Back to Journals » OncoTargets and Therapy » Volume 11
SATB1 siRNA-encapsulated immunoliposomes conjugated with CD44 antibodies target and eliminate gastric cancer-initiating cells
Authors Yang F, Zheng Z, Zheng L, Qin J, Li H, Xue X, Gao J, Fang G
Received 2 August 2018
Accepted for publication 13 September 2018
Published 11 October 2018 Volume 2018:11 Pages 6811—6825
DOI https://doi.org/10.2147/OTT.S182437
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Feng Yang,1 Zhi Zheng,1 Luming Zheng,2 Jianmin Qin,1 Haijia Li,1 Xuchao Xue,3 Jie Gao,4 Guoen Fang3
1Department of General Surgery, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai 201805, People’s Republic of China; 2Department of General Surgery, General Hospital of Jinan Military Area, Jinan 250031, People’s Republic of China; 3Department of General Surgery, Changhai Hospital, Second Military Medical University, Shanghai 200433, People’s Republic of China; 4Department of Pharmaceutical Science, College of Pharmacy, Second Military Medical University, Shanghai 200433, People’s Republic of China
Purpose: Gastric cancer, the cancer initiated from the stomach, is ranked as the third most frequent reason of cancer death worldwide. Gastric cancer-initiating cells (CICs) are one of the crucial causes for the metastasis and recurrence of gastric cancer, and CD44 is considered to be one marker for gastric CICs. Special AT-rich sequence binding protein 1 (SATB1) is a protein that promotes cancer progression, metastasis, and invasion and also participates in the maintenance of CICs. In this study, we investigated the therapeutic effect of SATB1 siRNA against gastric CICs and we constructed SATB1 siRNA-encapsulated immunoliposomes conjugated with CD44 antibodies (CD44-SATB1-ILs) to enhance the therapeutic effect of SATB1 siRNA against gastric CICs.
Methods: We investigated the therapeutic effect of the SATB1 suppression by SATB1 siRNA on CD44+ gastric CICs. CD44-SATB1-ILs were developed by the lyophilization/hydration approach. The targeting and cytotoxic effect of CD44-SATB1-ILs toward gastric CICs were evaluated in vitro.
Results: In this study, for the first time, we confirmed that SATB1 suppression by SATB1 siRNA preferentially eliminated CD44+ gastric CICs. The results showed that CD44-SATB1-ILs could efficiently and specifically promote the SATB1 siRNA delivery to CD44+ gastric CICs, achieving superior therapeutic effects against CD44+ gastric CICs than non-targeted liposomes.
Conclusion: As far as we know, our report is the first research that indicated the promotion of siRNA delivery via nanoparticles to gastric CICs and achievement of superior therapeutic effect against gastric CICs by utilization of CD44 antibody. Therefore, CD44-SATB1-ILs represent an up-and-coming approach for eliminating gastric CICs and also a promising treatment for therapy of gastric cancer.
Keywords: gastric cancer, SATB1, cancer-initiating cells, immunoliposomes, CD44
Introduction
Gastric cancer, which is initiated from the stomach, is the 15th most common cancer in USA and also the third most common cause of cancer death.1,2 Since many patients are at advanced stage of gastric cancer, it is difficult to cure gastric cancer. For gastric cancer therapy, we have not achieved remarkable improvements in the relapse-free and overall survival of gastric cancer, due to its treatment failure, recurrence, and metastasis.3,4 Thus, gastric cancer is considered to be a big mortality burden for global human health.
Gastric cancer-initiating cells (CICs), which are the seed of gastric cancer, are considered to be responsible for treatment failure, recurrence, metastasis, and multi-drug resistance of gastric cancer.5,6 Therefore, the elimination of gastric CICs could contribute greatly to the eradication of gastric cancer. CD44, a cell-surface glycoprotein associated with cell adhesion, interactions, and migration, is considered to be one marker for gastric CICs.5,6 Takaishi et al showed that CD44+ gastric CICs are more aggressive compared with CD44− gastric cancer cells, as reflected by their increased self-renewal, proliferative, clonogenic, and tumorigenic properties.5,6
Gene therapy, which is the removal or alteration of genes in cells for disease therapy, is regarded as an up-and-coming approach for various diseases, especially for cancer.7,8 The advantages of gene therapy include high specificity, low side effects, and curability in some cases.7,8 Currently, there have been several studies that reported the therapeutic effect of gene therapy against CICs using adenovirus and peptide complex as gene vectors.9,10 For example, Long et al reported that adenovirus-mediated truncated Bid overexpression induced significant cell apoptosis in CD133-positive ovarian CICs.10 It is noteworthy that siRNA-based gene therapy is regarded a good therapeutic approach for various diseases.11–13 Since siRNA molecules alone are not able to cross cellular barriers, nanoparticles have emerged as effective approach to significantly increase the cellular delivery of siRNA and facilitate translational use of siRNA.11–13
Among siRNA-loaded nanoparticles, cationic liposomes represent as one efficient siRNA delivery system.14 However, the application of siRNA-loaded cationic liposomes has severely been hampered by their short shelf life, since siRNA in aqueous solution is easy to be degraded by nuclease, and cationic liposomes also have a risk to lose their entrapped siRNA.7 Notably, cationic liposomes developed with the lyophilization/rehydration approach could conquer the problem of short shelf life. The lyophilization/rehydration method consists of two steps, including the lyophilization of drug-free liposomes and drug encapsulation by rehydration of drug-free liposomes with the drug suspended in aqueous solutions.15 Like all lyophilized liposomes, siRNA-loaded cationic liposomes developed with the lyophilization/rehydration approach make siRNA in its aqueous form only when in the use of liposomes, resulting in the risk of being degraded by nuclease and premature release of siRNA from liposomes minimal.16 Currently, Gao et al, Peer and Margalit developed lyophilized liposomes for siRNA delivery and demonstrated that they exhibited long shelf life, superior transfection efficacy, and gene-silencing activity.15,17
Special AT-rich sequence binding protein 1 (SATB1) is a protein participating in tissue-specific gene expression and higher-order chromatin organization.18 SATB1 has been reported to promote cancer metastasis and invasion and contribute to the unfavorable clinicopathological characteristics and poor prognosis of various cancers, such as gastric, liver, breast, and colorectal cancers.18,19 The knockdown of SATB1 significantly prevented the cancer growth and metastasis.20,21 Notably, SATB1 participates in the maintenance of stem cells and CICs.22,23 Therefore, SATB1 represents not only a superior candidate target in cancer cells but also in CICs.
To promote drug delivery to cancer cells, currently a considerable interest has been paid to antibody-targeted nanoparticles that could obtain targeted cancer therapy.24,25 It is well-known that antibody-targeted nanoparticles have improved the therapeutic effect of chemotherapy in various cancers.26,27 Since CD44 is a marker of gastric CICs, we hypothesize that the CD44 antibody could be used to promote the delivery of siRNA-encapsulated cationic liposomes to gastric CICs.
In this study, aiming to target and eliminate CD44+ gastric CICs, we constructed SATB1 siRNA-encapsulated immunoliposomes conjugated with CD44 antibodies (CD44-SATB1-ILs). The characteristics, targeting, and therapeutic effect of CD44-SATB1-ILs toward gastric CICs were investigated.
Methods
siRNA, lipids, kits, antibodies, and cell medium
The synthesis of the siRNA targeting human SATB1, negative control siRNA (NC siRNA), and 6-carboxy-fluorescein phosphoramidate (FAM)-labeled NC siRNA (FAM is a green fluorescent dye) done by Shanghai GenePharma Co., Ltd. (Shanghai, People’s Republic of China) (the sequence of these siRNAs is shown in Table S1). 1,2-Dioleoyl-3-trimethylammonium-propane (DOTAP), cationic lipids), cholesterol, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine [methoxy(polyethylene glycol)-2000] (DSPE-mPEG), and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPE-PEG-Mal) were bought from Avanti Polar Lipids (Alabaster, AL, USA). 2-Iminothiolane (used for production of thiols, Traut’s Reagent) and organic reagents were all bought from Sigma-Aldrich (St Louis, MO, USA). Recombinant rat anti-human CD44 monoclonal antibody (mAb) was provided by the R&D Systems, Inc. (Minneapolis, MN, USA), and the CD44 Fab’ from CD44 mAb was prepared as described before.28 The rat derived anti-human CD44 antibody linked with Alexa Fluor® 488 was also from R&D Systems, Inc. The CD44 MicroBead Kit were purchased by Shanghai Miltenyi Biotec (Shanghai, People’s Republic of China). The Cell Counting Kit-8 (CCK-8) kit was bought from Dojindo Laboratories (Kumamoto, Japan). The Pierce BCA Protein Assay Kit, Roswell Park Memorial Institute 1640 (RMPI-1640) medium, fetal bovine serum (FBS), Dulbecco’s Modified Eagle’s Medium (DMEM)/F12, B27, epidermal growth factors (EGF), basic fibroblast growth factors (bFGF), and insulin-transferrin-selenium (ITS) were purchased from the Thermo Fisher Scientific (Waltham, MA, USA).
Cell culture
MKN-45 and NCI-N87 are two human gastric cancer cell lines purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were maintained in a humidified atmosphere of 5% CO2 in the RMPI-1640 medium at 37°C. The RMPI-1640 medium was supplemented with 10% FBS, hydroxyethyl piperazine ethanesulfonic acid (HEPES, 25 mM), streptomycin (antibiotics, 100 μg/mL), and penicillin (antibiotics, 100 U/mL).
Flow cytometry-based CD44 analysis and magnetic cell sorting-based separation
Flow cytometry was carried out to do the cellular CD44 expression analysis. In short, the gastric cancer cells dissociated into single cells were washed and incubated with the fluorescent antibody (anti-human CD44 Alexa Fluor® 488-conjugated antibody at 1 μg/mL) for 0.5 hour at 4°C. After the incubation, the cells were washed thrice with PBS, aiming to wash away unconjugated fluorescent antibody. In the end of the assay, the washed cells were suspended in PBS, and a FACS Calibur FCM (Becton Dickinson, Franklin Lakes, NJ, USA) was used to analyze the proportion of positively stained cells.
The separation of CD44+ cells from the gastric cancer cells was done according to the protocol provided by the CD44 MicroBead Kit from the Shanghai Miltenyi Biotec, and a FACS Calibur FCM was used to analyze the proportion of positively stained cells as described above.
Analysis of the formation of the tumorspheres
The formation of tumorspheres was analyzed to evaluate the self-renewal ability of gastric CICs when the single cells are suspended in serum-free medium. In short, gastric cancer cells suspended in stem cell medium were cultivated in ultra-low adherent 6-well dishes (Corning Incorporated, Corning, NY, USA). The density of the suspended cells was 5,000 cells/well, and the components of stem cell medium included DMEM/F12, B27 (1×), ITS (1×), EGF, and bFGF (both cytokines were of a concentration of 20 ng/mL). After the cells were cultivated in the stem cells medium for 7 days, the counting of the number of tumorspheres was carried out under a conventional microscope. For the acquisition of the second passage tumorspheres, the tumorspheres of the first passage were washed with PBS and then the washed tumorspheres were dissociated by the cell dissociation reagent (StemPro® Accutase®; Thermo Fisher Scientific). Then, the dissociated tumorspheres were then propagated.
Development of siRNA-encapsulated immunoliposomes
siRNA-encapsulated immunoliposomes were developed as described below according to a protocol established before.15,17 Briefly, a mixture of different lipids (DOTAP, cholesterol, DSPE-mPEG, and DSPE-PEG-Mal, 48:48:1:3, molar ratios) was dissolved in chloroform, and the evaporation of chloroform was performed to completely remove the chloroform, aiming to form a dry lipid film. Then, 2 mL PBS was added to hydrate the dried lipid film, resulting in the multilayer liposomes (MLL). After hydration, the extrusion of the resultant MLL into unilamellar liposomes (ULL) was carried out. The extrusion was done with a hand-held liposome extruder (Avestin, Ottawa, ON, Canada) and used decreasing 200 nm and 100 nm pore-sized membranes (Nuclepore, Whatman; GE Healthcare, Chicago, IL, USA). For per pore size, ten cycles were carried out. Furthermore, the thiolation of CD44 Fab’ was performed with 2-iminothiolane at a 1:100 molar ratio of Fab’ to 2-iminothiolane. The thiolated CD44 Fab’ was incubated with the obtained liposomes (the molar ratio of DSPE-PEG-Mal/Fab’ is 10/1) with 12 hours at room temperature, aiming to conjugate thiolated antibodies to liposomes. Next, Amicon centrifugal filters (MWCO: 100 kDa) were used to remove unconjugated Fab’. Nontargeted liposomes were developed similarly as described above without the addition of the Fab’. Lyophilized liposomes were obtained as follows. Briefly, liposomes were mixed with sucrose (the final sucrose concentration was 9%, w/v) and lyophilized for 18 hours using the freeze dryer (VirTis® AdVantage™ Benchtop, SP Scientific, Gardiner, NY, USA). After lyophilization, the lyophilized liposomes were hydrated by siRNA solution in DEPC-treated water (2 μM), as described below. Lyophilized liposomes (8 μg liposomes) were hydrated with 30 μL siRNA solution and incubated at 25°C for 1 hour. The siRNA encapsulation was performed immediately before use. FAM-siRNA-loaded immunoliposomes were prepared in the same way as siRNA-loaded immunoliposomes.
The following abbreviations are used to designate the liposomes: CD44-SATB1-ILs (SATB1 siRNA-encapsulated immunoliposomes conjugated with CD44 antibodies); CD44-NC-ILs (NC siRNA-encapsulated immunoliposomes conjugated with CD44 antibodies); SATB1-Lipo (SATB1 siRNA-encapsulated liposomes); and NC-Lipo (NC siRNA-encapsulated liposomes).
The ultrafiltration of the liposomes was used to evaluate the conjugation efficacy of antibodies on liposomes. Briefly, after the antibodies were conjugated to the liposomes, the mixture of antibodies/liposomes was centrifuged with the Amicon centrifugal filters (nominal molecular weight limit is 100 kDa) to remove unconjugated antibodies. The unconjugated antibodies were measured by the Pierce Protein Assay Reagent Kit. In the end, the conjugation efficacy of antibodies on liposomes was evaluated with the following equation: (Mt − Mu)/Mt (Mt: the mass of total antibodies; Mu: the mass of unconjugated antibodies).
The size, zeta potential, encapsulation efficacy (EE), and stability assay of liposomes
The siRNA encapsulation efficiency of liposomes was evaluated as described below, using the ultrafiltration method. In short, 2 mL FAM-siRNA loaded liposome solution was taken out and added into the Ultra-4 centrifugal filter devices (EMD Millipore, Billerica, MA, USA). Then, the centrifugation was performed, with the desk centrifuge (30 minutes at 3,500 × g), aiming to remove the unencapsulated siRNA. After centrifugation, 4 mL deionized water was added again, and the centrifugation was repeated. The ultrafiltration was performed for four times, and unencapsulated FAM-siRNA was gathered. In the end, the FAM-siRNA was quantified by the constructed calibration line of FAM-siRNA. Using the Synergy™ 4 (BioTek Instruments, Inc., Winooski, VT, USA), the fluorescence of FAM-siRNA was examined (wavelength of excitation: 495 nm, wavelength of emission: 525 nm). The siRNA encapsulation efficiency was calculated with the formula: (the mass of total siRNA minus the mass of unencapsulated siRNA)/the mass of total siRNA × 100%.
The evaluation of the stability of liposomes was performed as follows. First, the liposomes were suspended in various media, including PBS, PBS with 10% FBS, or 20% FBS. Then the solution was incubated at 25°C for 5 days. In each day, an aliquot of liposomes was taken out for the analysis of the size change of liposomes.
In vitro targeting of fluorescent liposomes to gastric cancer cells
We adopted flow cytometry to examine the in vitro targeting of fluorescent liposomes. Briefly, gastric cancer cells were cultured on 12-well cell culture plates overnight at 37°C. The density of gastric cancer cells was 5 × 105 cells per well. Then, the old medium of each well was replaced with fresh medium which was dissolved with liposomes (the final FAM-siRNA concentration was 200 nM). The incubation of the fluorescent liposomes with the gastric cancer cells was taken for hours. Then, the gastric cancer cells were washed thrice and trypsinized to be dissociated single cells. In the end, the washed cells were suspended in PBS for the analysis with a FACS Calibur FCM.
Reverse transcription of cDNA, quantitative polymerase chain reaction (PCR), and Western blot
With TRIzol reagent, the RNA of the gastric cancer cells was extracted. With the Reverse Transcription System kit, the reverse transcription was done to provide the first-strand cDNA. After transcription, using the SYBR™ Green PCR Master Mix, PCR was taken with a Roche Light Cycler (Hoffman-La Roche Ltd., Basel, Switzerland), and a PCR procedure described as follows: denaturation step: 95°C for 2 minutes; 40 cycles: 3 seconds denaturation at 95°C, 10 seconds annealing at 55°; extension step: 25 seconds at 72°C. By the 2ΔΔCq approach, the quantification of the expression of mRNA was carried out. The sequences of β-actin and SATB1 primers are listed in Table S1.
For the analysis of protein expression by Western blot, the protein was extracted from the cells and 50 μg protein was first run in SDS-PAGE. After SDS-PAGE running, the transfer of protein was done by electricity to the membrane made of polyvinylidene fluoride. After transfer, the antibodies were used to detect the protein: the primary antibody (polyclonal rabbit anti-human SATB1 antibody; Abcam PLC, Cambridge, MA, USA) and the secondary antibody (the horseradish peroxidase [HRP]-conjugated goat anti-rabbit antibody; Abcam PLC). The internal antibody was the β-actin antibody. The Amersham™ ECL Plus™ kit (GE Healthcare) was used to detect the bands.
The cytotoxic effect of liposomes toward the gastric cancer cell lines
The cytotoxic effect of liposomes toward the gastric cancer cell lines was examined with the CCK-8 assay using the protocol provided by the producer. Briefly, the gastric cancer cells were washed, trypsinized into single cells, and inoculated in the 96-well cell culture plates overnight. In the cell culture plates, the density of the gastric cancer cells was 3 × 103 cells/well. After inoculating overnight, the old medium was discarded and changed with fresh medium containing liposomes (200 nM siRNA) for 6 hours. After 6 hours, the drugs were discarded and fresh medium was replaced. Seventy-two hours later, the cell viability was measured as the protocol provided in the CCK-8 kit. The cell viability of the gastric cancer cells was examined by the absorbance obtained by a Microplate Reader (Multiskan MK3).
The impact of liposomes on the gastric CICs proportion
The examination of the tumorsphere formation and proportion of CD44+ cells were done to measure the impact of liposomes on the gastric CICs proportion of the gastric cancer cells. In brief, gastric cancer cells were washed, trypsinized into single cells, and inoculated overnight in 12-well cell culture plates. The density of gastric cancer cells was 5 × 104 cells per well. After overnight incubation, the old cell culture medium was discarded. After the removal of the medium, the cells were washed with PBS. After washing, the fresh medium dissolved with the liposomes (200 nM siRNA) was added to the cells, and the incubation took further 6 hours. Six hours later, the drugs were discarded, and fresh medium was replaced. The incubation was taken for 72 hours. Then the cells were washed and trypsinized to single cells, and the formation of tumorspheres measured as described in the “Analysis of the formation of the tumorspheres” section. The percentage of tumorsphere formed of the treated group was normalized to the untreated control group of which the percentage is defined as 100%. Alternatively, the percentage of CD44+ cells of the trypsinized cells was measured by flow cytometry as described above.
The colony formation assays
After the gastric cancer cells were enzymatically dissociated into single cells, the gastric cancer cells were washed and resuspended in serum-containing medium. Then, the gastric cancer cells were seeded in 12-well plates overnight. The cell density was 1 × 105 cells per well. After overnight incubation, the cells were washed and incubated with the fresh medium dissolved with the liposomes (200 nM siRNA). After 24 hours, the cells were trypsinized into single cells and seeded in six-well plates. The cell density was 2,000 cells per well, then grown in culture medium for 7 days at 37°C. Finally, the staining of colonies was performed using crystal violet (1%) for 20 minutes. The counting of colonies was performed to evaluate the effect of drugs on colony formation.
Statistical analysis
The difference between two groups was measured by the Student’s non-paired t-test, and the differences among three or more groups were measured by one-way ANOVA. P-values < 0.05 were statistically significant. All data were presented as mean ± SD, unless otherwise stated.
Results
Fabrication and properties of immunoliposomes
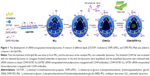
CD44-SATB1-ILs were fabricated by the three consecutive steps, including lipid film-based approach, lyophilization, and hydration. As shown in Figure 1, after the preparation of the liposomes, the thiolated antibodies were linked to the liposomes via the reaction of sulfhydryl groups on the antibody and maleimide groups. The size and other characteristics of liposomes are shown in Figure 2A. The size of immunoliposomes was a little larger than that of conventional liposomes. CD44-SATB1-ILs and CD44-NC-ILs were 159.3 and 156.8 nm, respectively, whereas NC-Lipo and SATB1-Lipo were 145.8 and 148.1 nm, respectively. The zeta potential of all the liposomes was positive and around 15–20 mV. The small polydispersity index (<0.2) of all liposomes showed the homogeneity of our prepared liposomes. Notably, the EE of all liposomes was higher than 90%, indicating that lyophilization/hydration approach represents an efficient way to encapsulate siRNA. The stability experiment, in which the liposomes were incubated in mediums for different periods, was evaluated in Figure 2B. An increase of about 15 nm was observed in the size of the liposomes, when they were in PBS + 10% FBS and PBS + 20% FBS, compared with that in PBS. However, the stability of liposomes was rather superior during the whole incubation period, suggesting that serum did not markedly affect the stability of liposomes.
The in vitro targeting of liposomes to gastric cancer cells
FAM-siRNA is an siRNA labeled with green fluorescence and could be used to track the in vitro targeting of siRNA-encapsulated liposomes in cells. As shown in Figure 3A, in CD44+ MKN-45 cells, the uptake of FAM-siRNA CD44-NC-ILs was prominently higher than that of FAM-siRNA NC-Lipo at every time points (at 1 hour, P < 0.05; at 2 hours, P < 0.01; at 4 and 6 hours, P < 0.001; at 12 hours, P < 0.01). However, in CD44− MKN-45 cells, FAM-siRNA CD44-NC-ILs showed similar uptake to FAM-siRNA NC-Lipo at every time points (Figure 3B). In the case of NCI-N87 cells, similar results were achieved (Figure 3C and D). FAM-siRNA CD44-NC-ILs showed increased uptake compared with FAM-siRNA NC-Lipo in CD44+ NCI-N87 cells at every time points, whereas it showed similar uptake compared with FAM-siRNA NC-Lipo in CD44− NCI-N87 cells. Taken together, the increased uptake of FAM-siRNA CD44-NC-ILs in CD44+ gastric cancer cells is dependent on the interaction of CD44 antibody and CD44 overexpressed on the CD44+ gastric cancer cells.
The SATB1 silencing ability of liposomes in gastric cancer cells
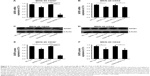
After the demonstration of the targeting activity of liposomes to gastric cancer cells, the SATB1 silencing ability of liposomes in gastric cancer cells was measured in both the mRNA and protein levels (Figure 4). We first evaluated the gene-silencing activity of liposomes in MKN-45 CD44+ cells (Figure 4A). As expected, NC-Lipo and CD44-NC-ILs barely affect the gene expression of SATB1. The gene-silencing activity of SATB1-Lipo is poor and only inhibited the gene expression of SATB1 by ~30%. On the contrary, CD44-SATB1-ILs remarkably inhibited the gene expression of SATB1 by ~80%, showing superior gene-silencing activity than SATB1-Lipo and CD44-NC-ILs (P < 0.001). We next evaluated the gene-silencing activity of liposomes in MKN-45 CD44− cells. Notably, CD44-SATB1-ILs exhibited similar poor gene-silencing activity to SATB1-Lipo, and both liposomes only suppressed the SATB1 mRNA expression by ~25% (Figure 4B). The analysis of the SATB1 protein expression after liposome treatment obtained the similar results shown in Figure 4C–F. In MKN-45 CD44+ cells (Figure 4C and E), NC-Lipo and CD44-NC-ILs barely affect the protein expression of SATB1. The protein suppression activity of SATB1-Lipo is poor and only inhibited the gene expression of SATB1 by ~30%, whereas CD44-SATB1-ILs markedly inhibited the gene expression of SATB1 by ~80%, showing superior gene-silencing activity than SATB1-Lipo and CD44-NC-ILs (P < 0.001). In MKN-45 CD44− cells, CD44-SATB1-ILs exhibited similar poor protein suppression activity to SATB1-Lipo, and both liposomes only suppressed the SATB1 protein expression by ~30% (Figure 4D and F). Therefore, these results confirmed that CD44-SATB1-ILs had potent SATB1-suppressing activity in CD44+ but not CD44− gastric cancer cells.
The cytotoxic effect of liposomes against the gastric cancer cells
The cytotoxic effect of liposomes on the proliferation of gastric cancer cells is investigated in Figure 5. NC-Lipo and CD44-NC-ILs barely affected the cell viability of MKN-45 CD44+ cells. SATB1-Lipo moderately inhibited the cellular proliferation of MKN-45 CD44+ cells by 40%. It is noteworthy that CD44-SATB1-ILs remarkably inhibited the cellular proliferation of MKN-45 CD44+ cells by ~80%, and the cellular proliferation inhibitory effect of CD44-SATB1-ILs was superior than SATB1-Lipo (P < 0.01). However, in MKN-45 CD44− cells, CD44-SATB1-ILs showed similar proliferation inhibitory effect to SATB1-Lipo, and both liposomes only suppressed the proliferation of MKN-45 CD44− cells by ~30%. As for NCI-N87 cells, similar results were achieved. In CD44+ NCI-N87 cells, the cytotoxic effect of CD44-SATB1-ILs was remarkably higher compared with SATB1-Lipo (P < 0.001), whereas its cytotoxic effect was similar to SATB1-Lipo CD44− NCI-N87 cells. In summary, CD44-SATB1-ILs showed preferential cytotoxic effects toward CD44+ gastric cancer cells.
The impact of liposomes on the gastric CICs proportion
Figure 6 shows the impact of liposomes on the percentage of gastric CICs, using the examination of tumorspheres formation and percentage of CD44+ cells. In MKN-45 cells, the treatment with SATB1-Lipo moderately decreased the number of tumorspheres (P < 0.05), whereas NC-Lipo and CD44-NC-ILs barely affect the number of tumorspheres (Figure 6A). Notably, the number of tumorspheres after CD44-SATB1-ILs treatment is significantly decreased compared with SATB1-Lipo (P < 0.01) and CD44-NC-ILs (P < 0.001). In NCI-N87 cells, similar results were achieved (Figure 6B). Although SATB1-Lipo moderately decreased the number of tumorspheres (P < 0.05), CD44-SATB1-ILs exhibited the best therapeutic efficacy in inhibiting the number of tumorspheres in NCI-N87 cells.
Consistent with the above results, after CD44-SATB1-ILs treatment, the percentage of CD44+ MKN-45 cells was remarkably reduced to 20%, from ~55% of CD44+ cells found in the original MKN-45 cell line (Figure 6C). The percentage of CD44+ MKN-45 cells moderately decreased to ~40% after treatment with SATB1-Lipo. Similar results were obtained in the case of NCI-N87 cells (Figure 6D).
Therefore, CD44-SATB1-ILs possessed the best efficiency toward the inhibition of formation of tumorspheres and percentage of CD44+ cells, thus presents a promising treatment of elimination of gastric CICs.
The effect of liposomes on colony formation in vitro
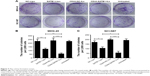
The effect of liposomes on colony formation in vitro was evaluated by crystal violet staining. After gastric cancer cells were incubated with liposomes for 24 hours and grown in fresh culture medium for 7 days, the number of colonies was counted (Figure 7). The representative images of the colony formation after treatment are shown in Figure 7A, and the number of the colonies was counted (Figure 7B and C). In MKN-45 cells, SATB1-Lipo reduced the formation of colonies compared with NC-Lipo (P < 0.01) (Figure 7B). Notably, the formation of colonies was significantly reduced by CD44-SATB1-ILs compared with SATB1-Lipo (P < 0.01) and CD44-NC-ILs (P < 0.001). Similar results were obtained in NCI-N87 cells (Figure 7C). SATB1-Lipo reduced the formation of colonies compared with NC-Lipo (P < 0.05) in NCI-N87 cells. Once again, the number of colonies after the treatment of CD44-SATB1-ILs was significantly lower than SATB1-Lipo (P < 0.05) and CD44-NC-ILs (P < 0.001).
Discussion
Since gastric CICs are considered to be the seed of gastric cancer, the eradication of gastric CICs could achieve better cancer therapeutic effect. Considering that CD44 is the marker of gastric CICs, hereby, we constructed SATB1 siRNA-encapsulated immunoliposomes conjugated with CD44 antibodies (CD44-SATB1-ILs), to target gastric CICs. In this study, CD44-SATB1-ILs showed significantly improved targeting and therapeutic effects toward gastric CICs.
Gene therapy is regarded as an up-and-coming treatment for various diseases such as cancer.7,8 Also, gene therapy is thought to be an alternative with much reduced effects to chemotherapy that are associated with lots of side effects.7,8 The gene vector is one of the main challenges encountered in application of gene therapy in clinic.7 Gene vectors are broadly classified into two categories: viral vectors and non-viral vectors. The non-viral vectors, including liposomes, and other nanoparticles, have several superior advantages than viral vectors. The advantages of non-viral vectors over viral vectors include biodegradability, low toxicity, and ease of synthesis. Therefore, non-viral vectors achieved extensive research interests.29 Particularly, cationic liposomes composed of DOTAP/cholesterol is one of the most efficient gene vectors.30 Nevertheless, the development of cationic liposomes was significantly impeded by their short circulation lifetime.31 The modification of poly(ethylene glycol) (PEG) in cationic liposomes could significantly prolong the short circulation lifetime of cationic liposomes.32 In our studies, we prepared PEGylated cationic liposomes, aiming to prolong the in vivo circulation time of cationic liposomes. However, siRNA in aqueous solution is susceptible to nuclease and the easy degradation of siRNA severely hampers the application of siRNA in clinic.7 Since the widespread use of siRNA-loaded liposomes is impeded by their instability, lyophilization is a very effective approach to increase the stability of liposomes.33 Only when in use, lyophilized liposomes are hydrated with siRNA solution and immediately used for therapeutic application.15,17 The rather short time of siRNA in aqueous solution avoids the long-time interaction of siRNA with nuclease.15,17 Although the current liposomes in the market are generally prepared by the active-loading approach, such as ammonium sulfate gradient method, the reason is that most chemotherapy drugs such as doxorubicin are rather stable and not easy to be degraded.34 In the case of siRNA, which is susceptible to be degraded by nuclease, the lyophilization approach represents a practical method for preparation of siRNA-encapsulated liposomes.35 Therefore, our lyophilized siRNA-encapsulated liposomes represent a very up-and-coming approach in the facilitation of siRNA-based therapy in clinic.
To enhance the targeting and gene-silencing activity of cationic liposomes, the development of immunoliposomes is a practical way.11,12 The most commonly adopted ligands is antibody, and the Fab’ of mAb was often used due to its better penetration into solid tumors, reduced immunogenicity, and improved pharmacokinetic profiles, compared with the mAb.25,26 In fact, antibody-conjugated nanoparticles represent a promising strategy toward various cancers because they can significantly enhance the therapeutic efficacy of chemotherapy drugs.25,26 It is noteworthy that there have been three antibody-conjugated nanoparticles loaded with chemotherapy drugs (doxorubicin and docetaxel), which have already successfully translated into early-phase clinical trials.36,37 Those results of the early-phase clinical trials demonstrated the good safety and efficacy of antibody-conjugated nanoparticles.36,37 In our study, the selection of CD44 antibodies is very critical for the specific targeting of our developed immunoliposomes to gastric CICs. The results showed that, in CD44+ gastric CICs, CD44-SATB1-ILs showed significantly increased targeting compared with SATB1-Lipo, resulting in increased cytotoxic effects and tumorsphere inhibitory effects. However, in CD44− gastric cancer cells, the cytotoxicity and tumorsphere inhibitory effects of CD44-SATB1-ILs did not differ from that of SATB1-Lipo. These data firmly demonstrated that CD44-SATB1-ILs were able to exert increased targeting and therapeutic effect toward gastric CICs, and CD44 antibodies could promote the targeting of nanoparticles to gastric CICs. As far as we know, this is the first research that the promotion of siRNA delivery to gastric CICs has been achieved by immunoliposomes.
The selection of gene target is important for the superior activity of our gene therapy. SATB1 is a validated anticancer target. Han et al reported that the overexpression of SATB1 has been found in aggressive breast cancer cells, and SATB1 expression level also had significant prognostic significance.38 In highly aggressive cancer cells, gene silencing of SATB1 altered the expression of over 1,000 genes, reversed tumorigenesis, and inhibited tumor metastasis and growth in vivo.38 In contrast, in non-aggressive cancer cells, ectopic SATB1 expression increased the expression of aggressive-tumor phenotype-associated genes.39 Zheng also showed that SATB1 expression is higher in aggressive cancer than in non-aggressive cancer.20 Similarly, forced overexpression of SATB1 in non-metastatic cells enhanced the invasion ability of tumors in mice, whereas suppression of SATB1 expression prevented tumor growth and metastasis in many tumors.40,41 Notably, SATB1 participates in maintenance of stem cells and CICs, suggesting that SATB1 represents a superior candidate target in both cancer cells and CICs.22,23 To our knowledge, there have been no studies that reported therapeutic effect of the inhibition of SATB1 expression against CICs. In this study, we confirmed that SATB1 suppression preferentially eliminated CD44+ gastric CICs in the cytotoxicity assay. In the tumorsphere formation assay, SATB1 suppression could also inhibit the number of tumorspheres. The percentage of CD44+ cells in gastric cancer cells was significantly decreased after SATB1 suppression. Consistently, CD44-SATB1-ILs could also reduce the formation of colonies in gastric cancer cells. To our knowledge, this is the first report that demonstrated that SATB1 suppression is able to exhibit potential activity toward gastric CICs.
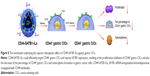
Taken together, the mechanism underlying the superior therapeutic effect of CD44-SATB1-ILs against gastric CICs is elucidated in Figure 8. CD44-SATB1-ILs could efficiently target CD44+ gastric CICs and repress SATB1 expression, resulting in the proliferation inhibition of CD44+ gastric CICs and also the decrease of the percentage of CD44+ gastric CICs and tumorsphere formation in gastric cancer cells.
Conclusion
This study is the first report of the potential activity of SATB1 suppression toward gastric CICs and also the first report of the promotion of siRNA delivery via nanoparticles to gastric CICs by utilization of targeting ligands. CD44-SATB1-ILs were confirmed to selectively target and eliminate CD44+ gastric CICs. Therefore, CD44-SATB1-ILs represent an up-and-coming approach for the therapy of gastric CICs.
Disclosure
The authors report no conflicts of interest in this work.
References
World Health Organization [webpage on the Internet]. Cancer: Fact Sheet No 297. WHO. Available from: http://www.who.int/mediacentre/factsheets/fs297/en/. Accessed May 21, 2015. | ||
Surveillance, Epidemiology, and End Results Program [webpage on the Internet]. SEER Stat Fact Sheets: Stomach Cancer. National Cancer Institute. Available from: http://seer.cancer.gov/statfacts/html/stomach.html. Accessed May 21, 2015. | ||
Yoo CH, Noh SH, Shin DW, Choi SH, Min JS. Recurrence following curative resection for gastric carcinoma. Br J Surg. 2000;87(2):236–242. | ||
Alberts SR, Cervantes A, van de Velde CJ. Gastric cancer: epidemiology, pathology and treatment. Ann Oncol. 2003;14(Suppl 2):31–36. | ||
Takaishi S, Okumura T, Tu S, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27(5):1006–1020. | ||
Takaishi S, Okumura T, Wang TC. Gastric cancer stem cells. J Clin Oncol. 2008;26(17):2876–2882. | ||
Niidome T, Huang L. Gene therapy progress and prospects: nonviral vectors. Gene Ther. 2002;9(24):1647–1652. | ||
Li SD, Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13(18):1313–1319. | ||
Suh JS, Lee HJ, Nam H, et al. Control of cancer stem cell like population by intracellular target identification followed by the treatment with peptide-siRNA complex. Biochem Biophys Res Commun. 2017;491(3):827–833. | ||
Long Q, Yang R, Lu W, et al. Adenovirus-mediated truncated Bid overexpression induced by the Cre/LoxP system promotes the cell apoptosis of CD133+ ovarian cancer stem cells. Oncol Rep. 2017;37(1):155–162. | ||
Gao J, Liu W, Xia Y, et al. The promotion of siRNA delivery to breast cancer overexpressing epidermal growth factor receptor through anti-EGFR antibody conjugation by immunoliposomes. Biomaterials. 2011;32(13):3459–3470. | ||
Gao J, Yu Y, Zhang Y, et al. EGFR-specific PEGylated immunoliposomes for active siRNA delivery in hepatocellular carcinoma. Biomaterials. 2012;33(1):270–282. | ||
Xu C, Lee SA, Chen X. RNA interference as therapeutics for hepatocellular carcinoma. Recent Pat Anticancer Drug Discov. 2011;6(1):106–115. | ||
Gao J, Ochyl LJ, Yang E, Moon JJ. Cationic liposomes promote antigen cross-presentation in dendritic cells by alkalizing the lysosomal pH and limiting the degradation of antigens. Int J Nanomedicine. 2017;12:1251–1264. | ||
Gao J, Sun J, Li H, et al. Lyophilized HER2-specific PEGylated immunoliposomes for active siRNA gene silencing. Biomaterials. 2010;31(9):2655–2664. | ||
Cushing BL, Kolesnichenko VL, O’Connor CJ. Recent advances in the liquid-phase syntheses of inorganic nanoparticles. Chem Rev. 2004;104(9):3893–3946. | ||
Peer D, Margalit R. Physicochemical evaluation of a stability-driven approach to drug entrapment in regular and in surface-modified liposomes. Arch Biochem Biophys. 2000;383(2):185–190. | ||
Cheng C, Lu X, Wang G, et al. Expression of SATB1 and heparanase in gastric cancer and its relationship to clinicopathologic features. APMIS. 2010;118(11):855–863. | ||
Lu X, Cheng C, Zhu S, et al. SATB1 is an independent prognostic marker for gastric cancer in a Chinese population. Oncol Rep. 2010;24(4):981–987. | ||
Zheng J. Is SATB1 a master regulator in breast cancer growth and metastasis? Womens Health (Lond). 2008;4(4):329–332. | ||
Neznanov N, Kohwi-Shigematsu T, Oshima RG. Contrasting effects of the SATB1 core nuclear matrix attachment region and flanking sequences of the keratin 18 gene in transgenic mice. Mol Biol Cell. 1996;7(4):541–552. | ||
Sun Z, Zhang C, Zou X, et al. Special AT-rich sequence-binding protein-1 participates in the maintenance of breast cancer stem cells through regulation of the Notch signaling pathway and expression of Snail1 and Twist1. Mol Med Rep. 2015;11(5):3235–3242. | ||
Chen B, Xue Z, Yang G, et al. Akt-signal integration is involved in the differentiation of embryonal carcinoma cells. PLoS One. 2013;8(6):e64877. | ||
Gao J, Xia Y, Chen H, et al. Polymer-lipid hybrid nanoparticles conjugated with anti-EGF receptor antibody for targeted drug delivery to hepatocellular carcinoma. Nanomedicine. 2014;9(2):279–293. | ||
Gao J, Chen H, Song H, et al. Antibody-targeted immunoliposomes for cancer treatment. Mini Rev Med Chem. 2013;13(14):2026–2035. | ||
Gao J, Feng SS, Guo Y. Antibody engineering promotes nanomedicine for cancer treatment. Nanomedicine. 2010;5(8):1141–1145. | ||
Wang J, Wu Z, Pan G, et al. Enhanced doxorubicin delivery to hepatocellular carcinoma cells via CD147 antibody-conjugated immunoliposomes. Nanomedicine. 2018;14(6):1949–1961. | ||
Gao J, Kou G, Wang H, et al. PE38KDEL-loaded anti-HER2 nanoparticles inhibit breast tumor progression with reduced toxicity and immunogenicity. Breast Cancer Res Treat. 2009;115(1):29–41. | ||
Yang SY, Zheng Y, Chen JY, et al. Comprehensive study of cationic liposomes composed of DC-Chol and cholesterol with different mole ratios for gene transfection. Colloids Surf B Biointerfaces. 2013;101:6–13. | ||
Kim SY, Lee SJ, Lim SJ. Formulation and in vitro and in vivo evaluation of a cationic emulsion as a vehicle for improving adenoviral gene transfer. Int J Pharm. 2014;475(1–2):49–59. | ||
Salvati A, Ciani L, Ristori S, Martini G, Masi A, Arcangeli A. Physico-chemical characterization and transfection efficacy of cationic liposomes containing the pEGFP plasmid. Biophys Chem. 2006;121(1):21–29. | ||
Dass CR, Choong PF. Selective gene delivery for cancer therapy using cationic liposomes: in vivo proof of applicability. J Control Release. 2006;113(2):155–163. | ||
Ghanbarzadeh S, Valizadeh H, Zakeri-Milani P. The effects of lyophilization on the physico-chemical stability of sirolimus liposomes. Adv Pharm Bull. 2013;3(1):25–29. | ||
Samad A, Sultana Y, Aqil M. Liposomal drug delivery systems: an update review. Curr Drug Deliv. 2007;4(4):297–305. | ||
Yadava P, Gibbs M, Castro C, Hughes JA. Effect of lyophilization and freeze-thawing on the stability of siRNA-liposome complexes. AAPS PharmSciTech. 2008;9(2):335–341. | ||
Mamot C, Ritschard R, Wicki A, et al. Tolerability, safety, pharmacokinetics, and efficacy of doxorubicin-loaded anti-EGFR immunoliposomes in advanced solid tumours: a phase 1 dose-escalation study. Lancet Oncol. 2012;13(12):1234–1241. | ||
Miller K, Cortes J, Hurvitz SA, et al. HERMIONE: a randomized Phase 2 trial of MM-302 plus trastuzumab versus chemotherapy of physician’s choice plus trastuzumab in patients with previously treated, anthracycline-naïve, HER2-positive, locally advanced/metastatic breast cancer. BMC Cancer. 2016;16:352. | ||
Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452(7184):187–193. | ||
Zimmerman MA, Rahman NT, Yang D, et al. Unphosphorylated STAT1 promotes sarcoma development through repressing expression of Fas and bad and conferring apoptotic resistance. Cancer Res. 2012;72(18):4724–4732. | ||
Hubackova S, Krejcikova K, Bartek J, Hodny Z. Interleukin 6 signaling regulates promyelocytic leukemia protein gene expression in human normal and cancer cells. J Biol Chem. 2012;287(32):26702–26714. | ||
Moore F, Cunha DA, Mulder H, Eizirik DL. Use of RNA interference to investigate cytokine signal transduction in pancreatic beta cells. Methods Mol Biol. 2012;820:179–194. |
Supplementary material
  | Table S1 Sequence of the primers and siRNA |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.