor immunity. Cancer Lett. 2022;547:215880. doi:10.1016/j.canlet.2022.215880
55. Hou B, Xu S, Xu Y, et al. Grb2 binds to PTEN and regulates its nuclear translocation to maintain the genomic stability in DNA damage response. Cell Death Dis. 2019;10(8):546. doi:10.1038/s41419-019-1762-3
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.