Back to Journals » Cancer Management and Research » Volume 12
Sal-Like Protein 4 Transcription Factor: A Significant Diagnostic Biomarker Involved in Childhood ALL Resistance and Relapse
Authors Ohadi F, Rahgozar S , Ghodousi ES
Received 29 November 2019
Accepted for publication 8 February 2020
Published 4 March 2020 Volume 2020:12 Pages 1611—1619
DOI https://doi.org/10.2147/CMAR.S240469
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Farzaneh Ohadi, Soheila Rahgozar, Elaheh Sadat Ghodousi
Department of Cell and Molecular Biology & Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, Iran
Correspondence: Soheila Rahgozar
Department of Cell and Molecular Biology & Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Hezar Jarib Street, Isfahan 81746-73441, Iran
Tel +98 313 793 2455
Fax +98 313 793 2456
Email [email protected]
Purpose: Sal‐like protein 4 transcription factor (SALL4) is a stem cell transcription factor that plays an essential role in the maintenance and self-renewal of embryonic and hematopoietic stem cells, functioning as an oncogene in several cancers. However, the role of SALL4 in the biological behavior of childhood acute lymphoblastic leukemia and its relationship with multidrug resistance and relapse has remained largely unknown.
Patients and Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was used to characterize the expression pattern of SALL4 in the bone marrow samples of 43 patients with Philadelphia negative ALL and 18 children in the non-cancer control group. The presence of minimal residual disease was measured a year after the initial therapy using SSCP (single-strand conformation polymorphism). In addition, the correlation between the expression of SALL4 and ABCA3 in relapsed patients was analyzed statistically.
Results: Results showed an overexpression of SALL4 in de novo patients compared with the control group (P=0.0001, AUC= 0.93), indicating the importance of this gene in the induction of leukemia. A significant increase in the ABCA3 expression levels was revealed in the relapsed patients, in comparison with the drug-sensitive group (P = 0.0005). The leukemogenetic effect of SALL4 can be related to the effect of this gene on the maintenance of pluripotency in cancer stem cells. Results also suggest that the expression of SALL4 can be considered as a diagnostic marker for pediatric ALL. Moreover, SALL4 expression levels in the minimal residual disease positive (mrd+) ALL group was significantly higher than those in the mrd− group (p=0.0001, AUC= 0.92).
Conclusion: These data demonstrate the prognostic impact of SALL4 in childhood ALL. Our findings also indicated a direct correlation between the mRNA expression levels of SALL4 and ABCA3 transporter in the relapsed group of ALL patients (r=0.7). These results describe a possible mechanism by which SALL4 may lead to the development of multidrug resistance.
Keywords: childhood acute lymphoblastic leukemia, multidrug resistance, SALL4, ABC transporters, relapse, ABCA3
Introduction
Acute lymphoblastic leukemia (ALL), a heterogeneous disease, is the most frequent type of cancer among all hematological malignancies and accounts for 75–80% of all pediatric leukemia.1–3 Clinically, ALL is characterized by excess T- or B-lymphoblasts in the bone marrow and peripheral blood, while its etiology is multi-factorial, with endogenous and exogenous exposures, genetic susceptibility, and chance all playing a part. Chemotherapy and radiotherapy are the standard first-line therapy for ALL.4 Despite the increasing efficiency of the involved anti-leukemic drugs over the past decades, multidrug resistance (MDR) remains a great obstacle for successful treatment, leading to relapse in 15–20% of the patients. MDR is a multifactorial phenomenon, but a subfamily of ABC transporters may be critically involved as they are responsible for the majority of drug efflux in human cancers, and past studies by our team have revealed prognostic roles for ABCA2 and ABCA3 in childhood ALL drug resistance.2,5
Recently, there has been an accumulation of evidence indicating that Sal‐like protein 4 transcription factor (SALL4), a stem cell gene, is an oncogene that has a central function in carcinogenesis, chemotherapeutic resistance, and relapse in a variety of cancers.6–8 In some cancers, SALL4 can promote the expression of a subfamily of ABC transporters, through direct or indirect interaction with the promoter, and affect sensitivity to the chemotherapy drug.9 As a member of the Sal-like (SALL) gene family (SALL1 to SALL4), SALL4 encodes for a C2H2 zinc-finger transcription factor.6 This is an essential factor that governs the pluripotent and self-renewal properties of embryonic stem cells through its interaction with other stemness-related genes, such as the Octamer-Binding Transcription Factor 4 (OCT-4), the Nanog Homeobox (NANOG), and the Sex Determining Region Y-Box 2 (SOX2).10–12 The SALL4 gene is located on 20q13, involving four coding exons and its expression is elevated in embryonic stem cells, and gradually decreases during embryo-fetal development and is almost entirely absent in fully adult tissues, with the exception of expression in germ line cells and the hematopoietic stem cells.13,14 In normal bone marrows (BMs), it is highly expressed in hematopoietic stem cells (HSPCs) but is not present after hematopoietic differentiation. However, re-expression and aberrant expression of SALL4 has been detected in a subset (30%) of solid tumors, lymphoma and acute/chronic myeloid leukemia, demonstrating its oncogenic capabilities.15,16 It is suggested that SALL4 can play roles in B cell lineage acute lymphoblastic leukemia (B-ALL) and acute myeloid leukemia (AML) pathogenesis.17 Moreover, in the AML patients, increased expression level of SALL4 results in drug resistance and poor prognosis through regulating the expression of the ATP-binding cassette (ABC) drug transports, such as ABCG2 and ABCA3.9,18 Nevertheless, with the general lack of studies about SALL4 expression and its correlation with ABC transporter genes, there is little information about the role of this gene in the biological behavior of childhood ALL and its correlation with MDR and relapse.
In the present study, we determine the expression levels of SALL4 in the bone marrow and peripheral blood mononuclear cell samples of children with ALL. In addition, we demonstrate a correlation between the expression of SALL4 and ABCA3 in relapsed patients.
Materials and Methods
Patients and Control Samples
In this study, children that were referred to Sayed-ol-Shohada Hospital (Isfahan, Iran) in 2015–2017 for bone marrow evaluation and were then clinically diagnosed and pathologically confirmed to be Philadelphia negative ALL patients, were selected for the study. The project was approved by the University of Isfahan review board under the agreement number 94/9684. The diagnoses of all patients were based on cytomorphology, cytoimmunology, immunohistochemistry and cytogenetic analysis. The type of leukemia was diagnosed through phenotype evaluation with flowcytometry (kits provided by Daco and the flow cytometer purchased from Partec) alongside routine tests of CBC count and morphology assessment.
The study was performed on 43 ALL Philadelphia negative, Burkitt leukemia/lymphoma excluded patients (15 females, 28 males, 0.8–14 years of age); 32 patients were untreated-newly diagnosed, and 11 were patients with bone marrow relapse (11 B-cell lineage). Eighteen cases with leukemia cell-free bone marrow were considered as the age- and sex-matched control group. Other details of the samples are listed in Table 1.
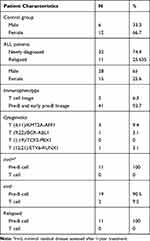 |
Table 1 Primary Data of the Patients Included in the Study |
Sampling was achieved with full written informed parent’s consent and in compliance with the ethical protocol and standards of Sayed-ol-Shohada Hospital, Isfahan, which is in accordance with the Declaration of Helsinki. Two to five milliliters of bone marrow sample or peripheral blood in heparin was obtained and sent on ice to the cellular-molecular biology laboratory of University of Isfahan for cell isolation and RNA extraction.
Isolation of Mononuclear Cells from Bone Marrow
Extraction of mononuclear cells from bone marrow samples of patients and controls was accomplished using density gradient LymphoprepTM (Axis-Shailed Diagnostics Ltd, Oslo, Norway) according to the protocol provided by the manufacturer and based on the buoyant density gradient of blood constituents.
RNA Extraction from Fresh-Frozen Samples
Total RNA was extracted from the mononuclear cells using the Trizol kit (Invitrogen, CA, USA). The extracted RNA was dissolved in 30μL RNase-free water and the amount and quality of the extracted RNA were assessed using a bio-photometer (Eppendorf) and gel electrophoresis.
cDNA Synthesis
Complementary DNA (cDNA) synthesis for SALL4 was carried out by transformation of 100 nanograms/microliter of total RNA, according to the protocol provided by the PrimeScript™RT reagent cDNA synthesis kit (Takara, Japan), using random hexamers and oligo dT primers in a specific temperature plan (sequentially: 60 min at 42 °C, 5 min at 95 °C and quickly at 4°C). The resulting cDNAs were preserved at −20 °C for future steps.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The expression levels of SALL4 and ABCA3 were determined by SYBR Premix Ex TaqII (Exiqon, Denmark) and melting curve analysis using Chromo 4 (BIO-RAD, USA). The quantitative RT-PCR was performed in a 10μL total volume containing 1 μL of cDNA, 8.2μL of PCR master mix (Exiqon, Denmark), and 0.8μL primer pairs. Forty cycles were done using the following protocol: 10 min pre-incubation at 95 °C, 10 s denaturation at 95 °C, 60 s annealing at 58–60 °C and 1 min product expansion at 72 °C. The housekeeping gene used in this study for the internal control was glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primers used included GADPH, and ABCA3 extracted from previous studies. Primer sequences of the SALL4, ABCA3 and GAPDH genes are summarized in Table 2.
 |
Table 2 Primer Sequences of the SALL4, ABCA3 and GAPDH |
A no-reverse transcription (no-RT) control was used to detect any potential non-specific amplification of genomic DNA and no template controls (NTC) were run to evaluate contamination. All reactions were performed in duplicates. The data were presented as the relative expression of the genes of interest relative to the internal control gene as determined by the 2(−ΔΔCT) Method. Additionally, the specific amplification of the PCR products was analyzed by melting curve analysis and agarose gel electrophoresis. The results were presented as fold change patient samples group relative to the normal samples group. The primers used for real-time PCR for all gene amplifications were synthesized by Pishgam Biotec Co.
Assessing Response to Therapy
Samples were isolated on the basis of Immunophenotype, and patients were treated based on the Australian and New Zealand Children’s Haematology/Oncology Group, consisting of induction (VCR, ADR, l-asparaginase, prednisolone), consolidation (6MP, CPM, cytarabine), protocol M (4 injections of High dose MTX), reinduction, reconsolidation, maintenance (6MP, weekly MTX, monthly CPM and VCR), and intrathecal injections (https://www.anzctr.org.au/Trial/Registration). In order to determine response to therapy, samples were sent to the Payvand Clinical and Specialty Laboratory (Tehran, Iran), the presence of minimal residual disease (mrd) was studied using PCR-SSCP (Polymerase chain reaction coupled single-strand conformation polymorphism) analysis for immunoglobulin heavy chain (IgH) and T-cell receptor gamma (TcRγ) gene rearrangements performed a year after treatment. This is generally believed that most malignancies are clonal in origin. Besides, TcR rearrangement analysis is a method used to determine clonality in a T-cell population. Therefore, PCR-SSCP is a technique inspecting the association between the number of amplified clones and band patterns of the amplified products using consensus primers for the IgH and TcRγ genes and a thermostatically controlled electrophoresis apparatus in case that one single band is detected on the gel.19 Monoclonality, referred to as mrd+, was reflected on as a poor response to therapy and drug resistance (Table 1).
Statistical Analysis
All statistical analyses were conducted using the SPSS 21.0 and GraphPad Prism 6. Differences in mRNA expression between two groups were analyzed using the Mann–Whitney U-test. Data were presented as median. Spearman’s rank correlation analysis was used to analyze the SALL4 and ABCA3 mRNA levels in different samples. Differences were considered statistically significant at P< 0.05.
Results
The high amplification efficiency of the SALL4 and ABCA3 genes was consistent with that of the GAPDH reference gene. The PCR products from all of the genes of interest were confirmed using 3% agarose gel electrophoresis (data not shown). The SALL4 and ABCA3 genes were detected in all of the bone marrow mononuclear cell samples from the non-cancer individuals and those with acute lymphoblastic leukemia.
Higher Expression Levels of SALL4 in Newly Diagnosed Children with ALL (T-ALL & B-ALL)
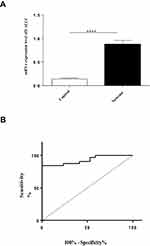
SALL4 overexpression was found in de novo ALL patients. The relative mRNA expression of SALL4 in ALL patients at diagnosis was increased compared with the control group (0.8718 ± 0.08479, n=32 vs 0.1390 ± 0.02075, n=18, P<0.001) (Figure 1A). Considering the statistically significant relative difference in mRNA expression of SALL4 between patients and the control group, Receiver Operator Characteristics (ROC) curves were made across various cut-off levels of the predicted probabilities and showed that the increased expression of this gene had a positive predictive value for ALL diagnosis, with remarkable sensitivity and specificity of 84.38% and 100%, respectively. The area under the ROC curve (AUC) for expression of SALL4 gene was 0·93 (Figure 1B).
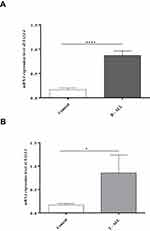
Further, a significant relationship was found between the phenotype of ALL (B cell and T cell) and SALL4 expression so that it demonstrated a higher expression in B and T ALL patients compared with the control group (P<0.0001 and 0.02, respectively). Even though the examined T-ALL population is too small, SALL4 expression in T ALL patients was higher than the cutoff point (Figure 2).
Evaluating SALL4 as a Prognostic Factor
Patients with positive mrd (persisting monoclonal band of IgH or TCγR gene rearrangements after one year of therapy) were considered as the drug resistant group. Interestingly, comparing the mRNA expression levels of the studied gene within the aforementioned group and the mrd−/drug-sensitive patients (those with elimination of the monoclonal band) indicated a significantly 4 fold increased expression of SALL4 in the resistant ALLs (0.9456 ± 0.1215, n=22 vs 0.2390 ± 0.07217, n=21, respectively, P<0.0001) (Figure 3A), proposing this gene as a prognostic biomarker for drug resistance in ALL. ROC curve analysis showed a notable AUC of 0.92, indicating SALL4 level as a negative prognostic biomarker which may discriminate between resistant ALL patients and drug-sensitive cases. More analysis also revealed a significant increase in SALL4 expression levels in relapsed patients compared with the mrd− group (0.6110 ± 0.1156, n=22 vs 0.2390 ± 0.07217, n=21, respectively, P=0.0002) (Figure 3B).
Correlation of Relative Expression of SALL4 and ABCA3 in Relapsed and mrd+ ALLs
A recent study by our research team determined the poor prognostic role of ABCA2 and ABCA3, from the subfamily of ABCA transporters, in multidrug-resistant childhood ALL. In accordance with our previous investigations, results showed that both transporters expression levels were increased significantly in relapsed patients compared with the mrd− group (P = 0.007 and P = 0.0005, respectively).20
Correlation analysis of the relative expression levels of SALL4 and ABCA3 was performed using Spearman’s rank correlation analysis of the relapsed patients (n=11). A positive expression correlation level for SALL4 and ABCA3 genes was found in the relapsed group (rs=0.7, P<0.0088) (Figure 4). In addition, Fisher’s exact test showed that when the SALL4 expression levels are higher than a cutoff point of 0.27, the risk of drug resistance or positivity of mrd will be 69-fold higher (P<0.0001) (Table 3). In contrast, the correlation between SALL4 and ABCA2 expression levels was not significant (P>0.05) (data not shown).
 |
Table 3 Relation Between SALL4 Gene Expression and mrd |
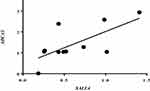 |
Figure 4 Correlation analysis of the relative SALL4 and ABCA3 expression levels in relapsed group (r= 0.7, P< 0.0088). |
Discussion
Despite improved cure rates for newly diagnosed childhood ALL, approximately 20% of patients are still classified as high-risk and resistant to current combined therapy.21,22 MDR is one of the main reasons for chemotherapeutic failure and consequent relapse in cancer therapy, with different mechanisms contributing to resistance; among these, the overexpression of ATP binding cassette (ABC) transporters is most important.23 Previous studies by our group had revealed a prognostic role for some of these transporters in childhood ALL drug resistance.20,24 However, the actual mechanism of failure has remained poorly understood.
SALL4 is a zinc-finger transcriptional factor and a stem cell gene that plays a critical role in the maintenance and self-renewal of embryonic and hematopoietic stem cells and its expression is normally restricted to embryonic and somatic stem cells.7,8 While SALL4 plays a key role in hematopoietic differentiation, it is also overexpressed in various human cancers, including hematologic malignancies and subsets of liver, gastric, lung, endometrial, and breast cancers.25,26 In addition, multiple studies have demonstrated that higher expression levels of SALL4 is associated with drug resistance in some malignancies.14,27,28 However, the role of SALL4 in the biological behavior of leukemia and its relation with MDR and relapse has remained largely unknown. Indeed, previous studies have not established an expression profile for the gene in childhood ALL. In the present study, we have demonstrated, for the first time, the SALL4 mRNA expression profile in ALL patients’ primary cancer cells. Our study showed that SALL4 expression levels were increased significantly in newly diagnosed patients compared with non-cancer individuals. Although the examined population was rather small, SALL4 could be detected with 84.38% sensitivity and 100% specificity, leading to the hypothesis that SALL4 might be used as a reliable diagnostic biomarker in pediatric ALL. On the other hand, it has been demonstrated that patients with increased expression levels of SALL4 at the time of diagnosis possessed a remarkably higher likelihood of being resistant to the therapy and a higher clinical risk classification. Therefore, SALL4 can be introduced as an important and promising potential cancer biomarker to predict the prognosis of childhood ALL. Moreover, SALL4 mRNA expression was increased in relapsed group, demonstrating a potential role of this gene in acquired ALL drug resistance.
In accordance with our investigation, several studies, using different analyses, have found higher expression levels of SALL4 in several hematological cancer cell lines.11,29 It is suggested that SALL4 can play a key role in B-ALL cell survival.17 A constitutive expression of SALL4 has been shown to be sufficient in inducing AML, and the SALL4B transgenic mouse model exhibits deregulated hematopoiesis and develops myelodysplastic syndrome (MDS)-like features.11 Moreover, knockdown or deletions of SALL4 in the leukemic cells of mouse bone marrow have led to a significant decrease in the expression of Bmi-1. This is significant, as Bmi-1, a target gene for SALL4 in both hematopoietic and leukemic cells, is a putative oncogene that modulates stem cell pluripotency and plays a role in leukemogenesis.30 Together, these data emphasize the role of SALL4 in leukemia. It is reasonable to suggest that SALL4 expression can be reactivated in pediatric ALL; and because of its high sensitivity, SALL4 detection may have therapeutic value for these patients. In agreement with this suggestion, recent studies have shown that SALL4 is a novel sensitive and specific diagnostic marker of ovarian primitive germ cell tumors, hepatoid gastric carcinoma from hepatocellular carcinoma, and testicular germ cell tumors.7,14,31,32 Therefore, SALL4 oncofetal proteins have great potential to become therapeutic target spots.
Another assertion by previous studies has been that SALL4 is highly expressed in B, but not, T ALL cell lines.33,34 However, our findings have revealed higher expressions in not only B-ALL, but T-ALL groups as well (Figure 2). Although the examined T-ALL population is too small, but all samples showed increased expression levels of the gene, that was higher than the cut-off point (P<0.02). Further studies are warranted to investigate the molecular mechanisms underlying this novel phenomenon in T-ALL patients.
It has been demonstrated that higher expression levels of SALL4 in several cancers, such as endometrial, liver and small cell lung carcinoma, is associated with drug resistance and relapse.14,27,35-37 In accordance with SALL4’s role in drug resistance, our results showed that the mRNA expression profile of the gene is significantly higher in MDR patients than drug-sensitive/mrd− individuals (P<0.0001), suggesting that its expression profile has failed to turn off in drug-resistant patients. The different SALL4 expression patterns in mrd− and mrd+ (Figure 3A) patients suggest that these two phase entities may have different biological characteristics and/or modulator mechanisms, at least concerning the SALL4 expression. It is even possible that decreased expression levels of SALL4 could be associated with complete disease remission.
SALL4 molecule co-operatively plays a role in MDR by regulating the expression of ABC transporters in patients.3,27 It has been shown that various ABC transporters contribute to drug resistance in diverse cancers. ABC transporters pump the drugs out of the cancer cells, and this efflux mostly governs the defense against chemotherapeutic drugs. Of note is that some of these drug resistance ABC transporters can be induced by SALL4, specially in leukemic cells.3 Based on previous reports, ABC transporter ABCB1 is critical for SALL-4-induced drug resistance in endometrial cancer. Furthermore, SALL4 can influence the dis-regulation of ABCG2 and ABCA3 in myeloid leukemia.14,27,28 They indicated that SALL4 directly binds to the promoter region of ABCA3, promoting drug resistance by up-regulation of this transporter, and affecting the formation of SP cells in AML and leukemic stem cells.3 Our study supports this by demonstrating a direct correlation between the mRNA expression levels of SALL4 and ABCA3 transporter in the relapsed pediatric ALL group. These findings suggest the therapeutic potential of inactivating ABCA3 expression through targeting SALL4 in ALL to conquer drug resistance.
A growing body of evidence has shown that reoccurring cancers may be initiating from cancer stem cells.38 SALL4 is a stem cell factor which has been demonstrated to be required for the self-renewal of cancer stem cells, side population (SP) cells, and various cancer cell types, while the knockdown of SALL4 has led to a reduced frequency of SP cells, eventually giving rise to disease relapse.39 On top of that, relapse could arise from acquisition of a resistant phenotype in response to therapy, presumably as a result of obtaining sequential genetic mutations after diagnosis and during chemotherapy.40,41 In the current study, based on Spearman correlation test, the abundance of ABCA3 transcript levels was significantly correlated with the excess levels of SALL4 in the patients with relapsed ALL (r=0.70, P< 0.0088) (Figure 4). Moreover, our results showed that the mRNA expression profile of SALL4 is significantly higher in MDR patients than drug-sensitive/mrd− individuals (P<0.0001) (Table 3). Besides, our data demonstrated a significant increase in SALL4 expression levels in relapsed patients compared with the mrd− group (Figure 3B). The results of Fisher exact test also revealed that the SALL4 expression levels higher than a cutoff point of 0.27 notably elevated the risk of positivity of mrd so that the significant increased risk was 69-fold higher than those whose mrd status were negative (Table 3). Bearing in mind that the positivity of mrd is correlated with high probability of relapse,41 it can be concluded that SALL4 overexpression can be introduced as a risk factor for relapse. Data also suggest a possible correlation between SALL4 expression and acquired resistance which can be mediated by induced increased expression levels of ABCA3 transcripts. This assertation is in agreement with the results of an investigation by Hupfeld et al which showed that CML cell lines exposure to a chemotherapy agent promotes increased expression of ABCA3 together with SALL4.42
In conclusion, the current study is the first to report the predictive significance of SALL4 in pediatric ALL patients. ROC curve analysis confirmed this impact with 100% sensitivity. Moreover, SALL4 is demonstrated to show a prognostic role for multidrug resistance and relapse in childhood ALL. A possible mechanism underlying this correlation is suggested to be the negative impact of SALL4 on the expression of ABCA3 transporter, which leads to the gross efflux of the administered chemotherapy drugs from the cancer cell cytoplasm, and thus contributes to chemoresistance and relapse. These results may open novel and exciting avenues for further research and development of improved therapies regarding ALL and MDR.
Acknowledgments
The authors are indebted to professor Alireza Moafi for kindly providing the samples, and the children and parents who participated in the study, without whom this research would not have been feasible.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541–1552. doi:10.1056/NEJMra1400972
2. Cooper SL, Brown PA. Treatment of pediatric acute lymphoblastic leukemia. Pediatr Clin North Am. 2015;62(1):61. doi:10.1016/j.pcl.2014.09.006
3. Jeong H-W, Cui W, Yang Y, et al. SALL4, a stem cell factor, affects the side population by regulation of the ATP-binding cassette drug transport genes. PLoS One. 2011;6(4):e18372. doi:10.1371/journal.pone.0018372
4. Antillón FG, Blanco JG, Valverde PD, et al. The treatment of childhood acute lymphoblastic leukemia in Guatemala: biologic features, treatment hurdles, and results. Cancer. 2017;123(3):436–448. doi:10.1002/cncr.v123.3
5. Moorman AV. New and emerging prognostic and predictive genetic biomarkers in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2016;101(4):407–416. doi:10.3324/haematol.2015.141101
6. Xiong J. SALL4: engine of cell stemness. Curr Gene Ther. 2014;14(5):400–411. doi:10.2174/1566523214666140825125138
7. Zhang X, Yuan X, Zhu W, Qian H, Xu W. SALL4: an emerging cancer biomarker and target. Cancer Lett. 2015;357(1):55–62. doi:10.1016/j.canlet.2014.11.037
8. Yang L, Liu L, Gao H, et al. The stem cell factor SALL4 is an essential transcriptional regulator in mixed lineage leukemia-rearranged leukemogenesis. J Hematol Oncol. 2017;10(1):159. doi:10.1186/s13045-017-0531-y
9. Shen Q, Liu S, Hu J, et al. The differential expression pattern of the BMI-1, SALL4 and ABCA3 genes in myeloid leukemia. Cancer Cell Int. 2012;12(1):42. doi:10.1186/1475-2867-12-42
10. Liu C, Wu H, Li Y, et al. SALL4 suppresses PTEN expression to promote glioma cell proliferation via PI3K/AKT signaling pathway. J Neurooncol. 2017;135(2):1–10.
11. Ma Y, Cui W, Yang J, et al. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood. 2006;108(8):2726–2735. doi:10.1182/blood-2006-02-001594
12. Ma J-C, Qian J, Lin J, et al. Aberrant hypomethylation of SALL4 gene is associated with intermediate and poor karyotypes in acute myeloid leukemia. Clin Biochem. 2013;46(4):304–307. doi:10.1016/j.clinbiochem.2012.11.018
13. Zhang J, Tam W-L, Tong GQ, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8(10):1114–1123. doi:10.1038/ncb1481
14. Oikawa T, Kamiya A, Zeniya M, et al. Sal‐like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57(4):1469–1483. doi:10.1002/hep.26159
15. Kohlhase J, Heinrich M, Schubert L, et al. Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet. 2002;11(23):2979–2987. doi:10.1093/hmg/11.23.2979
16. Deloukas P, Matthews LH, Ashurst J, et al. The DNA sequence and comparative analysis of human chromosome 20. Nature. 2001;414(6866):865–871.
17. Ueno S, Lu J, He J, et al. Aberrant expression of SALL4 in acute B cell lymphoblastic leukemia: mechanism, function, and implication for a potential novel therapeutic target. Exp Hematol. 2014;42(4):307–316.e308. doi:10.1016/j.exphem.2014.01.005
18. Ghodousi ES, Rahgozar S. MicroRNA-326 and microRNA-200c: two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. Jul. 2018;119(7):6024–6032.
19. Signoretti S, Murphy M, Cangi MG, Puddu P, Kadin ME, Loda M. Detection of clonal T-cell receptor gamma gene rearrangements in paraffin-embedded tissue by polymerase chain reaction and nonradioactive single-strand conformational polymorphism analysis. Am J Pathol. 1999;154(1):67–75. doi:10.1016/S0002-9440(10)65252-2
20. Rahgozar S, Moafi A, Abedi M, et al. mRNA expression profile of multidrug-resistant genes in acute lymphoblastic leukemia of children, a prognostic value for ABCA3 and ABCA2. Cancer Biol Ther. 2014;15(1):35–41. doi:10.4161/cbt.26603
21. Bhatla T, Jones CL, Meyer JA, Vitanza NA, Raetz EA, Carroll WL. The biology of relapsed acute lymphoblastic leukemia: opportunities for therapeutic interventions. J Pediatr Hematol Oncol. 2014;36(6):413. doi:10.1097/MPH.0000000000000179
22. Irving JA. Towards an understanding of the biology and targeted treatment of paediatric relapsed acute lymphoblastic leukaemia. Br J Haematol. 2016;172(5):655–666. doi:10.1111/bjh.13852
23. Li W, Zhang H, Assaraf YG, et al. Overcoming ABC transporter-mediated multidrug resistance: molecular mechanisms and novel therapeutic drug strategies. Drug Resist Updat. 2016;27:14–29. doi:10.1016/j.drup.2016.05.001
24. Aberuyi N, Rahgozar S, Moafi A. The role of ATP-binding cassette transporter A2 in childhood acute lymphoblastic leukemia multidrug resistance. Iran J Pediatr Hematol Oncol. 2014;4(3):118.
25. Nicolè L, Sanavia T, Veronese N, et al. Oncofetal gene SALL4 and prognosis in cancer: a systematic review with meta-analysis. Oncotarget. 2017;8(14):22968. doi:10.18632/oncotarget.v8i14
26. Yang J. SALL4 as a transcriptional and epigenetic regulator in normal and leukemic hematopoiesis. Biomarker Res. 2018;6(1):1. doi:10.1186/s40364-017-0115-6
27. Liu L, Zhang J, Yang X, Fang C, Xu H, Xi X. SALL4 as an epithelial-mesenchymal transition and drug resistance inducer through the regulation of c-Myc in endometrial cancer. PLoS One. 2015;10(9):e0138515. doi:10.1371/journal.pone.0138515
28. Yanagihara N, Kobayashi D, Kuribayashi K, Tanaka M, Hasegawa T, Watanabe N. Significance of SALL4 as a drug-resistant factor in lung cancer. Int J Oncol. 2015;46(4):1527–1534. doi:10.3892/ijo.2015.2866
29. Gao C, Kong NR, Chai L. The role of stem cell factor SALL4 in leukemogenesis. Crit Rev Oncogenesis. 2011;16:1–2. doi:10.1615/CritRevOncog.v16.i1-2
30. Yang J, Chai L, Liu F, et al. Bmi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad Sci. 2007;104(25):10494–10499. doi:10.1073/pnas.0704001104
31. Sun C, Lan P, Han Q, et al. Oncofetal gene SALL4 reactivation by hepatitis B virus counteracts miR-200c in PD-L1-induced T cell exhaustion. Nat Commun. 2018;9(1):1241. doi:10.1038/s41467-018-03584-3
32. Zhang L, Xu Z, Xu X, et al. SALL4, a novel marker for human gastric carcinogenesis and metastasis. Oncogene. 2014;33(48):5491. doi:10.1038/onc.2013.495
33. Cui W, Kong NR, Ma Y, Amin HM, Lai R, Chai L. Differential expression of the novel oncogene, SALL4, in lymphoma, plasma cell myeloma, and acute lymphoblastic leukemia. Mod Pathol. 2006;19(12):1585–1592. doi:10.1038/modpathol.3800694
34. Tang P, Sun H, Liu Y, Wang G, Yin Y. Expression of SALL4 and BMI-1 mRNA in acute leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2008;16(6):1271–1274.
35. Yang M, Xie X, Ding Y. SALL4 is a marker of poor prognosis in serous ovarian carcinoma promoting invasion and metastasis. Oncol Rep. 2016;35(3):1796–1806. doi:10.3892/or.2016.4545
36. Han SX, Wang JL, Guo XJ, et al. Serum SALL4 is a novel prognosis biomarker with tumor recurrence and poor survival of patients in hepatocellular carcinoma. J Immunol Res. 2014;2014:262385. doi:10.1155/2014/262385
37. Peng HX, Liu XD, Luo ZY, et al. Upregulation of the proto-oncogene Bmi-1 predicts a poor prognosis in pediatric acute lymphoblastic leukemia. BMC Cancer. 2017;17(1):76. doi:10.1186/s12885-017-3049-3
38. Setoguchi T, Taga T, Kondo T. Cancer stem cells persist in many cancer cell lines. Cell Cycle. 2004;3(4):412–413. doi:10.4161/cc.3.4.795
39. Yang J, Chai L, Gao C, et al. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood. 2008;112(3):805–813. doi:10.1182/blood-2007-11-126326
40. Choi S, Henderson MJ, Kwan E, et al. Relapse in children with acute lymphoblastic leukemia involving selection of a preexisting drug-resistant subclone. Blood. 2007;110(2):632–639. doi:10.1182/blood-2007-01-067785
41. Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer. 2009;9(9):665–674. doi:10.1038/nrc2714
42. Hupfeld T, Chapuy B, Schrader V, et al. Tyrosine kinase inhibition facilitates cooperation of transcription factor SALL4 and ABC transporter A3 towards intrinsic CML cell drug resistance. Br J Haematol. 2013;161(2):204–213. doi:10.1111/bjh.2013.161.issue-2
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.